Abstract
Glandular trichomes play important roles in protecting plants from biotic attack by producing defensive compounds. We investigated the metabolic profiles and transcriptomes to characterize the differences between different glandular trichome types in several domesticated and wild Solanum species: Solanum lycopersicum (glandular trichome types 1, 6, and 7), Solanum habrochaites (types 1, 4, and 6), Solanum pennellii (types 4 and 6), Solanum arcanum (type 6), and Solanum pimpinellifolium (type 6). Substantial chemical differences in and between Solanum species and glandular trichome types are likely determined by the regulation of metabolism at several levels. Comparison of S. habrochaites type 1 and 4 glandular trichomes revealed few differences in chemical content or transcript abundance, leading to the conclusion that these two glandular trichome types are the same and differ perhaps only in stalk length. The observation that all of the other species examined here contain either type 1 or 4 trichomes (not both) supports the conclusion that these two trichome types are the same. Most differences in metabolites between type 1 and 4 glands on the one hand and type 6 glands on the other hand are quantitative but not qualitative. Several glandular trichome types express genes associated with photosynthesis and carbon fixation, indicating that some carbon destined for specialized metabolism is likely fixed within the trichome secretory cells. Finally, Solanum type 7 glandular trichomes do not appear to be involved in the biosynthesis and storage of specialized metabolites and thus likely serve another unknown function, perhaps as the site of the synthesis of protease inhibitors.
Trichomes are epidermal structures widely conserved across the plant kingdom (Kim and Mahlberg, 1991; Wagner, 1991; Alonso et al., 1992; Yu et al., 1992; Kolb and Muller, 2003; Valkama et al., 2003; Giuliani and Bini, 2008). These structures perform important biological functions, such as discouraging herbivory, attracting pollinators, and maintaining a boundary layer (Nihoul, 1993; Van Dam and Hare, 1998; Kennedy, 2003; Moyano et al., 2003; Simmons and Gurr, 2005; Liu et al., 2006; Horgan et al., 2007; Gonzalez et al., 2008; Romero et al., 2008; Nonomura et al., 2009; Kang et al., 2010). Many of these functions are the result of the specialized nature of glandular trichomes (glands) as sites for the synthesis and storage of biologically active specialized metabolites (Alonso et al., 1992; Antonious, 2001; Iijima et al., 2004; Siebert, 2004; Deschamps et al., 2006; Nagel et al., 2008; Wang et al., 2008; Biswas et al., 2009; Sallaud et al., 2009). Comparisons between domesticated crop species and their wild progenitors have revealed that many of the more potent, glandular trichome-derived specialized metabolites have been lost during domestication (Rodriguez et al., 1993; Oghiakhe, 1997; Medeiros and Tingey, 2006; Zhang et al., 2008; Besser et al., 2009). The loss of these important compounds has led to an increased susceptibility of domesticated crops to pathogen and herbivore attack compared with their wild counterparts (Rodriguez et al., 1993; Puterka et al., 2003; Chao et al., 2006; Nonomura et al., 2009), and reintroduction of such compounds into crop species may prove to be an effective way to combat crop loss due to insects and disease.
The genus Solanum possesses two general categories of trichomes: nonglandular and glandular. The nonglandular trichomes are generally linear trichomes that lack secretory/storage cells at their tips (Schwab et al., 2000) and thus are not of interest for this investigation. The glandular trichomes of Solanum, on the other hand, first described by Luckwill (1943) and later reviewed extensively by Simmons and Gurr (2005), have been described as consisting of four distinct classes (types 1, 4, 6, and 7). The density of these glandular trichome types can vary according to species, cultivar, tissue, and environmental conditions (Luckwill, 1943; Wilkens et al., 1996; Economou et al., 2006; Maluf et al., 2007; Kang et al., 2010). The glandular trichome types differ according to the size and length of the supporting stalk as well as the number of secretory cells making up the glandular tip. In addition, other differences, such as in chemical content, have been proposed between most of these four types of glandular trichomes. Indeed, type 6 glandular trichomes are known to be the major site of terpenoid production in cultivated tomato (Solanum lycopersicum; Schilmiller et al., 2009; Kang et al., 2010).
To determine the biological roles and chemical contents of individual glandular trichome types in various Solanum species, we have taken a comparative functional genomics approach with particular emphasis on analysis of the glandular trichome secretory cell transcriptome and metabolic profile. This analysis has allowed us to address several important questions. What are the roles of the different glandular trichome types? Do some plants make more than one type of glandular trichome? What specialized functions do different glandular trichome types possess? And where do specific specialized metabolic processes occur (e.g. terpenoid versus acyl sugar production)?
RESULTS AND DISCUSSION
Isolation and Analysis of Different Trichome Types in Different Solanum Species
Accessions from five Solanum species were chosen for analysis: Solanum habrochaites (LA1777), Solanum pimpinellifolium (LA1589), Solanum pennellii (LA0716), Solanum arcanum (= Solanum peruvianum; LA1708), and S. lycopersicum (= Lycopersicon esculentum; M82). Within these species, several types of glandular trichomes are present and vary in abundance (Fig. 1; Table I). Of specific interest were secretory cells of type 1, 4, 6, and 7 glandular trichomes, which remain largely uncharacterized at the individual level across Solanum species. A recent report (Slocombe et al., 2008) describes the analysis of total trichome preparations from S. pennellii, which formed a reference point for our analysis, but did not analyze individual trichome type secretory cells.
Figure 1.
Glandular trichome density and distribution among four of the five Solanum species used in this study. A, S. pennellii accession LA0716 leaf. B, A closer look at S. pennellii glandular trichomes. C, S. pimpinellifolium accession LA1589 leaf. D, A closer look at S. pimpinellifolium glandular trichomes. E, S. habrochaites accession LA1777 leaf. F, A closer look at S. habrochaites glandular trichomes. G, S. arcanum accession LA1708 leaf. H, A closer look at S. arcanum glandular trichomes. I, S. lycopersicum accession LA3475 (M82) leaf. J to M, S. lycopersicum glandular trichomes. J, Type 1. K and L, Type 6. M, Type 7.
Table I. The accessions used in this study along with information on the glandular trichome density, metabolite profile, and EST approach.
A, Abundant; EP, EST sequencing using the 454 pyrosequencing approach, more than 100,000 resulting ESTs; ES, EST sequencing using the Sanger approach, less than 1,000 resulting ESTs; M, metabolite profile was performed; S, sparse; s, sticky residue/profuse secretion.
| Species (Accession No.) | Glandular Trichome Type |
|||
| Type 1 | Type 4 | Type 6 | Type 7 | |
| S. lycopersicum (M82/LA3475), domesticated | A, M, ES | S, –, – | A, M, EP | S, –, ES |
| S. pimpinellifolium (LA1589), current | –, –, – | –, –, – | A, M, EP | –, –, – |
| S. arcanum (LA1708), wild | A, –, – | –, –, – | A, M, – | –, –, – |
| S. habrochaites (LA1777), wild | Ss, M, ES | As, M, ES | A, M, EP | S, M, – |
| S. pennellii (LA0716), wild | Ss, –, – | As, M, ES | S, M, ES | S, –, – |
The trichomes referred to as types 1 and 4 glandular trichomes, as shown in Figure 1, A, B, I, and J, appear to be physically similar to nonglandular hairy trichomes but terminate with globe- or oblong-shaped one- to two-cell storage/secretion units that may or may not be covered by a waxy cuticle. One hypothesis explaining the physical similarities between these two glandular trichome types is that type 1 and type 4 trichomes represent different stages of development (Kang et al., 2010). However, both are present on very young leaves as well as on fully mature leaves. Despite their apparent physical differences, both types accumulate small droplets on their tips that contain sticky acyl sugar compounds (Rodriguez et al., 1993; Alba et al., 2009; Nonomura et al., 2009). The accumulation of such compounds is pronounced in S. pennellii and S. habrochaites, which have abundant type 4 trichomes (Weston et al., 1989; Snyder et al., 1998), resulting in a sticky residue when leaves of these species are handled. The opposite is true for S. arcanum and S. lycopersicum, which possess type 1 trichomes but lack the profuse secreting nature of S. habrochaites and S. pennellii type 1 or 4 trichomes.
The other dominant glandular trichome in Solanum species is the type 6 gland, shown in Figure 1, F, K, and L. Composed of four disc cells at the end of a two-celled stalk, these glands are found on all species used in this study but are present in low abundance on S. pennellii leaves and stems. Unlike the secreting type 1 and 4 glands of S. habrochaites and S. pennellii, type 6 glands appear to be specialized to produce metabolites and then store them under a waxy cuticle, as has been described for other “peltate” glandular trichomes found in mint (Mentha × piperita), basil (Ocimum basilicum), and other species (Croteau, 1991; Turner et al., 2000; Gang et al., 2001; Deschamps et al., 2006; Gunnewich et al., 2007). In Solanum type 6 glands, the area under the cuticle is filled completely and ready to release its contents given either the correct environmental conditions or physical contact (Lin et al., 1987; Maluf et al., 2007; Ben-Israel et al., 2009).
The final glandular trichome type of interest is the type 7 glandular trichome depicted in Figure 1M. Of all glandular trichomes analyzed, this gland is both of low abundance (Simmons et al., 2003) and the least characterized, due not only to its scarcity but also to its general physical properties. Located in close proximity to the epidermis, this glandular trichome consists of a small multicellular glandular head situated on a short, single-celled stalk, making it very difficult to isolate. Like the type 6 gland, type 7 glandular trichomes also possess a waxy cuticle that has been observed in our studies to be removable with harsh abrasive treatment. The exact content of the mixture under the waxy cuticle of type 7 glands remains unclear and is discussed below.
To collect enough glandular secretory cells for metabolomic and transcriptomic analyses, a variety of methods were employed. To collect the type 1 and 4 glands from the various Solanum species, microscissors were used to clip the glandular heads off of the stalks. This leads to very pure fractions of just the glandular head cells. Two alternative approaches were used to isolate the type 6 gland heads. The first approach involved directly “picking” type 6 glandular trichome heads using stretched glass pipettes (Schilmiller et al., 2009; Kang et al., 2010), while the second approach utilized glass beads in buffer to abrasively remove trichomes from the leaf surface (Gang et al., 2001; Fridman et al., 2005). Fractions enriched in type 7 glandular trichome secretory cells were obtained using the bead-beater procedure. Methods employing liquid N2 or dry ice (Yerger et al., 1992) did not work well for our purposes, as these preclude the ability to separate different types of trichomes from each other and lead to trichome fractions that consist mostly of the nonglandular stalks. The methods used in this investigation allowed us to obtain very pure preparations consisting almost exclusively of the secretory cells of interest. Secretory cells collected for transcriptome profiling were flash frozen in liquid nitrogen, whereas cells destined for metabolite profiling were immediately suspended in ice-cold solvent before being further processed (see “Materials and Methods”).
Gross Comparisons of Trichome Metabolite Profiles in Solanum Species
Metabolite profiles obtained for the 10 species/type entries marked as abundant or sparse in Table I were produced using a liquid chromatography (LC)-time of flight-mass spectrometry (MS)-based method and revealed that the majority (79%) of the detected compounds were unknowns (Supplemental Table S1). However, 119 known compounds were detected and their relative quantities determined between gland types and between species. It was impossible to determine absolute levels for these compounds in our samples because of differences in ionization efficiencies and potential ion suppression due to variable matrix effects. Moreover, potential intrametabolite contamination from the type 1/4 trichome exudates for the type 6 trichome samples may exist, especially for the type 6 samples from S. habrochaites and S. pennellii. Thus, it was also not possible to determine accurate differences in abundance for compound/compound comparisons. Nevertheless, trends in compound class abundance between trichome types and between species, based on relative quantification values, were determined. The box plot in Figure 2 shows the distribution of normalized peak areas for aggregate metabolite classes for known compounds detected in the various Solanum species and trichome types, providing a general concept of the prevalence of various compound classes in different glandular trichome extracts. The results presented in this figure must be evaluated with caution, however, in light of the issues of nonuniform ionization efficiencies and potential ion suppression and matrix effects mentioned above. Nevertheless, these results suggest interesting trends regarding metabolite production in these trichomes, where metabolite modules appear to be present and imply mechanisms involved in controlling the production of specific compounds and groups of compounds (Fig. 3; Supplemental Figs. S1 and S2; Supplemental Table S2; Xie et al., 2009).
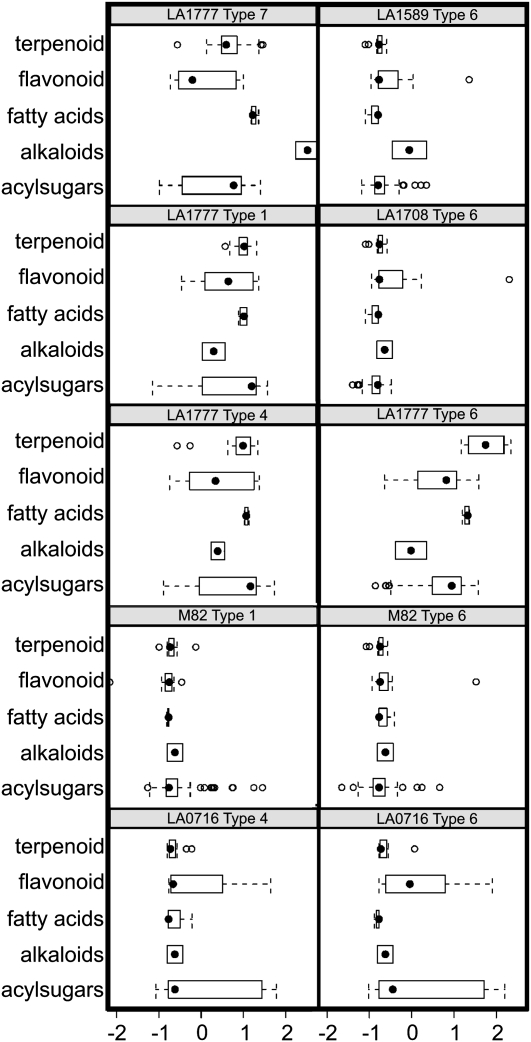
Figure 2.
Summary of major identifiable metabolites by species and trichome types. Each panel represents one species/trichome type. In each panel, the x axis is the normalized log2 (peak area), the y axis represents the five compound classes, and the panel title (at the top of each panel) lists the corresponding species/trichome type. Black circles indicate metabolite class medians, while the dotted lines indicate the limits of the 90th percentile and lower 10th percentile. White circles or semicircles indicate individual data points outside the range of 10th to 90th percentiles. For the purposes of this study, peak areas for each of the identified trichome metabolites were averaged within each species/trichome type. Using these averages, a log2 transformation was performed to decrease the effect of extreme values. Following log2 transformation, the resulting values were normalized across all 10 species/trichome types.
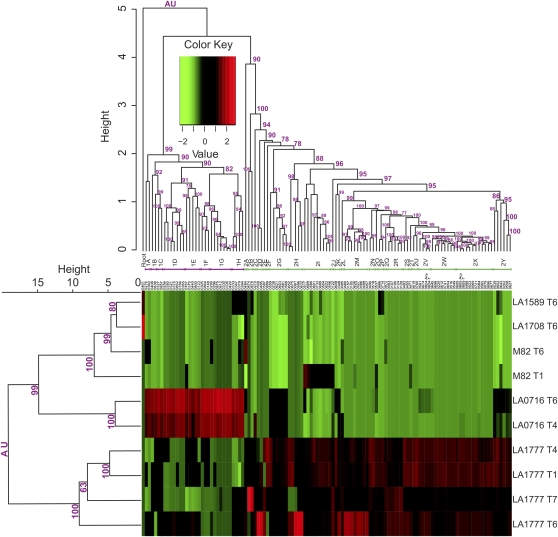
Figure 3.
Two-way cluster analysis of normalized LC-MS metabolite peak areas from Solanum glandular trichomes. In the heat-map plot, each column represents a feature, and the dendrogram of cluster tree for features is displayed at the top; each row represents a species/trichome type, and the dendrogram of cluster tree for types is displayed on the left margin. Dendrograms and bootstrap values (magenta numbers) are calculated using the R package pvclust (Shimodaira, 2002, 2004). Four-digit numbers along the top edge of the heat map indicate unique metabolite identifiers listed in Supplemental Figure S1. Two-digit alpha-numeric identifiers (1A–1H, 2A–2Y) immediately below the top dendrogram indicate metabolite clusters also listed in Supplemental Figures S1 and S2. Accession numbers represent the Solanum species examined: LA1589, S. pimpinellifolium; LA1777, S. habrochaites; LA0716, S. pennellii; LA1708, S. arcanum; M82, S. lycopersicum.
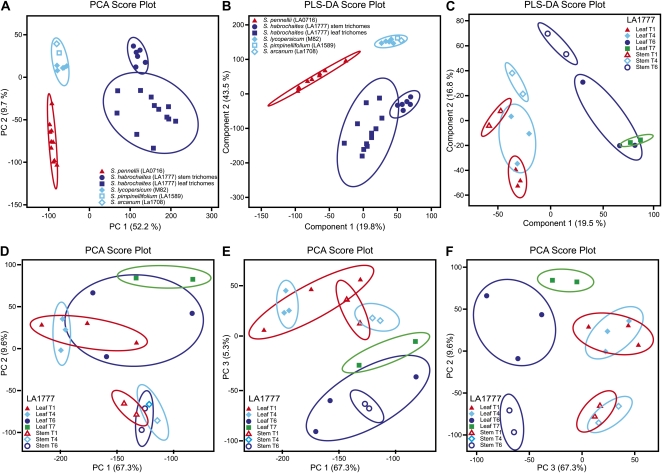
The normalized data used to produce Figure 3 revealed that the known metabolites observed in Solanum trichomes are differentially distributed between trichome types and species and that this variation is generally more evident between species than within. For example, S. habrochaites glands contain the most diverse set of known fatty acids, sesterterpenes, and acyl sucroses of all species and glandular trichome types (Supplemental Table S2; Supplemental Fig. S2, clusters 2A–2Y). Likewise, S. pennellii glands were dominated by acyl glucoses (Supplemental Fig. S2, clusters 1A–1H) compared with other species and gland types. It is also clear from Figure 2 that cultivated tomato (M82) lacks the chemical diversity of many of the wild species and is more similar to S. arcanum and S. pimpinellifolium than to S. pennellii and S. habrochaites, a conclusion further supported by the data shown in Figures 2 to 4 and Supplemental Figures S2 and S3. Figure 4, displaying results from additional multivariate statistical analyses (principal component analysis [PCA] and partial least squares-discriminant analysis [PLS-DA]) of the metabolite data set with individual samples from all species viewed separately instead of by analyzing means of replicates, clearly shows that S. lycopersicum, S. arcanum, and S. pimpinellifolium are closely related metabolically, regardless of trichome type. The similar metabolite profiles and morphologies of S. lycopersicum, S. arcanum, and S. pimpinellifolium is consistent with the close phylogenetic relationship among these species (Peralta and Spooner, 2001). These profiles are also illustrative of S. lycopersicum’s limited ability to produce and/or store the metabolites measured in our analysis within its glandular trichomes, although it does produce significant levels of a handful of monoterpenoids and sesquiterpenoids (Kang et al., 2010). This reduced chemical diversity relative to wild species may help to explain observations that the resistance of domesticated tomato cultivars to certain arthropod herbivores is less than that of many wild tomato species (Kumar et al., 1995; Pico et al., 1998; Martins Santana et al., 2001).
Figure 4.
PCA and PLS-DA of metabolite profiling data for comparison of trichome types of all species evaluated in this investigation (A and B) and of all trichome types from S. habrochaites (LA1777; C–F).
What was quite surprising was that greater differences were observed in overall metabolite profiles for the same glandular trichome type across species than were observed for comparisons of different gland types within the same species. For example, PLS-DA (Fig. 4C) and PCA (Fig. 4, C–F) analyses of metabolite data obtained for S. habrochaites (accession LA1777), the only accession that had readily isolatable glandular trichomes for all types (1, 4, 6, and 7), demonstrated that glandular trichome types 1 and 4 were indistinguishable from each other. Hierarchical cluster analysis (Supplemental Fig. S3A) also failed to separate type 1 and 4 trichomes based on overall or aggregate metabolite profiles, regardless of the method used for data normalization. The normalization used for the data presented above was log2-transformed data. Other normalization methods, such as autoscaling, pareto scaling, or range scaling, gave even poorer resolving power, where type 6 trichomes were also not readily differentiated from the type 1 or 4 trichomes. Interestingly, glandular trichomes from stems could be differentiated from glandular trichomes isolated from leaf surfaces (Fig. 4, D and F), depending on which principal components were considered, suggesting that significantly different metabolite profiles may exist for stems versus leaves. Such results only present an aggregate picture of a greater metabolomic content, however, and do not allow for closer inspection of how specific compounds that might be involved in specific plant defenses may be produced or regulated.
When specific classes of compounds were compared, instead of gross aggregate profiles, a few notable differences were observed among the different trichome types within species. For example, S. lycopersicum type 1 and 6 glandular trichomes exhibited markedly dissimilar profiles for certain subsets of metabolites. Whereas the levels of acyl glucoses in both types of S. lycopersicum glands were very low (although they were more abundant in the type 6 glands than in type 1), type 1 extracts from the same species were dominated by acyl sucroses (Supplemental Fig. S2, clusters 1A, 1E, 1G, 1H, 2I, 2O, 2T, and 2Y), but such compounds did not appear to be present in type 6 glands from this particular species. In addition, S. habrochaites type 1 and 4 trichomes had approximately 5-fold higher levels of methylated myricetin (a flavonoid) than did type 6 (A. Schmidt and E. Pichersky, unpublished data).
We observed great variability in relative amounts of acyl sugars between different preparations of the same gland type, especially for S. pennellii type 6 and 4 gland preparations. And, despite the possibility of variable matrix effects, the differences between preparations of a single gland type were very large compared with the likely variation resulting from matrix effects. Therefore, we are not convinced that further chemical analyses (even if acyl sugar standards were available) would enhance our understanding of the differences between gland chemistries.
Trichome Transcriptomes in Solanum Species
The transcriptomes of the secretory cells of various glandular trichome types from the different species were analyzed using a transcriptome sequencing approach, with six and eight libraries sequenced using Sanger and 454 approaches, respectively (Table I), resulting in 1.4 × 106 ESTs. The ESTs for the five species were separately assembled with PAVE (Soderlund et al., 2009) and annotated with UniProt (UniProt Consortium, 2010) and Rstat (Stekel et al., 2000). The ESTs from all trichome/species libraries were also assembled together, resulting in 32,261 contigs and 49,958 singletons. Annotation of these trichome/species contigs and singletons resulted in 80% of sequences being assigned a UniProt identifier, of which 33,092 had UniProt identifiers from plants as a top match. The resulting transcriptomic data can be viewed and analyzed at http://www.agcol.arizona.edu/pave/solanum/. This approach contrasts to what has been reported before for S. pennellii, where total trichomes, including stalks, were aggregately (trichome types were not distinguishable) analyzed for gene expression level by hybridization to the S. lycopersicum TOM2 microarray, an array containing sequences from a different Solanum species than what was used in the hybridization analysis (Slocombe et al., 2008). Such cross-species microarray experiments, while capable of differentiating rough relative expression levels between tissues within the same species for many (but not all) genes in the target species, are not capable of providing absolute expression levels or of being useful when comparing expression for any particular gene between species (Chain et al., 2008; Gilad et al., 2009). Thus, it is unfortunately practically impossible to draw comparisons between our approach and what has been previously published with regard to observed levels of expression for any particular gene, as our approach was fundamentally different from what has been described before.
Comparison of Type 6 Glandular Trichomes between Solanum Species
Analysis of metabolites from type 6 glandular trichomes across all five species resulted in the same trends discussed in the gross comparisons of metabolites across all species and glandular trichome types. Compared with the other species, S. habrochaites and S. pennellii type 6 glands have more in common with one another than they do with S. lycopersicum, S. arcanum, and S. pimpinellifolium (Figs. 2 and 3). Moreover, the S. habrochaites type 6 extracts also contain the most diverse sets of metabolites (which are specific to S. habrochaites type 6) compared with other species (Supplemental Fig. S2), being dominated by acyl sucroses, terpenoids, and fatty acids (Supplemental Fig. S2). S. pennellii type 6 glands possess the second most diverse complement of specialized metabolites specific to one species, containing the most diverse set of acyl glucoses in our comparisons (Supplemental Fig. S2). In contrast, type 6 gland extracts from S. lycopersicum, S. pimpinellifolium, and S. arcanum are very similar to each other (Supplemental Fig. S2), and both the diversity and quantity of metabolites in these three species are extremely limited. The unique type 6 glandular trichome metabolite profiles may play important roles in conferring to wild Solanum species resistance to disease and insect herbivory (Kumar et al., 1995; Pico et al., 1998; Martins Santana et al., 2001).
Gross analysis of Solanum type 6 transcriptomes revealed that transcripts for enzymes and proteins associated with photosynthesis light reactions, photosynthetic carbon fixation, glycolysis/gluconeogenesis, starch and Suc metabolism, and the citrate cycle were prevalent, in addition to many other downstream primary metabolic processes (Supplemental Table S3; Supplemental Figs. S4–S7; a reference iPath map can be accessed at http://pathways.embl.de/default_map.html for comparison), suggesting that Solanum type 6 glandular trichomes may be able to produce specialized compounds de novo, perhaps without the requirement for transport and uptake of source carbon from stalk cells. This contrasts somewhat with the report that in S. pennellii total trichomes, the expression level for photosynthesis-related genes was greatly reduced (Slocombe et al., 2008), based on hybridization to a nonspecies microarray platform (the TOM2 Affy array is based on a limited set of genes from a specific S. lycopersicum cultivar and not on S. pennellii). However, the trichome stalk cells provided the vast majority of the cell mass and presumably RNAs in that study (see Fig. 2, G and H, in Slocombe et al., 2008). In our study, we analyzed only the secretory cells for the majority of the trichome types analyzed, or only small portions of the stalks were included when microscissors were used to clip off the trichome heads at their connection to the stalk. This may have led to our ability to detect significant levels of expression of some photosynthesis-related genes, whereas the other report did not observe this.
Developing oil seed embryos from some species do not necessarily rely solely on imported Suc and the oxidative pentose phosphate pathway but rather also utilize an endogenous photosynthetic apparatus to provide both carbon skeletons and reducing power for oil (triglyceride) synthesis (Goffman et al., 2004; Alonso et al., 2007; Junker et al., 2007; Allen et al., 2009). Similarly, Wang et al. (2009) recently claimed that photosynthesis genes are expressed at very high levels in the glandular trichomes of Artemisia annua, with chlorophyll a/b-binding protein and ribulose bisphosphate carboxylase small subunit being among the top 10 most highly expressed transcripts in the trichomes of that species, and that in situ carbon fixation may be involved in the production of artemisinin in these trichome secretory cells. However, close inspection of figure 5 (isolation of glandular trichomes) from that report reveals significant contamination by green mesophyll cells in the final glandular trichome preparation used for 454 transcriptome analysis. The glandular trichomes shown in that figure appear to be colorless. Thus, it is not clear whether or not de novo carbon fixation in the secretory cells is indeed involved in the production of artemisinin, and the transcript levels observed may be due to expression not only in the trichome but also in underlying mesophyll cells that contaminated the trichome preparations. Indeed, species such as Glycine max, Mentha species, and basil possess glandular trichomes with plastids that lack developed thylakoid membrane systems and any apparent ability to perform photosynthesis (Franceschi and Giaquinta, 1983; Croteau, 1991; Gershenzon et al., 1992; McCaskill et al., 1992; McCaskill and Croteau, 1995; Lange et al., 2000; Turner et al., 2000; Gang et al., 2001; Turner and Croteau, 2004; Rios-Estepa et al., 2008; Xie et al., 2008). In these species, nonphotosynthetic plastids appear to be heavily involved in specialized metabolite synthesis. The glandular trichomes of A. annua appear to be of this type.
In contrast, work on tobacco (Nicotiana tabacum) glandular trichomes clearly suggested that some glandular trichomes in the Solanaceae have the ability to fix carbon and synthesize metabolites from sugar produced directly in the trichomes, although carbon imported into the trichome may also be involved (Keene and Wagner, 1985; Kandra and Wagner, 1988; Nielsen et al., 1991). The type 6 trichomes from S. lycopersicum, S. arcanum, S. pimpinellifolium, and S. habrochaites used for 454 sequencing in this analysis were isolated using the glass-bead-shaking method. However, the type 1/4 trichomes from all species and type 6 trichomes from S. pennellii used for both metabolite analysis and sequencing in this project were hand picked (Supplemental Fig. S20), leading to analysis of only the secretory cells and perhaps part of a stalk cell for those collected by the microscissors technique but no possible contamination by underlying mesophyll or other cell types. It must be noted that such photosynthesis-related genes were not among the most highly expressed in our analysis, as can be seen in Supplemental Tables S14 to S19, where many of them are listed. Therefore, the identification of transcripts for photosynthesis genes in the trichomes suggests the possibility that the secretory cells of Solanum species may possess the ability to have at least a rudimentary capacity for photosynthesis that may contribute to secreted metabolite synthesis. Thus, the role of imported versus de novo-synthesized sugars in glandular trichome secretory cell metabolism remains an area of intense interest.
Also observed in type 6 glands from all species, with the exception of S. pennellii, were transcripts representing the phenylpropanoid and flavonoid pathways. The lack of representation of the phenylpropanoid and flavonoid pathways in the transcriptome of S. pennellii type 6 glandular trichomes was likely the result of inadequate sampling of the transcriptome during sequencing (Supplemental Tables S4 and S5), as these trichomes were difficult to obtain due to the high densities of type 1/4 glands that obscured access, with the resulting production of only a small 454 library.
In further efforts to make quantitative comparisons between type 6 Solanum trichomes from these species, we utilized the methodology of Stekel et al. (2000) to identify differentially significantly expressed ESTs (Supplemental Tables S6 and S7) using summaries of three different annotations that were assigned as described in “Materials and Methods”: UniProt identification (UPID), Enzyme Commission (EC), and Gene Ontology. Supplemental Tables S6 and S7 provide interspecies and intraspecies comparisons for UniProt and EC, showing the normalized counts for up-regulated genes for each of the two species compared. The elements of the metabolic network present in Solanum type 6 trichome secretory cells are listed in Supplemental Table S3 and are pictorially reconstructed using the iPATH software (Letunic et al., 2008; Supplemental Figs. S4–S8). To ensure legitimate comparisons between library data sets, we took a conservative approach and only used library comparisons that were of similar size, collection method, sequencing technologies, and within the same sequence assembly (Supplemental Tables S4–S7). Thus, the type 6 gland comparisons that are listed in Supplemental Tables S6 and S7 excluded the S. arcanum and S. pennellii type 6 data sets. The three type 6 transcript comparisons that were performed reflect the metabolite profiles observed in the gross metabolite comparisons described above; this is shown in Supplemental Tables S8 to S15, which list overrepresented and underrepresented biological processes by Gene Ontology terms in each of the species. Interestingly, terpenoid-rich S. habrochaites appears to preferentially (compared with either S. pimpinellifolium or S. lycopersicum; Supplemental Table S14) express enzymes important for regulating flux into the terpenoid/isoprenoid pathway: 1-deoxy-d-xylulose 5-phosphate synthase 2 (annotated as Q68IP4_SOLHA), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (annotated as Q2XTB5_SOLTU, LYTB-like protein), and Z,Z-farnesyl pyrophosphate synthase (annotated as B8XA40_SOLHA). Also, S. habrochaites type 6 trichomes exhibit enriched expression of contig EC annotations implicated in processes associated with acyl sugar biosynthesis, including an acyl desaturase (EC 1.14.19.2) and an isopropylmalate synthase (EC 2.3.3.13; Supplemental Table S15; Slocombe et al., 2008). In contrast, it was not clear from this analysis why S. habrochaites type 6 glands contain limited quantities of Glc-derived acyl sugars compared with acyl sucroses, as ESTs for enzymes such as Suc synthase or gluconeogenesis pathway members were found in relatively equal abundance across species (Supplemental Tables S14 and S15). The mechanism responsible for regulating the production of these compounds remains unidentified, but based on our experiments, it is not likely to involve direct control by modulation of transcript levels.
In addition to the observations described above (i.e. that the specialized metabolites stored within the type 6 glandular trichome can be derived from imported, leaf-produced Suc or from carbon fixed within the trichome itself), the data also suggest that transcripts for general metabolic pathways that were detected were expressed at similar levels across species (Supplemental Tables S14 and S15). Additionally, it was evident that differences in type 6 gland biochemistry are likely due to differential up-regulation of select segments or branches of a pathway, not the whole pathway, as is suggested by Supplemental Figures S9 to S14. The differential regulation of select enzymes within type 6 glands is consistent with the biochemistries outlined in Figure 3, Supplemental Table S2, and Supplemental Figure S4, specifically with S. habrochaites compared with either S. lycopersicum or S. pimpinellifolium. It also appears that some differences in metabolite profiles between the species are most likely regulated at both the transcriptional and genomic levels. Unique EC annotations for the “same” enzyme between different species suggested that not only differences in expression levels for gene family members but also alterations in sequence leading to potentially differential activity may produce altered pathway functions.
A final determination was that type 6 glands also express genes not directly contributing to specialized metabolism. S. lycopersicum is the best example, and genes encoding enzymes and other proteins such as lipoxygenase, pathogenesis-related protein isoform b1, Arg decarboxylase, superoxide dismutase, various heat shock-related proteins, and of course polyubiquitin were apparently expressed at high levels (Supplemental Tables S14 and S15). It is likely that these proteins are important in preserving the viability of these cells, which are exposed to varied environmental conditions due to their location at the end of a long trichome stalk. Moreover, it is also notable that despite the presence of transcripts for transcription factor genes, none of these appear to be preferentially expressed between species, at least not in a manner that was detectable using our sampling technique, which we acknowledge can only provide an approximation of expression levels. The lack of preferentially expressed transcription factors between species is likely an indicator that those transcription factors detected had biological roles more in line with the development or maintenance of cell fate functions than with the regulation of specialized metabolism. More detailed measurements of expression and function for individual members of such protein classes is definitely warranted.
Comparison of Type 1 and 4 Trichomes
Comparisons across all species for the physically similar type 1 and type 4 trichomes revealed that there are substantial differences between species for these glandular trichome types. Again the S. pennellii and S. habrochaites type 1/4 glands contained the most diverse sets of acyl sugars, phenylpropanoids, terpenoids, and alkaloids compared with other species (Fig. 2; Supplemental Fig. S2). For instance, S. pennellii type 4 glands, despite the presence of a few acyl sucroses (Supplemental Fig. S2, cluster 2G), contain mostly acyl glucoses. Also, S. habrochaites type 1/4 trichomes contain largely acyl sucroses rather than acyl glucoses (Supplemental Fig. S2). As with the other trichome and species comparisons listed above, cultivated tomato type 1 glands are deficient in most of the specialized metabolites detected in this study.
Another trend evident in the type 1/4 comparison was the striking similarity between S. habrochaites type 1 and 4 trichomes at both the chemical and transcript levels (Figs. 3 and 4; Supplemental Fig. S2). While there were a few select compounds that exhibited differences in this comparison, such as a triacyl glucose with 20 acyl carbons, and two terpenoid metabolites tentatively assigned as glycosides of sesterterpene malonate esters, being more concentrated in type 1 than in type 4 (Supplemental Fig. S2, clusters 1A, 1B, and 2B), these differences were specific to only a few compounds and did not represent the overall metabolite profiles that were measured. As type 1 glandular trichomes are typically taller than type 4 trichomes, the relative differences in their metabolite contents may simply be a product of variation in cell size or trichome development.
Analysis of transcript levels (Supplemental Tables S6 and S7) demonstrated that the acyl sugar, phenylpropanoid, flavonoid, terpenoid, and specific alkaloid pathways are expressed in S. habrochaites type 1/4 glands, as are enzymes from core metabolism, as was observed for type 6 glands. Together, these data suggest that, like type 6 glands, S. habrochaites type 1/4 glands are most likely capable of both fixing carbon and metabolizing imported sugars. Not surprisingly, enzymes involved in the production of keto acids, fatty acids, lipids, and their subsequent derivatization are represented as well. Type 1/4 glands possess characteristic cuticle-enclosed heads that act as the site of the biosynthesis and storage of specialized metabolites, although these trichomes also apparently secrete the majority of the acyl sugars that are exuded from glands and coat the surface of Solanum leaves. The mechanism whereby these compounds are secreted from the gland cells has not yet been identified.
Comparison of transcript levels for annotated genes, whether from the EC or UPID standpoint, identified no significant differences between S. habrochaites type 1 and 4 glandular trichomes. Thus, the distinction of a trichome type as being either type 1 or 4 may be artificial. For this reason, comparisons to S. lycopersicum type 1 and S. pennellii type 4 were performed with a combined library referred as S. habrochaites type 1/4. Only one protein, a nonspecific lipid transfer protein, was preferentially expressed (in S. lycopersicum type 1 glands) in these comparisons. Not only was this the only gene with preferential expression in all of the Solanum type 1/4 comparisons, but it was also one of the most prevalent transcripts in all species sampled. It is no surprise that this particular protein is highly expressed, as it has been previously shown to be induced by both biotic and abiotic stress (Torres-Schumann et al., 1992; Ooi et al., 2008). Clearly, trichomes are exposed to “the weather” more than most cell types. In addition, the lack of differences in EST counts for UPID and EC annotations (Supplemental Figs. S15–S18) may be due to limited sampling of cells that were difficult to obtain. We expected to observe at least a few differences between species, considering the differences in metabolite content and quantities as shown in Figures 2 and 3 and Supplemental Table S2. An alternative explanation may be that regulation of compound production in this glandular trichome type may occur at levels not evaluated in this study, such as by protein turnover, biochemical control, substrate availability, or altered enzyme activity due to posttranslational modifications.
Comparison of Solanum Type 6 and Type 1/4 Trichomes
To test for processes specific to either type 6 or type 1/4 glandular trichomes, we made three intraspecies comparisons (S. lycopersicum type 6 versus type 1, S. habrochaites type 6 versus type 1/4, and S. pennellii type 6 versus type 4; Supplemental Tables S18–S20) using the method of Stekel et al. (2000). Comparisons of type 6 and type 1/4 transcriptomes identified a limited number of significant differences between the trichome types in each of the three species. The S. pennellii comparison between the Sanger type 6 and 4 libraries identified only one gene (A1XEL0_TOBAC) annotated as a cytochrome P450 of unknown function that was preferentially expressed in the type 6 gland (Supplemental Table S20). However, for the S. lycopersicum and S. habrochaites comparisons, which use two libraries of dissimilar size and sequencing methodologies, these comparisons must be more carefully considered. The results of the S. lycopersicum and S. habrochaites comparisons nevertheless demonstrate that a small number of known, annotated genes are preferentially expressed in either trichome type. Genes associated (Table II; Supplemental Tables S18 and S19) with the type 6 gland include several involved in terpenoid biosynthesis and stress response (B8XA40_SOLHA, Z,Z-farnesyl pyrophosphate synthase; Q9FQ28_SOLLC, sesquiterpene synthase 2; B8XA41_SOLHA, bergamotene/santalene synthase; EC 2.4.1.115, anthocyanidin 3-O-glucosyltransferase; EC 1.10.3.1, catechol oxidase; and EC 1.13.11.12, Q96573_SOLLC, lipoxygenase [Stekel et al., 2000]). We have used the observation that transcripts of the S. lycopersicum homologs of B8XA40, B8XA41, and Q9FQ28 are abundant in type 6 glands of S. lycopersicum, but not in other S. lycopersicum glands, as the starting point to identify the enzymes responsible for the synthesis of monoterpenes in the type 6 glands, but not in other glands, of this species (Schilmiller et al., 2009). In that work, we used biochemical assays and genetic analysis of near-isogenic lines to show that the B8XA40 homolog in S. lycopersicum encodes neryl diphosphate synthase. Neryl diphosphate is used as the substrate by the Q9FQ28 homolog, a terpene synthase, to synthesize β-phellandrene and other monoterpenes.
Table II. Selected pathways and known enzyme expression in different trichome types across Solanum species.
Per 10K, Parts per 10,000.
| UniProt Identifier/Pathway | Description | S. habrochaites Type 6 (per 10K) | S. habrochaites Type 1 (per 10K) | S. habrochaites Type 4 (per 10K) | S. habrochaites Type 1/4 (per 10K) | S. lycopersicum Type 6 (per 10K) | S. lycopersicum Type 1 (per 10K) | S. lycopersicum Type 7 (per 10K) | S. pennellii Type 4 (per 10K) | S. pimpinellifolium Type 6 (per 10K) |
| Terpenoid biosynthetic pathway | ||||||||||
| A9ZN10_HEVBR | 2-C-Methyl-d-erythritol 4-phosphate cytidylyltransferase | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| Q9XH50_SOLLC | 1-d-Deoxyxylulose 5-phosphate synthase | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| Q2XTB5_SOLTU | LYTB-like protein-like | 1,269 | 1 | 1 | 2 | 226 | 0 | 0 | 0 | 509 |
| Q1A746_SOLLC | Geranyl pyrophosphate synthase | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| B8XA40_SOLHA | Z,Z-Farnesyl pyrophosphate synthase (S. habrochaites)/neryl diphosphate synthase (S. lycopersicum) | 7,116 | 0 | 3 | 3 | 1,202 | 0 | 0 | 0 | 170 |
| B8XA41_SOLHA | Santalene and bergamotene synthase (S. habrochaites)/β-phelandrene synthase (S. lycopersicum) | 1,879 | 0 | 2 | 2 | 669 | 0 | 0 | 0 | 624 |
| Q9ZS34_TOBAC | Geranylgeranyl reductase | 42 | 0 | 0 | 0 | 94 | 2 | 1 | 0 | 78 |
| O64961_SOLLC | Epidermal germacrene C synthase (sesquiterpene synthase 1) | 39 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 423 |
| O64962_SOLLC | Germacrene C synthase | 0 | 0 | 0 | 0 | 445 | 0 | 0 | 0 | 5 |
| Q4A570_SOLLC | Putative squalene epoxidase | 2 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 1 |
| Q68IP4_SOLHA | Putative squalene epoxidase | 880 | 0 | 0 | 0 | 300 | 2 | 0 | 0 | 340 |
| Q9FQ28_SOLLC | Sesquiterpene synthase 2 (fragment) | 14 | 0 | 0 | 0 | 1,938 | 3 | 1 | 0 | 1 |
| Acyl sugar biosynthetic pathway | ||||||||||
| Q0VH87_GOSHI | 3-Ketoacyl-CoA reductase 2 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 2 |
| A9XU45_GOSHI | 3-Ketoacyl-CoA synthase 3 | 11 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 47 |
| KCS19_ARATH | 3-Ketoacyl-CoA synthase 19 | 7 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 6 |
| ILV5_ARATH | Ketol acid reductoisomerase, chloroplastic | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 |
| Q52QX9_MANES | Aldo/keto reductase | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| B5LAT2_CAPAN | Putative acetolactate synthase | 6 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 1 |
| Q9SMC2_NICPL | Acetolactate synthase small subunit | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| B5LAV3_CAPAN | Putative isopropylmalate synthase | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 1 |
| LEU1B_SOLPN | 2-Isopropylmalate synthase B | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 6 |
| Q9FEN7_SOLTU | Dihydrolipoyl dehydrogenase | 9 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 1 |
| IVD1_SOLTU | Isovaleryl-CoA dehydrogenase 1, mitochondrial | 29 | 0 | 0 | 0 | 52 | 0 | 1 | 0 | 36 |
On the other hand, both S. habrochaites and S. lycopersicum type 1/4 trichomes preferentially express a nonspecific lipid-transfer protein (NLTP1_SOLLC) when compared with type 6 glands; this gene may play an important role in the secretion of acyl sugars (Slocombe et al., 2008). Together, all three Solanum type 1/4 versus type 6 gland comparisons indicate that there appears to be no quantitative disparity in the expression of genes involved with specialized metabolite pathways, such as acyl sugar or terpenoid biosynthesis (Table II). In addition, the preferential expression of a nonspecific lipid-transfer protein in type 1/4 trichomes in two of the three species is not surprising given the secretory activity of this trichome type, as evidenced by acyl sugar exudate accumulation on the leaves of these species, and may implicate this gene in the secretory activity of these cells.
It is important to note that some of the individual type 1 and 4 gland EST databases are limited in size, so that the prevalence of some transcripts that are not highly represented could not be reliably compared with that in other types of glands. For example, we recently showed by quantitative PCR that a rare transcript found in the S. habrochaites type 4 trichome EST database is nevertheless present in type 1 and 4 trichomes at about a 10-fold higher level than in type 6 trichomes. Biochemical assays showed that the transcript encodes an enzyme that methylates myricetin, thus explaining the above-noted observation that type 1 and 4 glands of S. habrochaites contain severalfold higher levels of methylated myricetin than do type 6 glands (A. Schmidt and E. Pichersky, unpublished data).
Characterization of Solanum Type 7 Trichomes
The type 7 glandular trichome is a short, multicelled trichome (Fig. 1M) that has been largely uncharacterized. We were able to collect type 7 trichomes from S. habrochaites for metabolite and transcriptome profile analysis. However, this gland type was either absent or too difficult to obtain from the other species to be included in our analysis. Surprisingly, type 7 glands from S. habrochaites were found to possess considerably higher concentrations of most metabolite classes than many of the other trichome types from the other species (Figs. 2 and 3). However, most of the compounds observed were also present at similar levels in other trichome types from S. habrochaites, suggesting contamination by the exudates produced by type 1/4 glands. The exceptions were the alkaloids tomatine and dehydrotomatine (Supplemental Fig. S2, cluster 2B), which were present in S. habrochaites type 7 glands but essentially absent from all other glandular trichome types.
Further investigations into the biological roles of this particular glandular trichome type led us to sequence 1,980 ESTs using the Sanger method and analyze chemical extracts from individually isolated S. lycopersicum type 7 glands. This analysis revealed that this gland type contains few if any transcripts with gene ontologies associated with specialized metabolism (Supplemental Fig. S19; Supplemental Tables S21–S24). In contrast, Cys protease inhibitors were strongly up-regulated in this trichome type, suggesting a role separate from the other glandular trichome types in plant defense.
CONCLUSION
The type 6 and type 1/4 trichomes from the Solanum species analyzed in this study contain somewhat overlapping and modular sets of specialized metabolites, particularly acyl sugars, when compared within species. Despite this overlap, there are quantitative differences in metabolite profiles between Solanum species. These differences mirror previously determined Solanum phylogenies (Peralta and Spooner, 2001; Medeiros and Tingey, 2006). Glandular trichomes of S. habrochaites contain the most diverse sets of acyl sugars, fatty acids, alkaloids, and terpenoids compared with all other species, with sesterterpenes and acyl sucroses as the dominant forms of their respective metabolite classes (terpenoids and acyl sugars). The species with the second most diverse array of specialized metabolites is S. pennellii, dominated by the acyl glucose class of compounds. The remaining two wild species, S. arcanum and S. pimpinellifolium, have metabolite profiles that are very similar to S. lycopersicum and possess only modest differences when compared with their domesticated counterpart. Also, the transcriptomes of each of the trichome types generally mirror their respective biochemistries. Analysis of the glandular trichome EST collections reveals that all of the type 1/4 and 6 glands appear to contain most of the genes necessary for specialized metabolite biosynthesis, such as for the acyl sugar, terpenoid, or flavonoid pathways. More quantitative transcript comparisons (Supplemental Tables S14–S20) between species and gland types indicate that distinct chemical profiles may be due to differential regulation of specific genes or gene pathway segments on a transcriptional level or by other processes that are not directly connected to transcription, such as posttranslational modification of proteins, protein turnover, or biochemical control and cross-talk between pathways, among other possibilities.
Notable differences between different glandular trichome types were observed within species. For example, S. lycopersicum type 1 glandular trichomes are dominated by acyl glucoses (although even these compounds were present at relatively low levels in this trichome type from this species), while type 6 glands from this species are dominated by acyl sucroses. Complementary analysis of type 6 gland transcripts across species mirrored some of the traits observed in the different species’ metabolite profiles. Unfortunately, the comparison of metabolite levels with transcript levels was unable to identify specific genes whose expression levels could explain the differences in acyl sucrose and acyl glucose content for S. habrochaites or S. pennellii type 6 trichomes. Thus, unlike terpenoid biosynthesis in S. habrochaites, control of acyl sugar metabolism is likely exerted at a level not observable by the transcriptome analysis used in this study, a fact observed in the type 1/4-type 6 transcript comparisons of S. lycopersicum and S. habrochaites. However, this conclusion may be limited due to the small sampling size of the type 1/4 EST collections.
Many of the genes involved in photosynthesis and carbon fixation are expressed at significant (although not particularly high) levels in the Solanum trichome types investigated, in contrast to what has been observed in some other species (Turner et al., 2000; Gang et al., 2001; Deschamps et al., 2006; Gunnewich et al., 2007; Marks et al., 2009). Therefore, it is likely that at least some of the carbon required for the synthesis of the specialized metabolites found in Solanum glandular trichomes may be fixed within the trichome secretory cells. The relationship between de novo carbon fixation and import of Suc as the source for carbon skeletons, however, remains to be determined.
The least characterized of all glandular trichome types, the type 7 glandular trichome, appears to have a more limited metabolite profile than the other types. Because of the method required to collect type 7 glands, we cannot be sure that the type 7 transcriptome and metabolic profile sampled are completely exclusive to the type 7 gland. Nevertheless, it is clear that specialized metabolite production is limited in this glandular trichome type compared with the other types.
Finally, the distinction between type 1 and type 4 glandular trichomes, at least in S. habrochaites, may represent an artificial classification system, due to the lack of wholesale differences between the metabolic profile or the transcriptome of either cell type. Therefore, it may be more reasonable to consider type 4 glandular trichomes as simply a morphotype of a larger type 1 class.
MATERIALS AND METHODS
Plants and Growth Conditions
All Solanum germplasm used in this study was obtained from the C.M. Rick Tomato Genetics Resource Center. Seeds were treated with 10% sodium tripolyphosphate for 20 min and rinsed in 10% bleach for 1 min. After rinsing thoroughly with sterile water, seeds were spread out on moist 3M Whatman filter paper in a sealed container. After 4 to 5 d, the seeds were transferred to peat pellets. The plants were grown in a growth room under a 16-h-light (27°C)/8-h-dark (18°C) cycle with the light intensity at approximately 200 μE m−2 s−1. Plants in peat pellets were watered daily with Peter’s Professional general purpose 20:20:20 diluted to give 100 μL L−1 nitrogen. After 3 to 4 weeks, depending on the species, seedlings were transferred to 4-inch square pots and grown in the same growth room until analysis.
Production of 454 Libraries
Production of 454 libraries was exclusive to the type 6 trichome for all species except Solanum pennellii. For Solanum lycopersicum, Solanum arcanum, Solanum pimpinellifolium, and Solanum habrochaites, 15 to 20 g of young leaves approximately 1 cm wide was collected and covered for 15 min with 200 mL of ice-cold wash buffer (50 mm Tris-HCl, pH 7.5, 200 mm sorbitol, 20 mm Suc, 10 mm KCl, 5 mm MgCl2, 0.5 mm K2HPO4, 5 mm succinic acid, 1 mm EGTA, diethyl pyrocarbonate-treated water, 1 mm aurintricarboxylic acid, and 14 mm β-mercaptoethanol). Fifty milliliters of 0.5-mm glass beads (Biospec Products) was added to the beaker and then sealed shut using Parafilm. Once sealed, the beaker containing the glass beads and leaves was shaken by hand 300 times followed by 60 s on ice, repeated two more times. After shaking was complete, the leaf slurry was poured through a series of plastic funnels, each with an attached nylon mesh cloth with different pore sizes. The flow-through was collected in an ice-cold 1-L beaker at each step. The order of these meshes was as follows: 350 μm/105 μm/73 μm/54 μm/40 μm; with the type 6 trichomes collected on the final 40-μm mesh. The type 6 trichome fraction was immediately transferred to RNAlater and stored at −80°C until processed using the Qiagen RNEasy kit. cDNA was synthesized according to the protocol described by Kapteyn et al. (2010) and sequenced by 454 sequencing (GS FLX) at the Michigan State University Research Technology Support Facility.
Production of Sanger Libraries
In order to produce the remaining glandular trichome EST libraries from S. lycopersicum, S. pennellii, and S. habrochaites, a protocol utilizing stretched glass pipettes (Supplemental Fig. S20) or microscissors (for accessions where the glands would not stick to the glass pipettes) was used to collect samples consisting of 4,000 each of type 1 (S. lycopersicum, S. habrochaites), type 4 (S. pennellii, S. habrochaites), and type 6 (S. pennellii) trichomes. These were transferred immediately into RNAlater and stored at −80°C until processed using the Qiagen RNEasy kit. Again, the resulting cDNA synthesis was performed using the protocol described by Kapteyn et al. (2010) and then subcloned into the plasmid pCR2.1.
S. lycopersicum type 7 trichome fractions used for EST analysis were produced in a manner similar to those of the Solanum type 6 fractions described above; however, an additional set of filters were added to further remove as many type 6, type 1, and hairy trichomes as possible from the initial type 6 flow-through. Additional steps include further filtration through a 73-μm and a 33-μm mesh. The final, enriched type 7 fraction was collected on the 33-μm mesh and used to produce cDNA as described (Kapteyn et al., 2010).
LC-MS Analysis of Trichome Extracts
S. habrochaites type 7 trichomes were collected in the manner described above, transferred to a microfuge tube, spun down (3,000g, 30 s), and resuspended in extraction solvent after the wash buffer was removed. For the remaining trichome types, picked trichome fractions of approximately 1,500 glandular trichome secretory cell clusters (heads) were immediately transferred to 50 μL/50 glands ice-cold extraction solvent, 2-propanol:CH3CN:water (3:3:2, v/v/v). The gland/solvent mixture was then sonicated for 5 min followed by vortexing for 30 s and centrifugation at 5,000g for 10 min. The supernatant was then transferred to a glass autosampler vial equipped with a 100-μL glass insert. LC-MS analysis of isolated gland metabolites was performed using a model LCT Premier (Waters) mass spectrometer coupled to an LC-20AD pump and SIL-5000 autosampler (Shimadzu). All analyses utilized electrospray ionization in negative ion mode and employed a custom-packed Supelco Discovery Bio C18 column (1 × 150 mm, 3-μm particles). Mobile phase gradients were based on solvent A (0.15% aqueous formic acid) and solvent B (methanol), with a total flow rate of 0.05 mL min−1. The gradient consisted of initial condition 95% A, a linear gradient to 50% A at 5 min, a linear gradient to 5% A at 33 min, and then to 0% A at 35 min. The mobile phase was held constant until 38 min, when the composition returned to the original composition. Spectra were acquired using two quasisimultaneous conditions: high energy (aperture 1, 80 V) and low energy (aperture 1, 10 V) in the instrument source. Mass spectra were acquired at 1 Hz. Peak integration and alignment were performed using MarkerLynx software (Waters), and an array of peak areas was exported as a text file. Following acquisition of mass spectra, replicate peak areas for each metabolite were averaged within each species/trichome. Due to the calculated data being distributed dramatically over a wide range (0–800,000+), a log2 transformation was performed in order to compensate for the huge variation in the data. As log2(0) has no mathematical definition, log2(0+1) was used for transforming zero values. Transformed values were then normalized across 10 species/trichomes using the following equation: for values
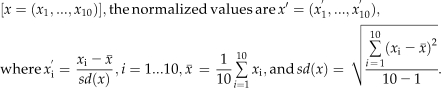 |
Once normalized, cluster analysis was performed on the known compound data that contains 119 annotated metabolites, as shown in Supplemental Table S2. To produce the box plot in Figure 2, these values were aggregated according to metabolite class, and the class medians, averages, and both upper and lower quartiles were calculated. The uncertainty in hierarchical cluster analysis was assessed using the bootstrap resampling method, which was implemented by the R package pvclust (Shimodaira, 2002, 2004). The P values listed are the approximately unbiased P values, which are computed by multiscale bootstrap resampling and are a better approximation to unbiased P value than those computed by normal bootstrap resampling using a P value cutoff chosen as 95%. PCA and PLS-DA were performed using MetaboAnalyst (http://www.metaboanalyst.ca).
Analysis and Comparisons of EST Libraries
ESTs were assembled and annotated using the PAVE system (Soderlund et al., 2009; http://www.agcol.arizona.edu/pave/solanum/). Annotation of the assembled contigs was primarily accomplished via the use of BLAST comparisons with first the UniProt (UniProt Consortium 2010 [www.uniprot.org]) database, followed by the lesser annotated GenBank (http://www.ncbi.nlm.nih.gov/GenBank/). The resulting annotation of the trichome contigs using either the UniProt or GenBank database permitted associated BLAST hits to be used as a basis for cross-species comparison of trichome transcriptomes on the basis of either EC numbers or UniProt identifiers. In addition, the Rstat values for each of the EC and UniProt identifiers were calculated as well as the minimum Rstat values necessary for the determination of a greater than 99% believability between libraries with a comparison (Stekel et al., 2000). Cross-species comparisons were specifically performed via PAVE annotation software and custom scripts, and only between EST collections of similar size and sequencing methodology. The results of these comparisons are summarized in Supplemental Tables S5 to S7, and the results of the assemblies and comparisons can be viewed at www.agcol.arizona.edu/pave/solanum. EC annotation of the contents of all available Solanum trichome metabolic pathways were pictorially reconstructed using the iPATH software (Letunic et al., 2008), which can be accessed at http://pathways.embl.de.
Sequence data from this article can be found in the GenBank EST and Sequence Read Archives under accession numbers GT157943 to GT161597, GT162273 to GT165033, GT166527 to GT168680, GT168709 to GT183821, SRR015435, SRR015436, and SRR027939 to SRR027943.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Two-way cluster analysis of normalized LC-MS metabolite peak areas from Solanum glandular trichomes and their associated clusters.
Supplemental Figure S2. Clusters of normalized LC-MS metabolite peak areas from Solanum glandular trichomes with corresponding accumulation patterns as identified by two-way cluster analysis.
Supplemental Figure S3. Multivariate analysis of metabolite data obtained for S. habrochaites, accession LA1777, the only accession that had readily isolatable trichomes for all types.
Supplemental Figure S4. Biochemical pathways expressed in S. habrochaites type 6 glandular trichomes.
Supplemental Figure S5. Biochemical pathways expressed in S. lycopersicum type 6 glandular trichomes.
Supplemental Figure S6. Biochemical pathways expressed in S. pennellii type 6 glandular trichomes.
Supplemental Figure S7. Biochemical pathways expressed in S. pimpinellifolium type 6 glandular trichomes.
Supplemental Figure S8. Biochemical pathways expressed in S. arcanum type 6 glandular trichomes.
Supplemental Figure S9. Biochemical pathways preferentially expressed in either S. habrochaites type 6 or S. lycopersicum type 6 glandular trichomes when queried by UniProt identifier.
Supplemental Figure S10. Biochemical pathways preferentially expressed in either S. habrochaites type 6 or S. lycopersicum type 6 glandular trichomes when queried by EC number.
Supplemental Figure S11. Biochemical pathways preferentially expressed in either S. habrochaites type 6 or S. pimpinellifolium type 6 glandular trichomes when queried by UniProt identifier.
Supplemental Figure S12. Biochemical pathways preferentially expressed in either S. habrochaites type 6 or S. pimpinellifolium type 6 glandular trichomes when queried by EC number.
Supplemental Figure S13. Biochemical pathways preferentially expressed in either S. lycopersicum type 6 or S. pimpinellifolium type 6 glandular trichomes when queried by UniProt identifier.
Supplemental Figure S14. Biochemical pathways preferentially expressed in either S. lycopersicum type 6 or S. pimpinellifolium type 6 glandular trichomes when queried by EC number.
Supplemental Figure S15. Biochemical pathways present in S. habrochaites type 1 and S. habrochaites type 4 glandular trichome EST libraries when queried by EC number.
Supplemental Figure S16. Biochemical pathways present in S. habrochaites type 1/4 composite and S. lycopersicum type 1 glandular trichome EST libraries when queried by EC number.
Supplemental Figure S17. Biochemical pathways present in S. lycopersicum type 1 and S. pennellii type 4 glandular trichome EST libraries when queried by EC number.
Supplemental Figure S18. Biochemical pathways present in S. habrochaites type 1/4 composite and S. pennellii type 4 glandular trichome EST libraries when queried by EC number.
Supplemental Figure S19. Biochemical pathways expressed in S. lycopersicum type 7 glandular trichomes when queried by EC number.
Supplemental Figure S20. S. habrochaites leaf showing how gland cells are removed from trichomes without disturbing other cell types.
Supplemental Table S1. Normalized LC-MS peak areas of known compounds from Solanum glandular trichome extracts.
Supplemental Table S2. Clusters of Solanum metabolites containing known compounds.
Supplemental Table S3. Summary of expressed, putative enzymes in Solanum type 6 trichomes.
Supplemental Table S4. Solanum species-specific trichome EST libraries and assemblies.
Supplemental Table S5. Solanum trichome EST libraries and assemblies.
Supplemental Table S6. Interspecies and intraspecies trichome-specific UPID comparisons used in this study.
Supplemental Table S7. Interspecies and intraspecies trichome-specific EC number comparisons used in this study.
Supplemental Table S8. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. habrochaites type 6 trichomes in comparison with S. lycopersicum type 6 trichomes.
Supplemental Table S9. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. lycopersicum type 6 trichomes in comparison with S. habrochaites type 6 trichomes.
Supplemental Table S10. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. habrochaites type 6 trichomes in comparison with S. pimpinellifolium type 6 trichomes.
Supplemental Table S11. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. pimpinellifolium type 6 trichomes in comparison with S. habrochaites type 6 trichomes.
Supplemental Table S12. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. lycopersicum type 6 trichomes in comparison with S. pimpinellifolium type 6 trichomes.
Supplemental Table S13. Overrepresented and underrepresented biological process Gene Ontology annotations of statistically significant overexpressed UniProt identifiers in S. pimpinellifolium type 6 trichomes in comparison with S. lycopersicum type 6 trichomes.
Supplemental Table S14. Summary of preferentially expressed, putative enzymes in Solanum type 6 trichomes and their expression in comparison with other Solanum species when queried by UniProt identifier.
Supplemental Table S15. Summary of preferentially expressed, putative enzymes in Solanum type 6 trichomes and their expression in comparison with other Solanum species when queried by EC number.
Supplemental Table S16. Summary of metabolic enzyme annotations expressed in Solanum type 1 and/or type 4 Solanum trichome libraries when queried using UniProt identifiers.
Supplemental Table S17. Summary of metabolic enzyme annotations expressed in Solanum type 1 and/or type 4 Solanum trichome libraries when queried using EC numbers.
Supplemental Table S18. Summary of metabolic enzyme annotations expressed in Solanum type 6 and type 1/4 Solanum trichome libraries when queried using identifiers.
Supplemental Table S19. Summary of preferentially expressed, putative enzymes in Solanum type 6 and type 1/4 trichomes within the species when queried by EC number.
Supplemental Table S20. Summary of metabolic enzyme annotations expressed in Solanum type 6 and type 1/4 Solanum trichome libraries when queried using UniProt identifiers.
Supplemental Table S21. Gene Ontology summary of the S. lycopersicum type 7 cDNA library.
Supplemental Table S22. UPID summary of the S. lycopersicum type 7 cDNA library.
Supplemental Table S23. Summary of metabolic enzyme annotations expressed in the S. lycopersicum type 7 trichome library when queried using EC numbers.
Supplemental Table S24. Summary of contig expression in S. lycopersicum type 7 trichomes.
Supplementary Material
Acknowledgments
We acknowledge the support of the staff of Michigan State University’s Research Technology Support Facility for performing the 454 sequencing and give special thanks to Jeff Landgraff, Shari Tjugum-Holland, and Kevin Carr. We thank Gregg Howe for suggestions to improve the manuscript and Robert Last and Curtis Wilkerson for helpful discussions.
References
- Alba JM, Montserrat M, Fernández-Muñoz R. (2009) Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp Appl Acarol 47: 35–47 [DOI] [PubMed] [Google Scholar]
- Allen DK, Ohlrogge JB, Shachar-Hill Y. (2009) The role of light in soybean seed filling metabolism. Plant J 58: 220–234 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y. (2007) Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J 52: 296–308 [DOI] [PubMed] [Google Scholar]
- Alonso WR, Rajaonarivony JIM, Gershenzon J, Croteau R. (1992) Purification of 4S-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha × piperita) and spearmint (Mentha spicata). J Biol Chem 267: 7582–7587 [PubMed] [Google Scholar]
- Antonious GF. (2001) Production and quantification of methyl ketones in wild tomato accessions. J Environ Sci Health B 36: 835–848 [DOI] [PubMed] [Google Scholar]
- Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E. (2009) Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol 151: 1952–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser K, Harper A, Welsby N, Schauvinhold I, Slocombe S, Li Y, Dixon RA, Broun P. (2009) Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol 149: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas KK, Foster AJ, Aung T, Mahmoud SS. (2009) Essential oil production: relationship with abundance of glandular trichomes in aerial surface of plants. Acta Physiol Plant 31: 13–19 [Google Scholar]
- Chain F, Ilieva D, Evans B. (2008) Single-species microarrays and comparative transcriptomics. PLoS ONE 3: e3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WS, Serpe MD, Anderson JV, Gesch RW, Horvath DP. (2006) Sugars, hormones, and environment affect the dormancy status in underground adventitious buds of leafy spurge (Euphorbia esula). Weed Sci 54: 59–68 [Google Scholar]
- Croteau R. (1991) Metabolism of monoterpenes in mint (Mentha) species. Planta Med (Suppl) 57: S10–S14 [DOI] [PubMed] [Google Scholar]
- Deschamps C, Gang D, Dudareva N, Simon JE. (2006) Developmental regulation of phenylpropanoid biosynthesis in leaves and glandular trichomes of basil (Ocimum basilicum L.). Int J Plant Sci 167: 447–454 [Google Scholar]
- Economou LP, Lykouressis DP, Barbetaki AE. (2006) Time allocation of activities of two heteropteran predators on the leaves of three tomato cultivars with variable glandular trichome density. Environ Entomol 35: 387–393 [Google Scholar]
- Franceschi VR, Giaquinta RT. (1983) Glandular trichomes of soybean leaves: cytological differentiation from initiation through senescence. Bot Gaz 144: 175–184 [Google Scholar]
- Fridman E, Wang JH, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E. (2005) Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang JH, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E. (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, McCaskill D, Rajaonarivony JIM, Mihaliak C, Karp F, Croteau R. (1992) Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem 200: 130–138 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Pritchard JK, Thornton K. (2009) Characterizing natural variation using next-generation sequencing technologies. Trends Genet 25: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani C, Bini LM. (2008) Insight into the structure and chemistry of glandular trichomes of Labiatae, with emphasis on subfamily Lamioideae. Plant Syst Evol 276: 199–208 [Google Scholar]
- Goffman FD, Ruckle M, Ohlrogge J, Shachar-Hill Y. (2004) Carbon dioxide concentrations are very high in developing oilseeds. Plant Physiol Biochem 42: 703–708 [DOI] [PubMed] [Google Scholar]
- Gonzalez WL, Negritto MA, Suarez LH, Gianoli E. (2008) Induction of glandular and non-glandular trichomes by damage in leaves of Madia sativa under contrasting water regimes. Acta Oecologica 33: 128–132 [Google Scholar]
- Gunnewich N, Page JE, Kollner TG, Degenhardt J, Kutchan TM. (2007) Functional expression and characterization of trichome-specific (−)-limonene synthase and (+)-alpha-pinene synthase from Cannabis sativa. Natural Product Communications 2: 223–232 [Google Scholar]
- Horgan FG, Quiring DT, Lagnaoui A, Pelletier Y. (2007) Variable responses of tuber moth to the leaf trichomes of wild potatoes. Entomol Exp Appl 125: 1–12 [Google Scholar]
- Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. (2004) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 134: 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Lonien J, Heady LE, Rogers A, Schwender J. (2007) Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68: 2232–2242 [DOI] [PubMed] [Google Scholar]
- Kandra L, Wagner GJ. (1988) Studies of the site and mode of biosynthesis of tobacco trichome exudate components. Arch Biochem Biophys 265: 425–432 [DOI] [PubMed] [Google Scholar]
- Kang JH, Shi F, Jones AD, Marks MD, Howe GA. (2010) Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J, He R, McDowell ET, Gang DR. (2010) Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genomics 11: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene CK, Wagner GJ. (1985) Direct demonstration of duvatrienediol biosynthesis in glandular heads of tobacco trichomes. Plant Physiol 79: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy GG. (2003) Tomato, pests, parasitoids, and predators: tritrophic interactions involving the genus Lycopersicon. Annu Rev Entomol 48: 51–72 [DOI] [PubMed] [Google Scholar]
- Kim ES, Mahlberg PG. (1991) Secretory cavity development in glandular trichomes of Cannabis sativa L. (Cannabaceae). Am J Bot 78: 220–229 [Google Scholar]
- Kolb D, Muller M. (2003) Different trichome types on the leaves of Styrian oil pumpkin. Phyton Ann Rei Bot 43: 365–379 [Google Scholar]
- Kumar NKK, Ullman DE, Cho JJ. (1995) Frankliniella occidentalis (Thysanoptera, Thripidae) landing and resistance to tomato spotted wilt tospovirus among Lycopersicon accessions with additional comments on Thrips tabaci (Thysanoptera, Thripidae) and Trialeurodes vaporariorum (Homoptera, Aleyrodidae). Environ Entomol 24: 513–520 [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Yamada T, Kanehisa M, Bork P. (2008) iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci 33: 101–103 [DOI] [PubMed] [Google Scholar]
- Lin SYH, Trumble JT, Kumamoto J. (1987) Activity of volatile compounds in glandular trichomes of Lycopersicon species against 2 insect herbivores. J Chem Ecol 13: 837–850 [DOI] [PubMed] [Google Scholar]
- Liu J, Xia KF, Zhu JC, Deng YG, Huang XL, Hu BL, Xu XP, Xu ZF. (2006) The nightshade proteinase inhibitor IIb gene is constitutively expressed in glandular trichomes. Plant Cell Physiol 47: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Luckwill LC. (1943) The Genus Lycopersicon: An Historical, Biological and Taxonomic Survey of the Wild and Cultivated Tomatoes. Aberdeen University Press, Aberdeen, UK [Google Scholar]
- Maluf WR, Inoue IF, Ferreira R, Gomes LAA, de Castro EM, Cardoso MDG. (2007) Higher glandular trichome density in tomato leaflets and repellence to spider mites. Pesquisa Agropecu Bras 42: 1227–1235 [Google Scholar]
- Marks MD, Tian L, Wenger JP, Omburo SN, Soto-Fuentes W, He J, Gang DR, Weiblen GD, Dixon RA. (2009) Identification of candidate genes affecting Delta9-tetrahydrocannabinol biosynthesis in Cannabis sativa. J Exp Bot 60: 3715–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins Santana F, Ribeiro SD, Moita AW, Moreira DJ, Giordano LD. (2001) Sources of resistance in Lycopersicon spp. to a bipartite whitefly-transmitted geminivirus from Brazil. Euphytica 122: 45–51 [Google Scholar]
- McCaskill D, Croteau R. (1995) Monoterpene and sesquiterpene biosynthesis in glandular trichomes of peppermint (Mentha × piperita) rely exclusively in plastid-derived isopentenyl diphosphate. Planta 197: 49–56 [Google Scholar]
- McCaskill D, Gershenszon J, Croteau R. (1992) Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha × piperita). Planta 187: 445–454 [DOI] [PubMed] [Google Scholar]
- Medeiros AH, Tingey WM. (2006) Glandular trichomes of Solanum berthaultii and its hybrids with Solanum tuberosum affect nymphal emergence, development, and survival of Empoasca fabae (Homoptera: Cicadellidae). J Econ Entomol 99: 1483–1489 [DOI] [PubMed] [Google Scholar]
- Moyano F, Cocucci A, Sersic A. (2003) Accessory pollen adhesive from glandular trichomes on the anthers of Leonurus sibiricus L. (Lamiaceae). Plant Biol 5: 411–418 [Google Scholar]
- Nagel J, Culley LK, Lu YP, Liu EW, Matthews PD, Stevens JF, Page JE. (2008) EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 20: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MT, Akers CP, Jarlfors UE, Wagner GJ, Berger S. (1991) Comparative ultrastructural features of secreting and nonsecreting glandular trichomes of 2 genotypes of Nicotiana tabacum L. Bot Gaz 152: 13–22 [Google Scholar]
- Nihoul P. (1993) Do light-intensity, temperature and photoperiod affect the entrapment of mites on glandular hairs of cultivated tomatoes. Exp Appl Acarol 17: 709–718 [Google Scholar]
- Nonomura T, Xu L, Wada M, Kawamura S, Miyajima T, Nishitomi A, Kakutani K, Takikawa Y, Matsuda Y, Toyoda H. (2009) Trichome exudates of Lycopersicon pennellii form a chemical barrier to suppress leaf-surface germination of Oidium neolycopersici conidia. Plant Sci 176: 31–37 [Google Scholar]
- Oghiakhe S. (1997) Trichomes and resistance to major insect pests in cowpea, Vigna unguiculata (L.) Walp.: a review. Discov Innov 9: 173–178 [Google Scholar]
- Ooi LSM, Tian L, Su MX, Ho WS, Sun SSM, Chung HY, Wong HNC, Ooi VEC. (2008) Isolation, characterization, molecular cloning and modeling of a new lipid transfer protein with antiviral and antiproliferative activities from Narcissus tazetta. Peptides 29: 2101–2109 [DOI] [PubMed] [Google Scholar]
- Peralta IE, Spooner DM. (2001) Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon). Am J Bot 88: 1888–1902 [PubMed] [Google Scholar]
- Pico B, Diez M, Nuez F. (1998) Evaluation of whitefly-mediated inoculation techniques to screen Lycopersicon esculentum and wild relatives for resistance to Tomato yellow leaf curl virus. Euphytica 101: 259–271 [Google Scholar]
- Puterka GJ, Farone W, Palmer T, Barrington A. (2003) Structure-function relationships affecting the insecticidal and miticidal activity of sugar esters. J Econ Entomol 96: 636–644 [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Turner GW, Lee JM, Croteau RB, Lange BM. (2008) A systems biology approach identifies the biochemical mechanisms regulating monoterpenoid essential oil composition in peppermint. Proc Natl Acad Sci USA 105: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AE, Tingey WM, Mutschler MA. (1993) Acylsugars of Lycopersicon pennellii deter settling and feeding of the green peach aphid (Homoptera, Aphididae). J Econ Entomol 86: 34–39 [Google Scholar]
- Romero GQ, Souza JC, Vasconcellos-Neto J. (2008) Anti-herbivore protection by mutualistic spiders and the role of plant glandular trichomes. Ecology 89: 3105–3115 [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, Jabès F, Duffé P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, et al. (2009) A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106: 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab B, Folkers U, Ilgenfritz H, Hulskamp M. (2000) Trichome morphogenesis in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 355: 879–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51: 492–508 [DOI] [PubMed] [Google Scholar]
- Shimodaira H. (2004) Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat 32: 2616–2641 [Google Scholar]
- Siebert DJ. (2004) Localization of salvinorin A and related compounds in glandular trichomes of the psychoactive sage, Salvia divinorum. Ann Bot (Lond) 93: 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AT, Gurr GM. (2005) Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric For Entomol 7: 265–276 [Google Scholar]
- Simmons AT, Gurr GM, McGrath D, Nicol HI, Martin PM. (2003) Trichomes of Lycopersicon spp. and their effect on Myzus persicae (Sulzer) (Hemiptera: Aphididae). Aust J Entomol 42: 373–378 [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J, et al. (2008) Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol 148: 1830–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Simmons AM, Thacker RR. (1998) Attractancy and ovipositional response of adult Bemisia argentifolii (Homoptera: Aleyrodidae) to type IV trichome density on leaves of Lycopersicon hirsutum grown in three day-length regimes. J Entomol Sci 33: 270–281 [Google Scholar]
- Soderlund CA, Johnson E, Bomhoff M, Descour A. (2009) PAVE: program for assembling and viewing ESTs. BMC Genomics 10: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F. (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res 10: 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Schumann S, Godoy JA, Pintor-Toro JA. (1992) A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant Mol Biol 18: 749–757 [DOI] [PubMed] [Google Scholar]
- Turner GW, Croteau R. (2004) Organization of monoterpene biosynthesis in Mentha: immunocytochemical localizations of geranyl diphosphate synthase, limonene-6-hydroxylase, isopiperitenol dehydrogenase, and pulegone reductase. Plant Physiol 136: 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GW, Gershenzon J, Croteau RB. (2000) Development of peltate glandular trichomes of peppermint. Plant Physiol 124: 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38: D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkama E, Salminen JP, Koricheva J, Pihlaja K. (2003) Comparative analysis of leaf trichome structure and composition of epicuticular flavonoids in Finnish birch species. Ann Bot (Lond) 91: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam NM, Hare JD. (1998) Biological activity of Datura wrightii glandular trichome exudate against Manduca sexta larvae. J Chem Ecol 24: 1529–1549 [Google Scholar]
- Wagner GJ. (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol 96: 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Tian L, Aziz N, Broun P, Dai XB, He J, King A, Zhao PX, Dixon RA. (2008) Terpene biosynthesis in glandular trichomes of hop. Plant Physiol 148: 1254–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang YJ, Zhang Q, Qi Y, Guo DJ. (2009) Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics 10: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston PA, Johnson DA, Burton HT, Snyder JC. (1989) Trichome secretion composition, trichome densities, and spider-mite resistance of 10 accessions of Lycopersicon hirsutum. J Am Soc Hortic Sci 114: 492–498 [Google Scholar]
- Wilkens RT, Shea GO, Halbreich S, Stamp NE. (1996) Resource availability and the trichome defenses of tomato plants. Oecologia 106: 181–191 [DOI] [PubMed] [Google Scholar]
- Xie Z, Kapteyn J, Gang DR. (2008) A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J 54: 349–361 [DOI] [PubMed] [Google Scholar]
- Xie Z, Ma XQ, Gang DR. (2009) Modules of co-regulated metabolites in turmeric (Curcuma longa) rhizome suggest the existence of biosynthetic modules in plant specialized metabolism. J Exp Bot 60: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerger EH, Grazzini RA, Hesk D, Cox-Foster DL, Craig R, Mumma RO. (1992) A rapid method for isolating glandular trichomes. Plant Physiol 99: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HF, Kowalski SP, Steffens JC. (1992) Comparison of polyphenol oxidase expression in glandular trichomes of Solanum and Lycopersicon species. Plant Physiol 100: 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Thacker RR, Snyder JC. (2008) Occurrence of 2,3-dihydrofarnesoic acid, a spidermite repellent, in trichome secretions of Lycopersicon esculentum × L. hirsutum hybrids. Euphytica 162: 1–9 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.