Abstract
Apple (Malus × domestica) represents an interesting model tree crop for studying fruit abscission. The physiological fruitlet drop occurring in this species can be easily magnified by using thinning chemicals, such as benzyladenine (BA), to obtain fruits with improved quality and marketability. Despite the economic importance of this process, the molecular determinants of apple fruitlet abscission are still unknown. In this research, BA was used to obtain fruitlet populations with different abscission potentials to be analyzed by means of a newly released 30K oligonucleotide microarray. RNAs were extracted from cortex and seed of apple fruitlets sampled over a 4-d time course, during which BA triggers fruit drop, and used for microarray hybridization. Transcriptomic profiles of persisting and abscising fruitlets were tested for statistical association with abscission potential, allowing us to identify molecular signatures strictly related to fruit destiny. A hypothetical model for apple fruitlet abscission was obtained by putting together available transcriptomic and metabolomic data. According to this model, BA treatment would establish a nutritional stress within the tree that is primarily perceived by the fruitlet cortex whose growth is blocked by resembling the ovary growth inhibition found in other species. In weaker fruits, this stress is soon visible also at the seed level, likely transduced via reactive oxygen species/sugar and hormones signaling cross talk, and followed by a block of embryogenesis and the consequent activation of the abscission zone.
Fruit development is an exquisitely plant-specific process under the control of a complex interplay of endogenous and environmental factors. Many molecular studies have focused on aspects of the last phases of fruit development and, mostly, for its important economical impacts, on the ripening process (Giovannoni, 2004). A large body of experimental data, obtained in tomato (Solanum lycopersicum) as a model system, supports the master role played by the hormone ethylene for the control of ripening in climacteric fruits (for review, see Giovannoni, 2004). A significantly minor body of research so far has been devoted to the molecular factors involved in fruit set and early fruit development. Despite the detailed characterization of growth dynamics and hormonal balance during the early steps of fruit development (Ozga and Reinecke, 2003; Nitsch et al., 2009), the molecular aspects underpinning these events have only recently begun to be unraveled.
The process of fruit set, defined as the commitment of the ovary tissues to undergo transformation into a fruit (Gillaspy et al., 1993), is gaining increasing interest also for its potential exploitation to control parthenocarpic fruit development in the absence of pollination/fertilization. Auxins and GAs play a pivotal role in the inductive phase of fruit set and the parthenocarpic development of fruits (Gillaspy et al., 1993; Pandolfini et al., 2007; de Jong et al., 2009). Several studies support the view that auxins may represent the master signal triggering cell division, and their interplay with GAs may be required for sustaining cell expansion (for review, see de Jong et al., 2009). In fact, data obtained from both tomato and Arabidopsis (Arabidopsis thaliana) have suggested that the transformation of the ovary into fruit is prevented by a negative control exerted by Auxin/Indole Acetic Acid (AUX/IAA) and Auxin Response Factor (ARF) proteins. The removal of this negative regulation, following pollination/fertilization or treatment with auxins, leads to cell proliferation and to fruit set. Consistently, the derepression of auxin responses through antisense inhibition of AUX/IAA9 (Wang et al., 2005) and ARF7 (de Jong et al., 2009) in tomato and loss of function of ARF8 in Arabidopsis (Goetz et al., 2006) lead to parthenocarpy.
Transcriptomic profiling studies carried out in tomato (Vriezen et al., 2008) have reinforced the view that fruit development appears to rely on the removal of a negative feedback regulation of ovary growth. This inhibition is established by a negative control exerted mainly by abscisic acid (ABA)- and ethylene-dependent pathways. In fact, as soon as fruit set is triggered, the molecular machineries of both ethylene and ABA biosynthesis and action appear to be significantly and promptly down-regulated and, concomitantly, those of auxin and GA biosynthesis and action are activated (Vriezen et al., 2008; Nitsch et al., 2009). These data are progressively giving a hint to the spatiotemporal regulation of the molecular factors involved in early steps of fruit set and development. However, very little or no information is available on how these factors could be modulated by the plant to restrain the development of a fraction of fruits in response to endogenous/environmental perturbations.
Fruit trees have evolved a system to control and adapt the size of the fruit population they bring to final maturity in relation to their nutritional status, thus allowing the plant to make efficient use of resources. This is accomplished by a process called “physiological drop,” consisting of the abscission of young fruitlets during the early phases of development. In apple (Malus × domestica), the physiological drop is eminently a correlative phenomenon and has to be distinguished from the senescence-driven abscission of ripe fruits (Bangerth, 2000). Therefore, drop of young fruits can be interpreted as a developmental arrest that the plant exerts selectively on fruitlets representing weaker sinks, during early phases of development, in response to nutritional shortage. In this scenario, studying the fruit physiological drop can provide important additional insights into the molecular mechanisms regulating early fruit development and the fruit developmental plasticity in response to endogenous and environmental changes. Apple trees are an interesting model system for such a study since they develop flower/fruit clusters in which a clear gradient of correlative dominance exists in relation to the position of the fruit within the cluster (Fig. 1). This dominance can be further exacerbated by means of shading or treatments with chemicals that can induce fruit drop (Greene et al., 1992; Bangerth, 2000), a practice called “fruit thinning.” Fruit thinning is adopted by horticulturists to reduce the number of fruits on the tree, therefore improving their final size and quality. Benzyladenine (BA) is a widely known chemical thinner exerting its action by stimulating shoot growth and, as a consequence, effecting fruit drop by exacerbating competition between shoots and fruit clusters, between the different clusters (intercluster competition), and, prominently, between fruits of the same cluster (intracluster competition; Bangerth, 2000; Bubán, 2000). A number of studies have elucidated this mechanism of action, showing that BA treatment in fact has no effect on fruit drop when applied directly only to fruits (Greene et al., 1992). Therefore, BA represents an interesting tool to evoke fruit developmental arrest and abscission in a controlled, inducible, and selective way through the enhancement of correlative inhibitions and for the identification of the molecular factors underpinning this developmental arrest.
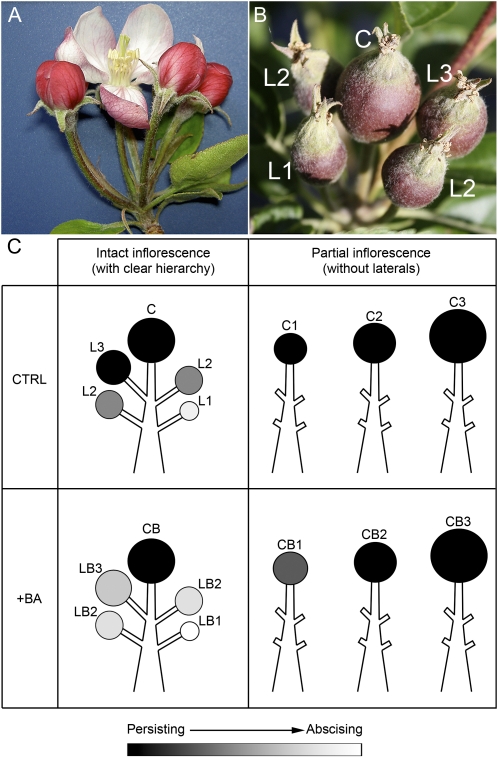
Figure 1.
A, The apple corymb with the central flower at bloom, whose anticipation with respect to the lateral flowers is clearly visible. B, Apple cluster with a clear hierarchy, as indicated by progressive numbers (C, central fruitlet; L1, small lateral fruitlets; L2, medium lateral fruitlets; L3, big lateral fruitlets). C, Schematic representation of the different abscission potentials ascribed to fruitlets within either intact or partial inflorescence (black, strongly persisting; white, strongly abscising) of control (CTRL; top panels) or BA-treated (+BA; bottom panels) trees. Sample fruitlets were labeled according to their size, position within the cluster, and eventual treatment (L, lateral; C, central; B, treated with BA; 1, small size; 2, medium size; 3, big size).
In this study, BA treatments on apple trees were exploited to selectively induce drop of lateral fruits that usually persist and develop. Their global transcriptional profiles were assessed by means of a new 30K microarray in both cortex and seed tissues and compared with those of persisting fruits. Candidate genes with a potential involvement in driving fruit developmental arrest, finally leading to fruit abscission, were identified. Although apple fruit is a pseudocarpic fruit, in which cells of the cortex arise mainly from the receptacle, our data suggest that the developmental arrest leading to fruit drop relies on the reestablishment of negative constraints based on the activation of ABA and ethylene signaling and the inhibition of GA biosynthesis, similar to those that are removed from the ovary to initiate fruit development. The involvement of sugars and reactive oxygen species (ROS) signaling may also be hypothesized.
RESULTS
Establishing Classes of Fruitlets with Different Abscission Potentials
In order to identify the molecular events responsible for apple fruitlet abscission, fruitlet subpopulations characterized by different abscission potentials and fruit drop dynamics were first obtained and sampled. Apple fruitlets develop in clusters, each including a central fruit (also called the “king fruit”) and four lateral fruits (Fig. 1). The position within the cluster is an important determinant of the hierarchy between competing fruits and, consequently, of their tendency to abscise, defined as abscission potential: the central fruit develops earlier, since it originates from an earlier flowering event (Fig. 1A), and exerts a correlative dominance over the lateral fruits, making the latter ones weaker sinks and significantly more prone to abscise (Bangerth, 2000; Fig. 1B). A hierarchy also exists between lateral fruitlets. In fact, those deriving from earlier blooming flowers (L3 fruitlets in Fig. 1) reach a bigger size, a stronger sink activity, and a lower abscission potential and exert a correlative dominance over the smaller ones, inserted below, that finally display the highest abscission potential (Fig. 1, B and C, L2 and L1). In the absence of external perturbations, the central (C) and biggest lateral (L3) fruits are less prone to abscise and virtually bound to stay on the plant, while L2 and L1 undergo shedding. Therefore, in apple clusters, a correlative reproductive dominance exists, starting from the central fruit toward the basal lateral ones, that, in turn, is reflected by an opposed increasing gradient of abscission potentials, as represented by gray to black scales in Figure 1C. As a consequence, fruit size and position within the cluster, being strongly correlated with the capacity of attracting assimilates (Bangerth, 2000), may be considered reliable parameters for predicting the fruitlet abscission potential. Taking these aspects into account, the experimental plan was aimed at triggering the induction of abscission mainly on L3 fruits by treating trees with BA (Fig. 1C, LB3), a well-known chemical thinner. BA exerts its action mainly by enhancing shoot growth and branching (Dal Cin et al., 2007) and therefore exacerbating the correlative competition between fruits, resulting in the abscission of an increased number of lateral fruits of the L3 class while leaving unaffected the L2 and L1 fruit abscission potential (Angeli et al., 2002). In parallel, a subpopulation of nonabscising persisting fruitlets (NAF) was obtained by removing from the cluster all lateral flowers at full bloom, as described by Dal Cin et al. (2005a, 2009a, 2009b), and leaving only the hand-pollinated central one (Fig. 1C, right top panel). However, since the smallest sized central fruits did show a minimal tendency to drop after BA treatment (less than 10%), these were considered “borderline” samples (Fig. 1C, right bottom panel). By means of this approach, eight populations were sampled and classified for transcript profiling and assigned to the following four predicted different abscission potentials (APs) based upon previous experiments (A. Botton, unpublished data): naturally abscising fruitlets (L1 fruitlets; 90% < AP < 100%), strongly abscising fruitlets (LB1 and LB3 fruitlets; 90% < AP < 100%), probably persisting fruitlets (CB1 fruitlets; AP < 10%), and strongly persisting fruitlets (L3, C1, C3, and CB3; AP ∼ 0%).
In order to test the reliability of the predicted abscission potentials of fruitlets and their actual representativeness (i.e. sample fruits with different destinies) for global transcriptomic analyses, the fruit drop dynamics was followed, in relation to the position and size of dropped fruits, and their ethylene biosynthesis and the expression of ethylene biosynthetic genes previously proved to be reliable diagnostic markers of the apple fruitlet destiny (Dal Cin et al., 2005a, 2009a, 2009b) were measured.
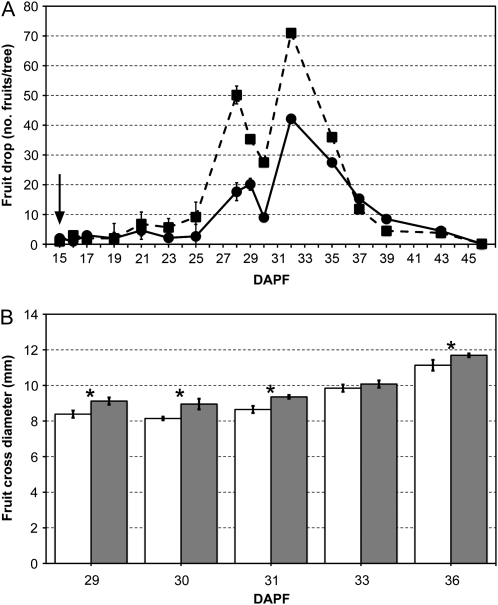
The fruit drop dynamics appeared biphasic, with a first peak occurring at 29 and 28 d after petal fall (DAPF) for control and BA-treated trees, respectively, and a second one at 32 DAPF in both groups (Fig. 2A). BA treatment did not result in changes of fruit shedding dynamics but on a magnification of the process. In fact, at the end of the fruit shedding period (around 46 DAPF), BA-treated trees showed overall about a 2-fold increase of fruit drop, measured throughout the entire experimental period, in comparison with that observed in the control ones (Fig. 2A). The average cross-diameter of abscised fruitlets was significantly higher (P < 0.01) in treated trees than in the untreated ones. In particular, the most significant differences were assessed at 29, 30, 31, and 36 DAPF, as shown in Figure 2B. This was also confirmed by the number of clusters with only one fruit left on the tree after BA treatment (data not shown). These data provide further evidence that BA-dependent magnification of abscission affected mainly the bigger lateral fruitlets (L3) that would normally persist on the tree. A sporadic nonsignificant fruit drop was also observed for the smallest central fruitlets of the BA-treated trees (CB1; data not shown).
Figure 2.
A, Fruit drop dynamics in control (circles and continuous line) and BA-treated (squares and dotted line) trees, expressed as number of drop fruitlets per tree. The arrow represents the time of BA treatment. B, Mean cross-diameter of dropped fruits in control (white bars) and BA-treated (gray bars) trees. Only the five most divergent dates are reported. Error bars represent sd, whereas asterisks show the statistically significant differences (P ≤ 0.05).
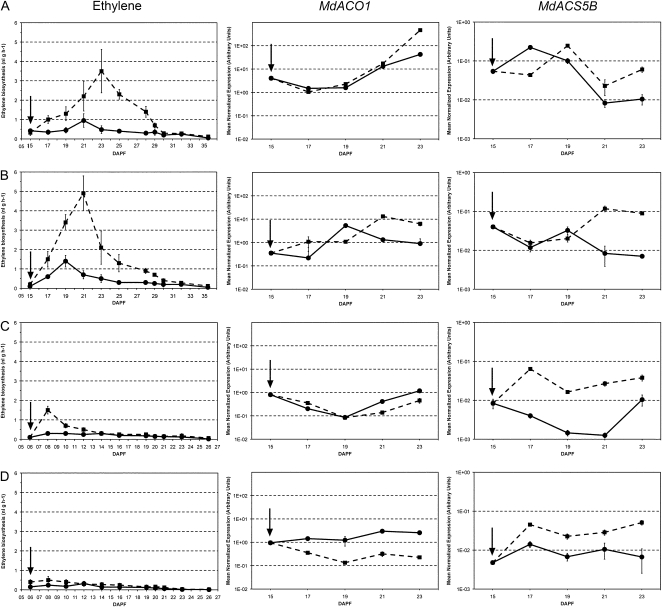
The drop potential of the fruit categories was verified by evaluating their different behaviors in terms of ethylene biosynthesis and 1-aminocyclopropane-1-carboxylate oxidase1 (MdACO1) gene expression, widely accepted indicators of an actual abscission induction and the earliest markers of fruitlet abscission in apple (Dal Cin et al., 2005a, 2009a, 2009b). In previous experiments, ethylene biosynthesis was shown to peak in abscising fruits, regardless of their size, around 3 d after BA treatment in abscising fruitlets and correlated well with the increase of MdACO1 transcripts in the fruit cortex (Dal Cin et al., 2005a).
Concerning ethylene biosynthesis, the small lateral fruitlets of control trees (L1) showed a minor peak of ethylene production at 21 DAPF that remained at basal levels thereafter. BA-treated fruits of the same size class (LB1) showed an increased ethylene biosynthesis already 2 d after the treatment, peaking at 23 DAPF, 8 d from the beginning of the experiments (Fig. 3A). Concerning the bigger laterals (L3), ethylene peaked at 19 DAPF in control samples (L3) and remained at basal levels thereafter, whereas the treated fruitlets (LB3) showed the highest ethylene biosynthesis at 21 DAPF with a fast decreasing trend thereafter (Fig. 3B). In central fruits, a different situation was pointed out in terms of ethylene production, which was lower than that found in the laterals, both in control and treated samples.
Figure 3.
Ethylene biosynthesis (left), expression of MdACO1 (center), and expression of MdACS5B (right) in small (A) and big (B) lateral fruitlets and in small (C) and big (D) central fruitlets, either untreated (circles and continuous lines) or treated with BA (squares and dotted lines). Error bars represent sd.
As far as MdACO1 expression in LB1 fruitlets is concerned, a significant divergence from the control was observed at 23 DAPF (Fig. 3A), correlated with ethylene levels. MdACO1 transcripts peaked earlier at 21 DAPF in LB3, paralleling ethylene production (Fig. 3B). In small central fruitlets, no significant difference was observed (Figs. 3C), whereas, concerning the big central ones, lower MdACO1 levels were measured in the treated samples (CB3), starting from 17 DAPF, throughout the experiment (Fig. 3D). The 1-aminocyclopropane-1-carboxylate synthase5B (MdACS5B) gene expression levels were also assessed and shown to correlate with those of MdACO1, although only in lateral fruitlets. In both C1 and C3, divergent expression trends were observed, most likely responsible for the slightly enhanced ethylene biosynthesis found upon treatments with BA, at least in C1 (Fig. 3).
The 30K Apple Microarray
The apple oligonucleotide microarray herein set up by means of the CombiMatrix platform represents one of the most complete transcriptomic tools available for this species, allowing one to analyze more than 30,000 transcripts with three technical replicates in a single experiment. Similar molecular tools were previously set up based upon apple sequence sets obtained almost exclusively by means of the publicly available TGICL tool (Pertea et al., 2003) and further empirical fine-tuning procedures such as elimination of short sequences and duplicates. Schaffer et al. (2007) set up a microarray with 15,720 sequences chosen among a total number of 42,938 nonredundant records, comprising 17,460 tentative contigs and 25,478 singletons, obtained from 151,687 ESTs from different tissues and cultivars (Newcomb et al., 2006). More recently, an apple oligonucleotide microarray with 55,230 sequences was built starting from 184,132 publicly available records (Jensen et al., 2009). In our research here, a total of 255,950 ESTs and mRNAs were retrieved from public databases, clustered, and assembled by means of a dedicated pipeline (Supplemental Fig. S1), allowing us to obtain 41,927 final nonredundant sequences, including tentative contigs and singletons, among which 30,419 with transcription orientation were chosen to be spotted on the slide. Different from previous research, additional steps were introduced both before and after the TGICL elaboration phase in order to improve contig reliability and to further decrease redundancy (see “Materials and Methods”). In particular, before carrying out the additional steps introduced at the end of the clustering/assembly pipeline, 26,658 tentative contigs and 50,382 singletons were obtained, for a total of 77,040 nonredundant sequences. At this stage, the proportion between the starting number of ESTs/mRNAs and the final nonredundant sequences (3.2:1) was closer to those previously achieved by Newcomb et al. (2006; 3.6:1) and Jensen et al. (2009; 3.3:1) than that by Park et al. (2006; 4.5:1). After the additional processing, the proportion was 6.2:1, indicating that a strong reduction occurred, most likely due to redundancy elimination. On the one hand, this approach was effective in decreasing redundancy based upon a “functional model,” since sequences contained in the same Unigene (i.e. with putative identical functions) were considered once by retaining just the longest record. In this way, different alleles and eventual duplicated genes were most likely clustered together. On the other hand, paralogs were not clustered together because of the high-stringency parameters adopted.
Annotation of apple sequences spotted on the microarray was based on similarity to Uniprot hits and transfer of their Gene Ontology (GO) annotation terms and descriptions to apple sequences. Among all spotted sequences, 39.1% were not annotated for the molecular function (MF) category, 45.4% for the biological process (BP), and 54% for the cellular compartment (CC) subvocabulary. The relative proportions of each GO category on the total within each subvocabulary (i.e. MF, BP, and CC) were well correlated with the annotation distribution found for other species, such as Arabidopsis, Vitis vinifera, Prunus persica, and Populus (Supplemental Fig. S3). The worst correlation was found for the BP subvocabulary, probably due to the known higher fragmentation existing in this GO section (Supplemental Fig. S3) generated by the higher total number of terms (18,189) than in the other two subvocabularies (8,671 in MF and 2,672 in CC; for more information, see www.geneontology.org).
The apple sequence set used by Jensen et al. (2009) had a relatively low coverage with respect to the Arabidopsis proteome, assessed as equal to 52.1% with homology to 14,266 unique proteins on a total of 27,379 records (The Arabidopsis Information Resource 9 database), as well as a high redundancy level. A total of 25,580 (84.1%) apple contigs and singletons used herein for the microarray construction were shown to match with 13,706 unique Arabidopsis proteins (BLASTx algorithm with 1E-3 cutoff), giving 50.1% coverage. Considering the total number of genes (57,386) recently predicted on the genome of domesticated apple (Velasco et al., 2010), 53% coverage is achieved. Based upon these data and taking into account that the majority of ESTs used to set up the microarray derive from fruit tissues (data not shown), it is likely that almost the whole fruit transcriptome is represented along with a relevant part of the genes expressed in the seed.
Globaltest Analysis of Apple Fruitlet Transcriptomes
The Globaltest package (Goeman et al., 2004) of Bioconductor was used to assess whether significant associations exist between global gene expression profiles, in cortex and seed, and “phenotypes” or physiological responses in terms of abscission potential (fruitlet destiny), fruit weight, and fruit position within the cluster (herein called the “response variables”). Globaltest analysis was performed either on the whole gene set (30,419 genes) or on the subset of significantly variable genes identified, separately in cortex and seed, as described in “Materials and Methods.” It has to be highlighted that this approach allows one to identify static associations between the transcriptome and a given variable/phenotype, regardless of the time course of expression profiles of the genes considered in the analysis. Therefore, a second complementary and confirmatory approach was also performed to identify genes displaying divergent kinetics (see following paragraph) related to different abscission potentials.
As far as the overall gene expression data in the cortex are concerned, no significant association with fruitlet weight was identified. Significant associations were found with the position within the cluster (P < 0.05) and the treatment (P < 0.07), whereas a highly significant association was pointed out with fruitlet destiny (P < 0.004). When only the significantly differentially expressed (DE) genes were considered, the significance level increased for all the considered response variables, except for “treatment,” reaching P = 0.001 for the association with abscission potential (for the overall statistics for the cortex, see Supplemental Table S1). Considering the seed transcriptome, highly significant and significant associations were detected only with fruitlet weight (P < 0.008) and abscission probability (P < 0.01). The significance level improved in all cases when the subset of DE genes was considered, indicating the reliability of the statistical analysis. In particular, it has to be pointed out that the association between gene expression data and weight reached P = 0.0008 (extremely significant), whereas the statistical test on fruitlet destiny was highly significant (P < 0.004). In order to test the time course association between transcriptomic data and response variables, gene expression data at each sampling date (beginning of the experiment [T0], after 2 d [T2], and after 4 d [T3]) were processed separately, taking into account that the lower number of samples analyzed in each test may have partially biased the statistical calculation, resulting in lower levels of significance. As far as the cortex is concerned, no significant association was reported at T0, whereas significant levels at T2 (P < 0.03) and T3 (P < 0.01) were pointed out for fruit destiny. At T3, also the response variable treatment showed a significant level (P < 0.05). A statistically relevant association was reported between the seed transcriptome and fruitlet weight (P < 0.08) already at T0, which became nonsignificant at T2 and again significant at T3 (P < 0.06). Also, the abscission potential was significantly associated with the seed transcriptome, but only later at T3 (P < 0.02). All the statistics for the seed are reported in Supplemental Table S2.
The association of the response variables with expression data of gene subsets encoding elements involved in hormone biosynthesis, metabolism, perception, signal transduction, and cross talk was also investigated. The statistical analyses were performed separately for the five major plant hormones (ABA, auxin, cytokinin, ethylene, and GA) and as a whole for minor plant growth regulators (jasmonates, salicylic acid, polyamines, and brassinosteroids).
As far as the overall gene expression data are concerned, the highest levels of significance were found again in the cortex. In fact, extremely significant associations with fruit destiny were found for genes related to ABA, cytokinin, and GAs, with P < 0.0005 in all cases, whereas for auxin and ethylene, the statistics were highly significant in both cases (P < 0.002 and P < 0.005, respectively). In the same tissue, GA-related genes were highly associated also with the position of the fruit within the cluster, with P < 0.005. Concerning the time course, the ABA-related genes were significantly correlated (P < 0.01) with the abscission potential already at T2 (2 d after the BA treatment). All the other major plant hormone-related genes showed a significant P level of association with the same response variable. Concerning the position within the cluster, GA-related genes were already correlated at T2, with P < 0.03. The group of genes related to the minor hormones showed highly significant and significant statistics at T2 concerning the association with the abscission potential (P < 0.007) and BA treatment (P < 0.09), respectively. At T0, only nonsignificant P levels were found. Statistics for all samples and gene subsets are shown in Supplemental Table S1.
Concerning the seed, a slightly different situation with respect to the cortex was pointed out, both in terms of significance levels in overall samples and the time course of the associations. The global test evidenced a highly significant association with fruit weight and destiny for the gene subsets related to ABA, auxin, and GAs. For ethylene and minor hormones, the statistic was significant only with respect to the former response variable, whereas for cytokinin, lower levels of significance were assessed (for P values, see Supplemental Table S2). Concerning the time course statistics, a significant P level was calculated already at T0, with the exception of genes related to GAs. For the major hormones, this level of significance was kept up to T3, when a highly significant test was reported for the association between auxin-related genes and abscission potential (P < 0.01), being earliest in the seed considering all the plant hormones with respect to fruit destiny. In Supplemental Table S2, all the statistics are reported for the seed.
Hierarchical Clustering of Abscission-Related Genes
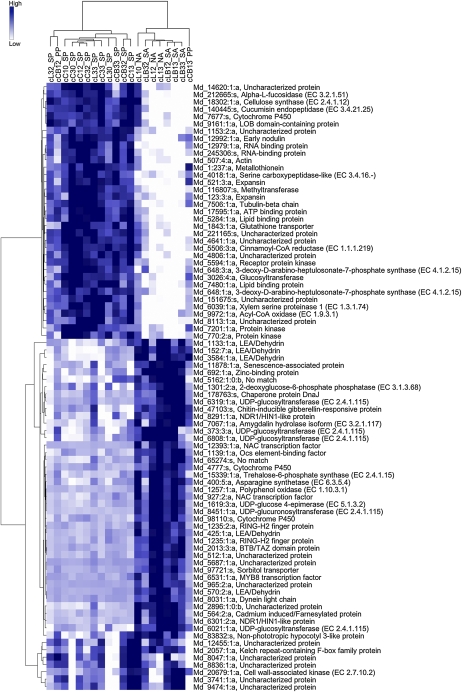
According to the Globaltests, a highly significant association exists between the expression of DE gene subsets in cortex and seed and the fruits’ probability of abscising. When the same genes were clustered according to their expression levels in all samples, these genes were not able to finely discriminate samples according to the corresponding abscission potential. This analysis instead pointed out that the BA treatment had a relevant weight on the overall gene expression profiles and, in turn, on the hierarchical clustering process, as evidenced in Supplemental Figure S6, thus masking the clustering of genes associated with fruitlet destiny. Therefore, further clustering analyses were carried out only with genes highly correlated with the fruit destiny, choosing a highly stringent cutoff score (Z > 7) and selecting among the whole set regardless of significant differential expression. In fact, samples with a high probability of persisting clustered together, whereas those with higher abscission potentials grouped in a distinct cluster, confirming Globaltest analyses (Figs. 4 and 5). Moreover, it is worth noting that BA-treated central fruitlets of small size (CB1) were split into two different clusters, at T2 (CB12) in the persisting group and at T3 (CB13) in the abscising one. This is consistent with the borderline condition of “probably persisting” fruitlets ascribed to this sample class. A temporal shift was shown for LB3 samples, in that LB32 clustered closer to naturally abscising fruits whereas LB33 grouped together with treated samples with equally high abscission potential.
Figure 4.
Hierarchical clustering of genes with expression levels in the cortex that are highly associated with the fruitlet abscission potentials. Only genes with a score of Z ≥ 7 are reported, according to the Globaltest analysis. Samples are reported on the top side of the heat map with the following codes: c, cortex; L, lateral; C, central; B, treated with BA. The first number indicates fruit size category: 1, small fruitlets; 3, big fruitlets. The second number indicates sampling time: 0, T0; 2, T2; 3, T3. NA, Naturally abscising; PP, probably persisting; SA, strongly abscising; SP, strongly persisting.
Figure 5.
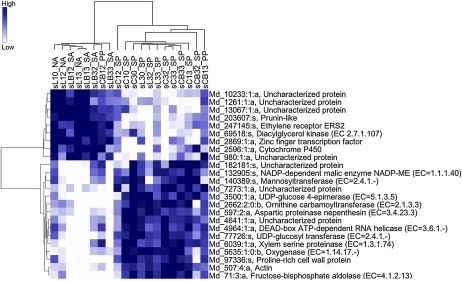
Hierarchical clustering of genes with expression levels in the seed that are highly associated with the fruitlet abscission potentials. Only genes with a score of Z ≥ 7 are reported, according to the Globaltest analysis. Samples are reported on the top side of the heat map with the following codes: s, seed; L, lateral; C, central; B, treated with BA. The first number indicates fruit size category: 1, small fruitlets; 3, big fruitlets. The second number indicates sampling time: 0, T0; 2, T2; 3, T3. NA, Naturally abscising; PP, probably persisting; SA, strongly abscising; SP, strongly persisting.
According to the Globaltest analyses, the seed transcriptomes appeared to be associated with fruitlet destiny to a lesser extent than those of the cortex. Consistently, only 24 genes in the seed compared with 83 in the cortex had a Z score higher than 7. Also in this case, a clear distinction was reported, when the genes were hierarchically clustered, in terms of expression levels in association with the abscission potential (Fig. 5). Indeed, the same borderline samples that were split into two distinct clusters in the cortex were separated also in the seed, although inversely (i.e. CB12 with the abscising samples and CB13 with the persisting ones), strengthening the hypothesis that the small central fruitlets may have a higher probability to abscise than the bigger ones. Remarkably, transcriptomes of L3 lateral fruitlets (untreated) clearly clustered together within the persisting ones, while BA treatment reverted this and forced their clustering together with abscising samples at all time points in both seed and cortex. This finding confirmed that BA treatment had a significant effect in inducing the abscission of L3 fruits, thus changing their developmental destiny, and this effect could be linked to transcriptional signatures in cortex and seed that are specifically associated with the induction of the abscission response. For microarray data validation, quantitative PCR experiments were performed on a subset of selected genes and revealed similar expression patterns and strong correlations (Supplemental Fig. S7).
Signatures of Fruitlet Abscission in Cortex
Genes representing the abscission-specific transcriptional signatures in cortex are clustered in Figure 4 and listed in Table I. Concerning those involved in metabolism, a marker of high abscission potential encodes a trehalose-6-phosphate synthase (Md_15339:1:a; EC 2.4.1.15), discriminating also the borderline CB13 sample. BLAST analysis pointed out a 62% identity with Arabidopsis AtTPS10 (At1g60140), a class II TPS gene induced by sugar starvation (Osuna et al., 2007), cytokinins (Brenner et al., 2005), and ABA (Paul, 2007). A transcript for a sorbitol transporter (Md_97721:s) coregulated with the previous one showed a high level of identity (77%) with MdSOT5 (accession no. BAD42345), functioning either in import or export of sorbitol in/from leaves (Watari et al., 2004). Five genes (Md_ 6319:1:a, Md_373:3:a, Md_6808:1:a, Md_8451:1:a, and Md_6021:1:a) encoding UDP-glucosyltransferases (EC 2.4.1.115) were highly expressed in abscising samples. In Arabidopsis and Beta vulgaris, the transcription of genes belonging to this family was induced during superoxide-dependent cell death (Mazel and Levine, 2002) and oxidative stress (Sepúlveda-Jiménez et al., 2005), respectively. A UDP-Glc-4-epimerase (EC 5.1.3.2) gene (Md_1619:3:a), coregulated with the previous ones, displayed a high degree of similarity (79% identity) with UGE5 of Arabidopsis (At4g10960), induced by ABA and coregulated with carbohydrate biosynthetic enzymes (Rösti et al., 2007). Md_400:5:a, encoding an Asn synthetase (EC 6.3.5.4) similar to AtASN1 of Arabidopsis (At3g47340), was up-regulated in abscising samples, although at low levels in LB32. In other species, genes encoding this class of enzymes are controlled by sugar starvation and involved in resource mobilization (Herrera-Rodríguez et al., 2004; Rook et al., 2006; Rose et al., 2006). A different gene set showed a high discriminating power in terms of higher expression in persisting fruitlets (Fig. 4). This set comprised genes coding for cellulose synthase (Md_18302:1:a; EC 2.4.1.12), cinnamoyl-CoA reductase (Md_5506:3:a; EC 1.1.1.219), acyl-CoA oxidase (Md_9972:1:a; EC 1.9.3.1), and 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (two genes: Md_648:3:a and Md_648:1:a; EC 4.1.2.15) that are typically expressed in developing organs (Lauvergeat et al., 2001; Pedersen and Henriksen, 2005; Sato et al., 2006), such as the persisting apple fruitlets. High transcription rates were reported in the persisting fruitlets also for a gene encoding a cucumisin endopeptidase (Md_140445:s; EC 3.4.21.25), which is expressed at high levels also during the early developmental stages in melon (Cucumis melo; Choi et al., 2004), and an α-l-fucosidase (Md_212665:s; EC 3.2.1.51) probably involved in fruit growth (Desveaux et al., 1998).
Table I. Genes with expression data in the cortex associated with abscission potential with a score of Z ≥ 7.
A tentative annotation, the influence on the whole association, the statistical score (Z), and the expression pattern (NA, high expression in naturally abscising fruitlets; SA, high expression in strongly abscising fruitlets; SP, high expression in strongly persisting fruitlets) are reported for each gene.
| Gene Idenitfier | Tentative Annotation | Influence | Z | Expression |
| Md_8451:1:a | UDP-glucosyltransferase (EC 2.4.1.115) | 1,987.67 | 8.80 | NA |
| Md_521:3:a | Expansin | 209.73 | 8.74 | SP |
| Md_245306:s | RNA-binding region-containing protein | 569.08 | 8.67 | SP |
| Md_9972:1:a | Acyl-CoA oxidase (EC 1.9.3.1) | 123.73 | 8.40 | SP |
| Md_4641:1:a | Uncharacterized protein | 167.99 | 8.35 | SP |
| Md_12979:1:a | RNA-binding protein | 323.19 | 8.34 | SP |
| Md_7480:1:a | Lipid-binding protein | 210.18 | 8.29 | SP |
| Md_570:2:a | LEA/dehydrin | 2,686.19 | 8.22 | NA |
| Md_8836:1:a | Uncharacterized protein | 26.56 | 8.08 | SA |
| Md_151675:s | Uncharacterized protein | 121.54 | 8.06 | SP |
| Md_4018:1:a | Ser carboxypeptidase-like (EC 3.4.16.-) | 112.73 | 8.05 | SP |
| Md_2896:1:0:b | Uncharacterized protein | 562.33 | 7.94 | NA |
| Md_152:7:a | LEA/dehydrin | 964.27 | 7.88 | SA |
| Md_15339:1:a | Trehalose-6-phosphate synthase (EC 2.4.1.15) | 375.72 | 7.87 | NA |
| Md_97721:s | Sorbitol transporter | 1,185.32 | 7.87 | NA |
| Md_1257:1:a | Polyphenol oxidase (EC 1.10.3.1) | 1,532.64 | 7.85 | NA |
| Md_116807:s | Generic methyltransferase | 164.21 | 7.79 | SP |
| Md_507:4:a | Actin | 231.25 | 7.78 | SP |
| Md_2057:1:a | Kelch repeat-containing F-box family protein | 55.08 | 7.73 | NA |
| Md_7506:1:a | Tubulin β-chain | 122.74 | 7.73 | SP |
| Md_1619:3:a | UDP-Glc 4-epimerase (EC 5.1.3.2) | 680.49 | 7.67 | NA |
| Md_12393:1:a | NAC transcription factor | 2,144.47 | 7.64 | NA |
| Md_8047:1:a | Uncharacterized protein | 23.37 | 7.64 | SA |
| Md_1133:1:a | LEA/dehydrin | 546.19 | 7.59 | SA |
| Md_648:3:a | 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase (EC 4.1.2.15) | 294.22 | 7.57 | SP |
| Md_6039:1:a | Xylem Ser proteinase 1 (EC 1.3.1.74) | 111.24 | 7.56 | SP |
| Md_3741:1:a | Uncharacterized protein | 35.15 | 7.55 | SA |
| Md_3026:4:a | Glucosyltransferase | 160.01 | 7.55 | SP |
| Md_4806:1:a | Uncharacterized protein | 242.23 | 7.54 | SP |
| Md_1235:2:a | RING-H2 finger protein | 806.57 | 7.52 | NA |
| Md_8291:1:a | NDR1/HIN1-like protein | 322.13 | 7.52 | SA |
| Md_12992:1:a | Early nodulin | 172.86 | 7.50 | SP |
| Md_7677:s | Cytochrome P450 | 86.16 | 7.50 | SP |
| Md_512:1:a | Uncharacterized protein | 962.45 | 7.49 | NA |
| Md_8113:1:a | Uncharacterized protein | 161.13 | 7.46 | SP |
| Md_47103:s | Chitin-inducible GA-responsive protein | 134.13 | 7.44 | SA |
| Md_3584:1:a | LEA/dehydrin | 1,491.95 | 7.43 | SA |
| Md_1153:2:a | Uncharacterized protein | 280.40 | 7.38 | SP |
| Md_770:2:a | Protein kinase family protein | 139.58 | 7.38 | SP |
| Md_20679:1:a | Cell wall-associated kinase (EC 2.7.10.2) | 101.45 | 7.35 | SA |
| Md_18302:1:a | Cellulose synthase (EC 2.4.1.12) | 104.04 | 7.34 | SP |
| Md_221165:s | Uncharacterized protein | 91.34 | 7.34 | SP |
| Md_11878:1:a | Senescence-associated protein | 114.63 | 7.33 | SA |
| Md_373:3:a | UDP-glucosyltransferase (EC 2.4.1.115) | 372.45 | 7.32 | SA |
| Md_1:237:a | Metallothionein | 348.11 | 7.32 | SP |
| Md_123:3:a | Expansin | 701.08 | 7.32 | SP |
| Md_425:1:a | LEA/dehydrin | 767.30 | 7.31 | NA |
| Md_140445:s | Cucumisin endopeptidase (EC 3.4.21.25) | 84.62 | 7.31 | SP |
| Md_5594:1:a | Receptor protein kinase | 154.01 | 7.28 | SP |
| Md_2013:3:a | BTB/TAZ domain protein | 711.00 | 7.27 | NA |
| Md_6319:1:a | UDP-glucosyltransferase (EC 2.4.1.115) | 255.14 | 7.24 | SA |
| Md_400:5:a | Asn synthetase (EC 6.3.5.4) | 561.06 | 7.23 | NA |
| Md_5284:1:a | Lipid-binding protein | 142.63 | 7.23 | SP |
| Md_6021:1:a | UDP-glucosyltransferase (EC 2.4.1.115) | 257.50 | 7.22 | SA |
| Md_1235:1:a | RING-H2 finger protein | 849.17 | 7.21 | NA |
| Md_17595:1:a | ATP-binding protein | 89.54 | 7.21 | SP |
| Md_6808:1:a | UDP-glucosyltransferase (EC 2.4.1.115) | 256.94 | 7.20 | SA |
| Md_6531:1:a | MYB8 transcription factor | 1,273.41 | 7.19 | NA |
| Md_7201:1:a | Protein kinase | 214.48 | 7.19 | SP |
| Md_927:2:a | NAC transcription factor | 693.00 | 7.16 | NA |
| Md_9474:1:a | Uncharacterized protein | 30.35 | 7.15 | SA |
| Md_1301:2:a | 2-Deoxyglucose-6-phosphate phosphatase (EC 3.1.3.68) | 83.26 | 7.14 | SA |
| Md_5162:1:0:b | No match | 141.10 | 7.13 | NA |
| Md_6301:2:a | NDR1/HIN1-like protein | 907.91 | 7.13 | NA |
| Md_9161:1:a | LOB domain-containing protein | 77.16 | 7.11 | SP |
| Md_7067:1:a | Amygdalin hydrolase isoform (EC 3.2.1.117) | 264.52 | 7.10 | NA |
| Md_65274:s | No match | 312.20 | 7.09 | NA |
| Md_178763:s | Chaperone protein DnaJ | 338.06 | 7.09 | SA |
| Md_14620:1:a | Uncharacterized protein | 41.32 | 7.09 | SP |
| Md_12455:1:a | Uncharacterized protein | 97.40 | 7.07 | NA |
| Md_4777:s | Cytochrome P450 | 764.14 | 7.07 | NA |
| Md_8031:1:a | Dynein light chain | 1,044.35 | 7.07 | NA |
| Md_98110:s | Cytochrome P450 | 977.28 | 7.07 | NA |
| Md_564:2:a | Cadmium-induced/farnesylated protein-like | 542.16 | 7.06 | NA |
| Md_5506:3:a | Cinnamoyl-CoA reductase (EC 1.1.1.219) | 133.48 | 7.06 | SP |
| Md_5687:1:a | Uncharacterized protein | 1,511.38 | 7.05 | NA |
| Md_1843:1:a | Glutathione transporter | 56.80 | 7.05 | SP |
| Md_83832:s | Nonphototropic hypocotyl 3-like protein | 144.01 | 7.03 | NA |
| Md_692:1:a | Zinc-binding protein | 207.75 | 7.03 | SA |
| Md_648:1:a | 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase (EC 4.1.2.15) | 179.34 | 7.03 | SP |
| Md_1139:1:a | Ocs element-binding factor | 466.11 | 7.02 | NA |
| Md_965:2:a | Uncharacterized protein | 1,487.27 | 7.00 | NA |
| Md_212665:s | α-l-Fucosidase | 118.65 | 7.00 | SP |
As far as the genes encoding structural elements are concerned, it is worthy to note that in persisting fruitlets an up-regulation of genes encoding actin (Md_507:4:a), tubulin (Md_7506:1:a), and expansins (Md_521:3:a and Md_123:3:a) was detected, consistent with the active growth characterizing this fruitlet class. Among the transcripts up-regulated in the abscising samples, no structural element was reported, except for a dynein gene (Md_8031:1:a) probably involved in the organization and control of vesicle trafficking (Lawrence et al., 2001). In the same samples, dehydrin/late embryogenesis abundant (LEA) protein genes represent a clear genetic signature. In fact, five transcripts related to this class of proteins (Md_1133:1:a, Md_152:7:a, Md_3584:1:a, Md_425:1:a, and Md_570:2:a), which are known to be expressed in senescing organs strictly upon ABA control (Hong-Bo et al., 2005; Rorat, 2006), are strongly up-regulated in abscising fruitlets also during the early stages of shedding induction, consistent with the destiny of these samples.
Genes encoding elements of signaling pathways were found among the most discriminating transcriptional signatures. A Lateral Organ Boundaries (LOB) gene (Md_9161:1:a) was expressed at much lower levels in the abscising fruitlets than in the persisting ones. A 75% identity was assessed between this gene and Asymmetric Leaves12/LOB21 of Arabidopsis (At3g11090), which is expressed at high levels in the silique and belongs to a gene family whose members promote lateral organ fate and polarity, thereby restricting the developmental potential of the organ-forming cells (Ha et al., 2007; Matsumura et al., 2009). Similar expression patterns were observed for metallothionein-like protein transcripts (Md_1:237:a), closely similar to senescence-induced Arabidopsis MT3 (At3g15353). Interestingly, two genes encoding protein kinases (Md_7201:1:a and Md_770:2:a) were very powerful in discriminating strongly abscising fruitlets showing very low expression levels. Abscising fruitlets showed higher expression levels for genes encoding a zinc-binding protein (Md_692:1:a), a chaperone protein DnaJ (Md_178763:s), and a GA-responsive protein (Md_47103:s), putatively involved in cell cycle regulation, senescence, and GA signaling, respectively (The Arabidopsis Information Resource data). A similar transcriptional profile was reported for an ocs element-binding factor gene (Md_1139:1:a) involved in ethylene signaling (Büttner and Singh, 1997; Singh et al., 2002) and for two coregulated NAM, ATAF, and CUC (NAC) genes (Md_12393:1:a and Md_927:2:a), which were shown to be involved also in senescence-associated mobilization of resources (Uauy et al., 2006). Specifically, the former was closely similar to cotton (Gossypium hirsutum) NAC5, promptly induced by exogenous ABA (Meng et al., 2009), whereas the latter showed a significant degree of identity with senescence-associated ANAC083 (At5g13180) of Arabidopsis (Ay et al., 2009). Among the other signaling elements, transcripts for a MYB transcription factor (Md_6531:1:a) and two RING-H2 finger proteins (Md_1235:2:a and Md_1235:1:a) were overexpressed in abscising fruits. The former is apple MdMYB8 (DQ267899), whereas the latter were both similar to the XERICO gene of Arabidopsis (At2g04240), a positive regulator of ABA signaling (Ko et al., 2006). Finally, a Broad-Complex, Tramtrack, and Bric-a-Brac/Transcriptional Adaptor Zinc finger (BTB/TAZ) domain protein was most likely encoded by the gene Md_2013:3:a, highly expressed in fruitlets with high abscission potential as well. This gene showed 63% identity with BTB/TAZ domain protein1 of Arabidopsis, promptly induced by treatments with hydrogen peroxide (Du and Poovaiah, 2004). In Figure 4, the transcriptional profiles of all the most discriminating genes are shown.
Signatures of Fruitlet Abscission in Seed
As far as the seed transcriptome is concerned, 24 highly discriminating genes were clustered, among which nine were up-regulated in the abscising samples and 15 in the persisting ones (Fig. 5; Table II). Among the former, Ethylene Response Sensor2 (MdERS2) (Md_247145:s) and a diacylglycerol kinase (DGK) gene (Md_69518:s; EC 2.7.1.107) were found. The second gene showed 62% identity with Arabidopsis ATDGK5 (At2g20900), induced by ozone, ethylene, and jasmonic acid (Tamaoki et al., 2003), and also MdERS2 was shown to be ethylene inducible (Tatsuki et al., 2009). A zinc finger protein (Md_2869:1:a) similar to Leech Zinc Finger1 of Arabidopsis (At1g78600) and a cytochrome P450 gene (Md_2596:1:a) similar to CYP714A1 (At5g24910), grouped in the same cluster. These genes were shown to be involved in seedling photomorphogenesis and seed development, respectively, of Arabidopsis (Kushiro et al., 2004; Chang et al., 2008; Datta et al., 2008). Persisting fruitlets expressed at higher levels some genes encoding proteins involved in metabolism, such as a NADP-dependent malic enzyme (Md_132905:s) similar to ATNADP-ME2 (At5g11670), a mannosyltransferase (Md_140389:s) similar to PEANUT1 (At5g22130), and a UDP-Glc-4-epimerase (Md_3500:1:a) similar to At4g20460. Their putative Arabidopsis orthologs were shown to have fundamental roles in embryo development (Gillmor et al., 2005; Wheeler et al., 2005; Gómez et al., 2006). Other coregulated genes involved in metabolism are reported in Figure 5. Two genes encoding structural elements (actin [Md_507:4:a] and a Pro-rich cell wall protein [Md_97336:s]) were also expressed at high levels in the seed of persisting fruitlets, consistent with an actively developing status. Also, the overexpression of a Asp-Glu-Ala-Asp (DEAD) box gene (Md_4964:1:a) similar to mitochondrial RNA helicase may be representative of a very active metabolism.
Table II. Genes with expression data in the seed associated with abscission potential with a score of Z ≥ 7.
A tentative annotation, the influence on the whole association, the statistical score (Z), and the expression pattern (NA, high expression in naturally abscising fruitlets; SA, high expression in strongly abscising fruitlets; SP, high expression in strongly persisting fruitlets) are reported for each gene.
| Gene Identifier | Tentative Annotation | Influence | Z | Expression |
| Md_140389:s | Mannosyltransferase (EC 2.4.1.-) | 39.91 | 9.37 | SP |
| Md_4641:1:a | Uncharacterized protein | 135.78 | 8.74 | SP |
| Md_6039:1:a | Xylem Ser proteinase (EC 1.3.1.74) | 95.01 | 8.25 | SP |
| Md_247145:s | Ethylene receptor ERS2 | 173.68 | 8.15 | SA |
| Md_77726:s | UDP-glucosyltransferase (EC 2.4.1.-) | 104.49 | 8.04 | SP |
| Md_507:4:a | Actin | 286.42 | 7.86 | SP |
| Md_203607:s | Prunin-like | 316.63 | 7.79 | SA |
| Md_10233:1:a | Uncharacterized protein | 224.81 | 7.64 | NA |
| Md_4964:1:a | DEAD box ATP-dependent RNA helicase (EC 3.6.1.-) | 75.52 | 7.48 | SP |
| Md_2869:1:a | Zinc finger transcription factor | 49.21 | 7.34 | NA |
| Md_182181:s | Uncharacterized protein | 67.95 | 7.32 | SP |
| Md_2662:2:0:b | Orn carbamoyltransferase (EC 2.1.3.3) | 68.18 | 7.31 | SP |
| Md_97336:s | Pro-rich cell wall protein | 317.78 | 7.28 | SP |
| Md_3500:1:a | UDP-Glc 4-epimerase (EC 5.1.3.5) | 176.60 | 7.27 | SP |
| Md_132905:s | NADP-dependent malic enzyme (EC 1.1.1.40) | 155.09 | 7.21 | SP |
| Md_980:1:a | Uncharacterized protein | 681.02 | 7.18 | SA |
| Md_1261:1:a | Uncharacterized protein | 800.80 | 7.13 | NA |
| Md_5635:1:0:b | Oxygenase (EC 1.14.17.-) | 146.93 | 7.13 | SP |
| Md_7273:1:a | Uncharacterized protein | 48.74 | 7.11 | SP |
| Md_597:2:a | Aspartic proteinase nepenthesin (EC 3.4.23.3) | 201.44 | 7.08 | SP |
| Md_69518:s | Diacylglycerol kinase (EC 2.7.1.107) | 38.81 | 7.07 | NA |
| Md_71:3:a | Fru-bisphosphate aldolase (EC 4.1.2.13) | 188.95 | 7.06 | SP |
| Md_13067:1:a | Uncharacterized protein | 58.76 | 7.03 | NA |
| Md_2596:1:a | Cytochrome P450 (EC 1.3.3.9) | 77.41 | 7.01 | NA |
It has to be noted that the transcriptional profiles of the above genes in the seed are clearly less discriminative than those found in the cortex. In fact, some samples (i.e. L10, L12, and LB32), despite the hierarchical clustering, showed expression levels somehow different from those of other members of the same cluster (Fig. 5). Considering only fruitlets at T3, the same genes were very reliable in discriminating a fruit’s destiny, therefore confirming the Globaltest results indicating an earlier association of the cortex transcriptome than the seed’s with fruit abscission potential.
Genes Involved in BA-Induced Abscission
A parallel approach was adopted along with the Globaltest by subtracting gene pools either developmentally or pharmacologically regulated by BA from the overall DE genes in the LB3 fruitlets. This “subtractive” approach is detailed in “Materials and Methods” and represents a validation of the Globaltest analysis. A detailed description of DE genes is reported below only for the most interesting categories. The number of DE genes for each comparison and an overall list along with a tentative annotation, molecular function classification, and expression pattern are reported in Supplemental Tables S2, S4, and S5, whereas in Tables III and IV only the most interesting genes are listed. For microarray data validation, quantitative PCR experiments were performed on a subset of selected genes and showed similar expression patterns and reliable correlations (Supplemental Fig. S7).
Table III. A selection of DE genes in the cortex.
The gene identifier is reported along with a short annotation, the pattern of expression from T0 to T2 and from T2 to T3, likely correlations with hormones, metabolites, or physiological events, and references reporting specific information about the genes. Categories are indicated as follows: H, hormone biosynthesis, metabolism, and action; H/S, hormone-sugar cross talk; P, protein synthesis and metabolism; R, ROS synthesis, metabolism, and signaling; TF, transcription factors; VT, vesicle trafficking. The complete list along with further details are available in Supplemental Table S4.
| Category | Gene Identifier | Tentative Annotation | T0–T2 | T2–T3 | Notesa | References |
| H | Md_1133:1:a | LEA/dehydrin | Up | - | +ABA | - |
| Md_131178:s | 14-3-3-like protein | Down | − | ABA/ET | Lancien and Roberts (2006) | |
| Md_14070:1:a | GA 2-oxidase | Up | − | −GA | - | |
| Md_140962:s | BRI1-associated receptor kinase 1 (BAK1) | Up | − | +ROS, BR/JA | Xia et al. (2009) | |
| Md_214104:s | GA 2-oxidase | Up | − | −GA | - | |
| Md_246936:s | Mitogen-activated protein kinase (MAPK) | Up | − | +ABA, ABA/ET | Xin et al. (2005) | |
| Md_25179:s | GA 2-oxidase | Up | − | −GA | - | |
| Md_2556:1:a | GASA4-like protein | Down | − | +GA | Chen et al. (2007) | |
| Md_2750:1:a | LEA/dehydrin | Up | − | +ABA | ||
| Md_4451:1:a | Jasmonate-induced protein | - | Down | +JA | – | |
| Md_4451:1:a | Jasmonate-induced protein | Up | − | +JA | – | |
| Md_5550:1:a | IAA-amino acid hydrolase, ILR1 | Up | − | +IAA | Seidel et al. (2006) | |
| Md_5793:1:0:b | Mitogen-activated protein kinase kinase (MAPKK) | Up | − | ET | – | |
| Md_7045:1:a | GA 2-oxidase | Up | − | −GA | – | |
| Md_74377:s | Cytokinin dehydrogenase | Up | − | −CK | Frébortová et al. (2004) | |
| Md_93:4:a | 14-3-3-like protein | Down | − | ABA/ET | Lancien and Roberts (2006) | |
| H/S | Md_12387:1:a | SNF1-related kinase 3.10 | Up | − | +ST | Purcell et al. (1998); Chikano et al. (2001) |
| Md_253006:s | AMP-activated protein kinase, γ-regulatory subunit | Up | − | +ABA | Genevestigator | |
| Md_9662:1:a | Suc synthase | Up | − | +SUC | Chikano et al. (2001) | |
| P | Md_20453:1:a | Aspartic proteinase nepenthesin | Up | − | − | – |
| Md_240669:s | Ubiquitin-protein ligase | Up | − | − | – | |
| Md_6142:1:a | Subtilisin-like protease | Up | − | − | – | |
| Md_66411:s | Ubiquitin-protein ligase | Up | − | − | – | |
| R | Md_288:2:a | Ferritin | Up | − | +ROS | Ravet et al. (2009) |
| Md_5375:1:a | Respiratory burst NADPH oxidase | Up | − | +ROS | Torres et al. (2002); Kwak et al. (2003) | |
| Md_67394:s | Peroxidase | Up | − | +ROS | Almagro et al. (2009) | |
| TF | Md_1109:1:a | WRKY53 transcription factor | Up | − | +JA, +ROS, +SEN | Miao et al. (2004); Miao and Zentgraf (2007); Pitzschke and Hirt (2009); Zentgraf et al. (2010) |
| Md_1122:1:a | NAC/NAM transcription factor | Up | − | +ABA | Fujita et al. (2004); Tran et al. (2004) | |
| Md_117252:s | WRKY53 transcription factor | Up | − | +JA, +ROS, +SEN | Miao et al. (2004); Miao and Zentgraf (2007); Pitzschke and Hirt (2009); Zentgraf et al. (2010) | |
| Md_119754:s | MYC1 transcription factor | Up | − | +STR | Smolen et al. (2002) | |
| Md_121294:s | Ethylene-responsive AP2/ERF transcription factor | Up | − | ET/JA | Lorenzo et al. (2003) | |
| Md_12393:1:a | NAC/NAM transcription factor | Up | − | +ABA | Uauy et al. (2006); Meng et al. (2009) | |
| Md_1709:2:a | EIL2 (EIN3-like) | Up | − | +ET, +ROS | Zhong et al. (2009); Huang et al., (2010) | |
| Md_19496:1:a | Zinc finger DHHC domain-containing protein | Up | − | − | – | |
| Md_200958:s | Zinc-finger C2H2 protein SERRATE | Up | − | − | – | |
| Md_249698:s | Ethylene-responsive AP2/ERF transcription factor | Up | − | ET/JA | Lorenzo et al. (2003) | |
| Md_2575:1:a | MdMYB6 transcription factor | Up | − | +SUC | Genevestigator | |
| Md_3896:1:0:b | WRKY4 transcription factor | Up | − | +JA | Fonseca et al. (2009) | |
| Md_40605:s | Ethylene-responsive AP2/ERF transcription factor | Up | − | +JA | Oñate-Sánchez and Singh (2002) | |
| Md_5724:1:a | Zinc finger protein CONSTANS-LIKE 5 | Down | − | − | – | |
| Md_6240:1:a | Zinc finger homeodomain protein SZF-HD1 | Down | − | − | – | |
| Md_6531:1:a | MdMYB8 transcription factor | Up | − | +SA | Yanhui et al. (2006) | |
| Md_7112:2:a | WRKY19 transcription factor | Up | − | − | – | |
| Md_9390:1:a | Auxin response factor 3, ARF3 | Up | − | +ROS, +ST, +SUC, +ABA | Genevestigator | |
| VT | Md_135689:s | ADP-ribosylation factor, ARF | Down | − | − | – |
| Md_16163:1:a | Protein transport protein Sec23 | Up | − | − | – | |
| Md_179063:s | Dynamin | Up | − | − | Bubán (2000); Jin et al. (2001); Zhang and Hu (2010) | |
| Md_6659:1:a | Synaptotagmin | Up | − | − | Schapire et al. (2008) | |
| Md_7571:1:a | Coatomer β-subunit | Up | − | − | Bassham et al. (2008) | |
| Md_8558:1:a | Clathrin assembly protein | Up | − | − | Legendre-Guillemin et al. (2004) |
+, Positive correlation; −, negative correlation; /, cross talk; BR, brassinosteroids; CK, cytokinin; ET, ethylene; JA, jasmonic acid; SA, salicylic acid; SEN, senescence; ST, starvation; STR, stress; SUC, Suc.
Table IV. A selection of DE genes in the seed.
The gene identifier is reported along with a short annotation, the pattern of expression from T0 to T2 and from T2 to T3, likely correlations with hormones, metabolites, or physiological events, and references reporting specific information about the genes. Categories are indicated as follows: H, hormone biosynthesis, metabolism, and action; P, protein synthesis and metabolism; R, ROS synthesis, metabolism, and signaling; TF, transcription factors; TR, transport. The complete list and further details are available in Supplemental Table S5.
| Category | Gene Identifier | Tentative Annotation | T0–T2 | T2–T3 | Notesa | References |
| H | Md_570:2:a | Dehydrin, ABA responsive | − | Up | +ABA | – |
| Md_74377:s | Cytokinin dehydrogenase | − | Up | −CK | – | |
| P | Md_11646:1:a | Protein synthesis inhibitor, DPH2 | Up | − | − | – |
| Md_13925:1:a | Subtilisin | − | Up | − | – | |
| Md_20453:1:a | Aspartic proteinase, nepenthesin-1 | − | Up | − | – | |
| R | Md_288:2:a | Ferritin | − | Up | +ROS | Ravet et al. (2009) |
| TF | Md_114537:s | Homeobox-Leu zipper protein ATHB-40 | − | Up | +ABA | Henriksson et al. (2005) |
| Md_19486:1:a | Bel1 homeotic protein | − | Up | +T6P | Ray et al. (1994); Dong et al. (2000); Schluepmann et al. (2004); Skinner et al. (2004) | |
| Md_3290:2:a | MADS-box transcription factor, STK/AGL11 | Down | − | − | Yao et al. (1999); Tani et al. (2009) | |
| Md_3329:1:a | Ethylene-responsive AP2/ERF transcription factor | − | Up | +ET | Genevestigator | |
| TR | Md_4628:1:a | High-affinity nitrate transporter | − | Up | +ST | Remans et al. (2006); Chopin et al. (2007) |
+, Positive correlation; −, negative correlation; /, cross-talk; CK, cytokinin; ET, ethylene; ST, starvation; T6P, treahalose-6-phosphate.
It is worthy to note that also in this case, the cortex showed the most relevant transcriptional response, at least in terms of number of genes up- or down-regulated during abscission induction because of BA-specific action and in a development-independent manner (JABS and KABS sets). Specifically, from T0 to T2, 218 DE genes included 26 and 182 down- and up-regulated transcripts, respectively. From T2 to T3, only 10 DE genes were detected, six and four of which were down- and up-regulated, respectively.
Twenty genes putatively encoding transcription factors of diverse families were differentially expressed in the cortex, most of them with an up-regulation pattern from T0 to T2. From T2 to T3, no transcription factor-encoding gene was either up- or down-regulated with statistical significance (Table III). Besides some of the transcription factors, additional hormone-related genes were differentially expressed during abscission induction, among them some indicating an ongoing recovery of auxin homeostasis and an extensive inactivation of GAs and cytokinins. ABA signaling seemed to be strongly affected by abscission induction as well, particularly concerning the cross talk with ethylene, ROS, and sugars. The expression of some jasmonate-inducible genes along with an indicator of active brassinosteroid-jasmonate cross talk may point toward the involvement of these two hormones. Ethylene signaling was clearly affected during abscission induction, as shown for some transcription factors. Two additional key elements of the ethylene signal transduction pathway were also found, coding for a mitogen-activated protein kinase and a mitogen-activated protein kinase kinase, the former probably involved in ethylene-ABA cross talk (Xin et al., 2005). Key elements were found among the DE genes, which are likely involved in ROS-sugar-hormone cross talk, and three up-regulated genes showed close similarity with ROS-induced, ROS-detoxifying, or ROS-producing elements. Another interesting category concerns the vesicular trafficking, since at least six DE genes may encode elements involved either in endocytosis or exocytosis. Finally, as far as protein degradation is concerned, four genes were up-regulated during abscission induction, indicating a likely remobilization of resources. According to this analysis, the Globaltest results were largely confirmed not only from a quantitative point of view but also qualitatively, especially concerning the involvement of ABA, sugars, and ROS (Table III).
Concerning the seed, a situation very close to that assessed by the Globaltest analysis was again reported, in that a consistent transcriptional response was detected later, but to a less overall extent, than in the cortex. From T0 to T2, only 31 genes were differentially expressed (13 down-regulated and 18 up-regulated), whereas 45 transcripts differentially accumulated from T2 to T3, most of them up-regulated.
Among the four DE seed genes that were shown to putatively encode transcription factors, Md_3290:2:a was the only one early down-regulated from T0 to T2. It showed a high identity (90%) with Agamous-like11 (AGL11)-like MdMADS10, expressed in apple after pollination (Yao et al., 1999), and with the MADS box transcription factor Seedstick (STK) of P. persica, thought to be important for embryo development (Tani et al., 2009). Although less relevant than in the cortex, the hormonal response of the seed herein pointed out seemed to resemble the Globaltest results, in that active ABA and ethylene signalings were found, along with a likely degradation of cytokinins, ongoing oxidative stress, and a probable inhibition of protein synthesis concurrent with a later increase of protein degradation (Table IV).
Carbohydrates and Peroxides
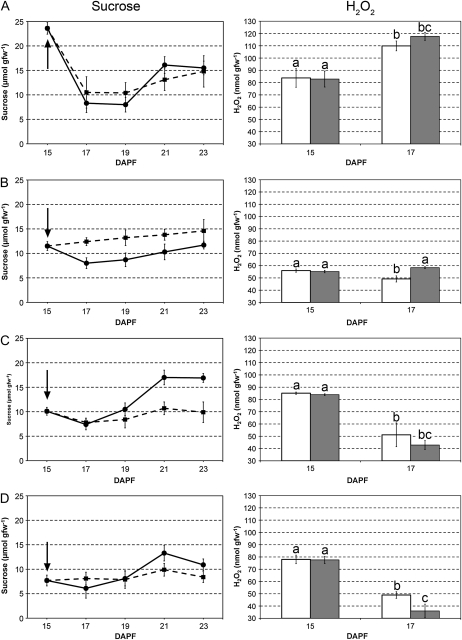
Since transcriptomic analyses pointed out components of sugar metabolism and genes related to responses to hydrogen peroxide, the main sugars along with hydrogen peroxide content were measured in persisting and abscising fruitlets. Suc, Glc, Fru, sorbitol, and starch levels were assessed in all samples up to 23 DAPF. Significant variations throughout the experiment were found only for Suc and starch, the former being correlated with abscission induction (Fig. 6), whereas the latter most likely depended on the BA treatments (Supplemental Fig. S4). Concerning Suc, a significant increase of its levels was observed immediately after the treatment only in LB3 fruitlets, remaining higher than in untreated samples throughout the experiment. In all the other samples, no significant variations were observed in this phase. Another relevant aspect of Suc behavior was pointed out in central fruitlets, both CB1 and CB3, in which a significant decrease was assessed later after abscission induction. The other carbohydrates did not show significant variations associated with abscission potential (Supplemental Fig. S4).
Figure 6.
Suc concentration (left) and hydrogen peroxide (H2O2) amount (right) in small (A) and big (B) lateral fruitlets and in small (C) and big (D) central fruitlets, either untreated (circles and continuous lines for Suc, white bars for hydrogen peroxide) or treated with BA (squares and dotted lines for Suc, gray bars for hydrogen peroxide). Letters indicate significant differences as pointed out by lsd test (P < 0.05). Error bars represent sd.
Since the majority of ROS-related genes were differentially expressed in the cortex from T0 to T2, hydrogen peroxide was measured only in this time lapse, which is crucial for abscission induction. Also in this case, divergent trends were observed upon BA treatment in LB3 fruits with respect to the central fruitlets. The latter showed a decreasing trend in untreated samples, with a magnifying effect of the treatment resulting in lower levels of peroxides. On the other hand, control L3 fruitlets displayed a decreasing trend in peroxide levels as in the previous ones, although to a lower extent, but had an opposite reaction when treated with BA. In fact, the significant increase observed in treated LB3 fruitlets at T2 was well correlated with expression data of ROS-related genes and, therefore, with the abscission potential. It is noteworthy that L1 fruitlets showed increasing levels of peroxide, being highest at T2, along with a positive effect of BA treatment, although less significant than in LB3 (Fig. 6).
DISCUSSION
The apple inflorescence is almost a unique model system for studying correlatively driven abscission, and the availability of chemical thinning tools able to selectively induce fruit drop allows the setting up of controlled experimental plans in the field aimed at magnifying the natural abscission potential. Indeed, the interfruitlet dominance relationships existing within the apple cluster can be assessed with good approximation in order to predict the destiny of each fruitlet in terms of probability to abscise. However, a deep knowledge of the molecular events occurring during the early phases of apple fruitlet abscission induction is still lacking. Previous studies carried out by Dal Cin et al. (2009a) addressed this issue with a preliminary approach, by using a cDNA-amplified fragment length polymorphism-based differential display, but without laying out an overall model of the early inductive events. A different study by the same authors (Dal Cin et al., 2009b) pointed out interesting evidence in terms of polar auxin transport element transcription in relation to the ethylene burst occurring in the postinduction phase. Therefore, our research here was focused on the earlier events occurring at the fruit level (the cortex and the seed), which is where the abscission signal is thought to be generated.
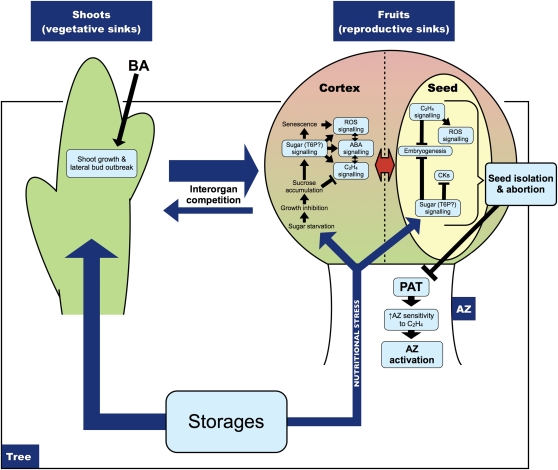
A model was devised for apple fruitlet abscission taking into account the overall transcriptomic data herein obtained and some key metabolic analyses aimed at strengthening and confirming the crucial steps pointed out by microarray experiments (Fig. 7). According to this model, apple fruitlet abscission takes place in four main steps, corresponding to the four structural levels where the key events may occur (i.e. the tree, the fruit cortex, the seed, and the abscission zone). The initial steps arise at the tree level, where a nutritional stress is established either naturally or upon a thinning treatment with BA. Such a condition is plausible, since at the beginning of the vegetative and reproductive season, the growth of shoots and fruitlets is supported to a large extent by stored assimilates. As a consequence, a strong competition for storage assimilate reallocation exists among shoots, between shoots and fruits, among fruits of different clusters, and among fruits of the same cluster. Since young growing shoots act as stronger sinks than fruits, the tree is unable to support all the growing fruitlets, causing the “weaker” ones to naturally abscise, thus generating the fruitlet physiological drop. When BA is used as a thinner, a magnification of the already existing nutritional stress occurs. This cytokinin is known to induce lateral bud outgrowth, thus enhancing the shoot sink activity and, consequently, the competition for assimilates (Bangerth, 2000; Bangerth et al., 2000; Bubán, 2000). At this point, how does this nutritional stress trigger fruitlet abscission? And how is this nutritional signal transduced into the abscission signal?
Figure 7.
Hypothetical model for immature fruit abscission in apple. The interorgan competition for stored assimilates existing within the tree is magnified by BA, which stimulates shoot growth and bud outbreak. This condition is perceived by weaker fruitlets as a nutritional stress, which is translated at both the cortex and seed level throughout cross talk signaling pathways, mainly involving sugars, ROS, ABA, and ethylene. When the seed perceives the situation as unrecoverable, a block of embryo development occurs, leading to seed isolation and abortion. This crucial step would determine the depolarization of auxin transport, the enhancement of abscission zone sensitivity to ethylene, and its activation. AZ, Abscission zone; CKs, cytokinins; PAT, polar auxin transport; T6P, trehalose-6-phosphate. The thickness of the arrows related to interorgan competition and storage partitioning is proportional to the strength of the organ as a sink. [See online article for color version of this figure.]
Our transcriptomic data suggest that the cortex is the primary response tissue perceiving this nutritional stress, at least in quantitative terms. Nutrient and sugar starvation affect its transcriptomic profiles already at 2 d after treatment, whereas significant changes related to abscission in the seed transcriptome appear later, at 4 d after BA spray (Supplemental Tables S1 and S2). During the early steps of abscission induction, a sugar signal, most likely involving trehalose-6-phosphate, induces a prompt reaction to nutritional stress. The involvement of trehalose-6-phosphate is suggested by the high expression levels of a class II TPS gene in abscising fruitlets, as found also during induced abscission of Citrus fruits (Alferez et al., 2007). A significant increase of Suc concentration in abscising fruitlets with respect to the NAFs was also found (Fig. 6) promptly after the treatment, as demonstrated previously by Stopar et al. (2001). Accumulation of Suc is often reported as a reaction to sugar starvation and has generally been considered to be an adaptive response to the stress condition (Roitsch, 1999). It is also associated with senescence, whose regulation in plants is known to be triggered by sugars (Wingler et al., 2009). Also, an increased ROS production may be linked to sugar starvation (Hooks et al., 1995; Contento et al., 2004), and Suc accumulation may also represent an oxidative stress balance mechanism (Couée et al., 2006). Moreover, the idea that high sugar (carbon)/low nitrogen conditions and not starvation would trigger changes in gene expression that are characteristic of developmental senescence is supported by experimental evidence, at least in leaf (Wingler et al., 2009). According to this view, sugar starvation would not directly trigger senescence-associated gene expression in the cortex of abscising fruitlets but rather would contribute to the instigation of the sugar signaling, causing, in turn, the transcriptomic reaction associated with abscission induction. This reaction would be most likely mediated by ROS accumulation, since a higher concentration of hydrogen peroxide in the abscising fruitlets than in the NAFs was herein assessed during early abscission induction (Fig. 6). These findings are further supported by ROS-related gene expression, as described in the previous section, especially concerning a NADPH oxidase gene highly similar to Arabidopsis Respiratory Burst Oxidase Homologue D involved in ROS production and a gene coding for a class III peroxidase with diverse possible roles (Cosio and Dunand, 2009), both up-regulated in the cortex of fruitlets induced to abscise. In this context, ABA signaling concurrently orchestrates sugar-ROS cross talk, as pointed out by transcriptomic data indicating typical signatures of ABA action. The TPS gene found overexpressed in abscising fruitlets may also regulate ABA signaling, as found in Arabidopsis (Avonce et al., 2004). During the early phases of abscission induction, an active resource mobilization is already established in the cortex, according to gene expression data. Moreover, persisting fruitlets show transcriptomic profiles typical of actively growing organs, in contrast with the abscising ones displaying expression levels for the same genes compatible with a block of their growth, especially in terms of transcripts encoding structural and metabolic elements.
As far as the involvement of transcription factors is concerned, some key elements were shown to be active during abscission induction in the cortex. However, most of these may possibly regulate downstream processes mostly related to ongoing senescence, rather than to the early inductive events. In fact, the NAC genes MdMYB8 and the two RING-H2 finger genes overexpressed in the abscising fruitlets are closely similar to senescence-associated or ABA-induced transcription factors found in other species and putatively involved in ABA signaling downstream of the abscission induction signal (Ko et al., 2006; Uauy et al., 2006; Ay et al., 2009; Meng et al., 2009). On the other hand, the subtractive approach focused on the fruitlets induced to abscise allowed the discovery of the likely involvement of some transcription factors during the earlier events, mostly with an up-regulation profile at T2, specifically, two WRKY genes possibly involved in ROS signaling (Pitzschke and Hirt, 2009), a MYB gene similar to Suc-induced transcription factors, a MYC putatively triggering stress-responsive genes (Smolen et al., 2002), and, interestingly, an ARF similar to Arabidopsis AtARF3, whose transcription is induced by hydrogen peroxide, nitrogen starvation, Suc, and ABA. All these genes may act as early regulators of the abscission induction, probably involved in the translation of the initial stress condition into abscission signal at the cortex level. A functional validation is in progress to elucidate their relative importance and roles in the generation of the signal cascade triggering fruit shedding.
Part of this signaling may also include specific kinase cascades, such as those found to be up-regulated in abscising fruitlets from T0 to T2. However, also in this case, the majority of these genes are most likely involved downstream of abscission signal generation, except for an Sucrose Non-Fermenting1 (SNF1)-related kinase gene closely similar to Arabidopsis SnRK3. The latter was shown to be induced by exogenous cytokinin (Chikano et al., 2001) and involved in Suc-dependent transcription stimulation of Suc synthase genes (Purcell et al., 1998) and in Suc synthase phosphorylation. Interestingly, a gene coding for a Suc synthase closely similar to Arabidopsis SUS3, induced by Suc and regulated by SnRK3 itself (Chikano et al., 2001), was coregulated in the same samples. These two elements, the SnRK3-like gene and the SUS3-like Suc synthase gene, may determine the early sugar sensing/signaling generating the abscission signal as a response to nutrient starvation, thus representing a key regulation point leading to Suc accumulation in the shedding fruitlets.
Gene expression data indicate not only an active resource mobilization but also active protein degradation and vesicular trafficking, all of which are most likely triggered later on when the abscission signal is fully installed.
Hormones seem to play a relatively important role during the early phases of abscission in the cortex, since the majority of the transcriptionally activated elements involved in hormone signaling seem to be downstream of the abscission induction. The earliest association with the abscission potential was found at T2 for ABA-related transcriptome (highly significant; P < 0.001). Beyond the genes discussed in the above paragraphs, early ABA signaling involves also a down-regulation of 14-3-3 genes in the abscising fruitlets. The related proteins may trigger ABA-ethylene cross talk and responses to sugar starvation (Lancien and Roberts, 2006). ABA-sugar cross talk may also involve a gene encoding an AMP-activated protein kinase similar to an Arabidopsis ABA-induced SNF1-related kinase, which was found to be up-regulated in the abscising fruitlets. Interesting data concern the hormone metabolic pathways, especially regarding auxin, GAs, and cytokinins. In fact, a gene for an IAA-amidohydrolase thought to disjoin IAA from specific amino acids was up-regulated at T2 in abscising fruitlets, probably as a homeostatic response. Concerning GAs, four overexpressed genes encoding deactivating enzymes (GA 2-oxidases) and a down-regulated Gibberellic Acid-Stimulated Arabidopsis4 (GASA4)-like transcript would indicate a decrease of active GA levels in the cortex. As far as cytokinins are concerned, the up-regulation of a deactivating gene coding for a cytokinin dehydrogenase was found during abscission induction, pointing also in this case toward a decrease of the active hormone amount. Finally, ethylene signaling was also found to be triggered, but only downstream of the abscission signal generation and as a consequence of the cross talk with ABA and ROS. Two elements are to be considered pivotal in this context, both up-regulated and putatively involved in ethylene signal transduction and cross talk with other transductive pathways. The first is an Arabidopsis Mitogen-Activated Protein Kinase11 (AtMPK11)-like gene, induced by ABA, putatively involved in ethylene-ABA cross talk and in modulation of ABA signaling (Xin et al., 2005), whereas the second is an Arabidopsis Mitogen-Activated Protein Kinase Kinase9-like gene, probably downstream of AtMPK11, involved in the up-regulation of ACS and Ethylene Responsive Factor (ERF) genes. Both elements are positioned in the same transductive pathway and most likely upstream of the regulation of ethylene biosynthesis induction occurring later in abscising fruitlets. Therefore, the ethylene burst usually found in abscising fruitlets may result from the cross talk between ABA and ROS, generated during the early inductive phases, immediately after the perception and signaling of the sugar starvation status. In this context, it is worthy to note that hormone-related transcriptomic signatures assessed in the cortex resemble those claimed to be responsible for the negative feedback regulation occurring before pollination and fertilization and preventing fruit set in tomato (Vriezen et al., 2008). In fact, our data show that ABA and ethylene signaling are strongly up-regulated concurrently, with a down-regulation of GA signaling specifically in fruits induced to abscise.
After the early reaction of the cortex, it may be hypothesized that a link is established with the seed when the abscission inductive process reaches an irreversible status. The seed appeared indeed affected at the transcriptional level at a later stage (Supplemental Tables S1 and S2), at least from a quantitative point of view. Furthermore, the seed is a structure with a stronger homeostasis than the cortex, since it represents the reproductive endeavor carried out by the tree, and thus is protected until the abscission process may become unrecoverable. Ethylene may function as the signal generated within the cortex and, through diffusion, carrying to the seed the abscission signal, as suggested by the transcription rates of several elements of its transductive pathway. In the seed, the signaling cascade activated by the abscission induction causes a block of embryogenesis, as suggested by the significant down-regulation of MdMADS10, an AGL11-like gene differentially expressed only in the lateral fruitlets induced to abscise by BA, whose role is strictly linked either to ovule or embryo development (Yao et al., 1999; Tani et al., 2009). Several other genes linked to embryo and seed development were differentially expressed in abscising fruitlets, among which are some ethylene-responsive genes, such as MdERS2, and an Apetala2 (AP2)/ERF, up-regulated in abscising fruitlets, and a series of genes involved in metabolism that are expressed at higher levels in the NAFs. Again, as in the cortex, persisting fruitlets show a more dynamic metabolism and the active transcription of genes coding for structural proteins. Interestingly, a nitrate transporter gene closely similar to AtNRT2.7 was overexpressed in LB3 fruits. In Arabidopsis, this gene controls nitrate content in the seed (Chopin et al., 2007) and is induced by nitrogen starvation (Remans et al., 2006). Moreover, actively growing organs, such as the young apple fruitlets, are a relevant source of auxin, whose main biosynthesis site is the seed. The hormone is actively transported from the fruit through the pedicel, and its continuous flow would keep the abscission zone insensitive to ethylene (Sexton and Roberts, 1982; Taylor and Whitelaw, 2001). ROS may play an important role also in this case, as demonstrated for leaf abscission by Sakamoto et al. (2008). In fact, some typical transcriptional signatures of high ROS levels were found also in the seed, although later than in cortex, such as a ferritin gene whose putative orthologs in Arabidopsis and rice (Oryza sativa) are induced by hydrogen peroxide as a protective mechanism (Ravet et al., 2009). Therefore, the oxidative atmosphere where the seed is constricted at this stage along with the nutritional stress and the signals coming from the cortex may contribute to the increase of ROS production, which in turn would disrupt metabolism and suppress the synthesis of IAA as described previously (Sakamoto et al., 2008). The reduced supply of auxin to the abscission zone concurrently with a likely depolarization of its transport would enhance its sensitivity to ethylene and the consequent activation of cell wall-degrading enzymes (Sexton and Roberts, 1982; Taylor and Whitelaw, 2001).
CONCLUSION
To the best of our knowledge, this study provides the first global monitoring of gene expression changes occurring during the early phases of apple fruitlet abscission induction. The model herein proposed takes into account both the temporal evolution of differential gene expression and its static association with abscission potential in both cortex and seed. According to this approach, the cortex would be the place where the primary abscission signal is generated, whereas the seed would function as a modulator of the physiological response, translating this signal to the abscission zone. However, as the time course of the inductive events in the two organs was based mainly upon massive transcriptomic data, a more targeted approach is now necessary to identify the actual key elements in charge of generating the abscission signal and the temporal sequence of these molecular events. It cannot be ruled out that the cortex reaction may be due to an amplification of biologically relevant transcriptomic changes occurring at the seed level, herein not detected because of the high stringency adopted in the statistical analyses. Future studies will be focused on the transductive pathways pointed out in this research to be responsible for early abscission induction, such as the ROS-sugar-ABA cross talk. Besides these aspects, the downstream effectors, especially at the seed level, will be investigated with particular attention devoted to MADS box and homeotic genes such as those evidenced in the transcriptomic analyses.
MATERIALS AND METHODS
Plant Material and Treatments
Experiments were carried out in 2008 on 8-year-old apple trees (Malus × domestica ‘Golden Delicious/M9’) trained with standard horticultural practices at the experimental farm of the Istituto Agrario San Michele all‘Adige. Populations of fruits with different abscission potentials (abscising fruitlets versus persisting fruitlets) were established as described by Dal Cin et al. (2005a, 2007, 2009a), Angeli et al. (2002), and other preliminary experiments (A. Botton, unpublished data). Briefly, the abscising population was made up of lateral fruitlets treated with BA at 200 μL L−1 (commercial name, Brancher-Dirado), when fruits had an average size of 13 mm (about 15 DAPF). The population of central persisting fruitlets was generated by removing all the laterals from each cluster at petal fall and leaving exclusively the central flower that had been hand-pollinated at full bloom with compatible pollen (cv Stark Red). Samples of the two populations were collected at defined time points from groups of 20 homogeneous trees randomly distributed in the orchard in four blocks. Fruits were collected and categorized into three classes of size (class 1, smaller fruits; class 2, medium fruits; class 3, bigger fruits), two classes related to the position within the clusters (lateral versus central fruits), and two classes on the basis of the treatment (BA-treated versus untreated fruits). Fruits of the intermediate size (labeled with number 2 in Fig. 1) were not considered for sampling and subsequent molecular analyses, and only fruits of the two more divergent size classes 1 and 3 were kept. This resulted in a combined categorization of fruits into four classes: (1) untreated lateral fruitlets, (2) lateral fruitlets treated with BA, (3) untreated central fruitlets, and (4) central fruitlets treated with BA (Fig. 1). Each class was further distinguished into the two size categories 1 (small fruits) and 3 (bigger fruits), for a total of eight experimental groups. Fruitlet shedding and ethylene evolution were monitored throughout the physiological drop from the beginning of the experiments to 46 DAPF in all fruitlet classes, separately. Seed and cortex (including epidermis) samples were collected from all classes of fruitlets at 0 (T0), 1 (T1), 2 (T2), 4 (T3), 6 (T4), and 8 (T5) d after the BA treatment from control and treated trees, and according to their position within the clusters (central versus lateral) and size (small versus big), as described. The latter parameter was decided at each sampling date based upon the mean cross-diameter of the whole population of lateral fruits, calculated over a sample of 100 fruits measured randomly on 20 trees. The small ones had a cross-diameter below the mean (−sd), whereas the big ones had a cross-diameter above the mean (+sd). Lateral fruitlets were collected from intact clusters showing a clear hierarchy in terms of fruit size (i.e. with a clearly distinguishable central fruit, bigger than any lateral). Acronyms were ascribed to samples according to the following code: the first letter(s) describes fruitlet position within the cluster and the presence of BA treatment (L = untreated lateral, C = untreated central, LB = treated lateral, CB = treated central), then a digit to describe the size (1 = small, 3 = big), and finally a digit to describe the time point (0 = time of the treatment, 1 = 1 d after treatment, 2 = 2 d after treatment, 3 = 4 d after treatment, etc.). For example, LB32 is a lateral fruit, treated with BA, big sized, 2 d after treatment. All samples were frozen in liquid nitrogen and stored at −80°C for later molecular analyses.
RNA Isolation
Total RNA was extracted from cortex and seed following the method of Ruperti et al. (2001), with a few adaptations due to differences between tissues. The extraction buffer volume was set at 10 mL for the cortex and 1 mL for the seed, and the starting amount of tissue was 0.60 and 0.02 g, respectively. In order to achieve the final quality of the extract, 30 μL (cortex) and 3 μL (seed) of a calcium hydroxide suspension at 60 g L−1 were added just before the first centrifugation step (Dal Cin et al., 2005b; Botton et al., 2008, 2009a, 2009b). Total RNA was quantified spectrophotometrically, and its integrity was checked by running 1 μg on a 1% agarose gel stained with SYBR Safe (Invitrogen).
Microarray Analysis
The 30K custom microarray was set up by means of the CombiMatrix technique starting from publicly available apple sequences (for a detailed description of the whole pipeline, see Supplemental Materials and Methods S1). For hybridizations, 1 μg of total RNA was amplified and 6 μg of antisense RNA was labeled using the RNA Ampulse amplification and labeling kit with Cy5 for CombiMatrix arrays (Kreatech Diagnostics), according to the manufacturer’s instructions, and were hybridized to arrays according to CombiMatrix protocols. Scanning was performed on a GenePix 4000B scanner. Data extraction was done using CombiMatrix Microarray Imager software.
Globaltest Analysis of Transcriptional Profiles and Hierarchical Clustering
Global transcriptional profile testing was carried out with the Globaltest package version 4.14.4 (Goeman et al., 2004) of R software version 2.9.1 (http://www.r-project.org/). This package tests the overall gene expression for significant association with a given variable. The test gives a unique P value for the whole group, therefore avoiding a multiple testing adjustment (Goeman et al., 2004). If the statistic is significant, the genes in the group are, on average, more associated with the response variable than would be expected. The strength of this association is given by a Z score, calculated for each gene. In this way, at least part of the variance of the response variable can be predicted from the gene expression measurements of the gene set, or vice versa. Raw intensity data were used as input for the package and normalized using the vsn2 function within R. Association was considered significant with P < 0.1, highly significant with P < 0.01, and extremely significant with P < 0.001. The response variables considered were as follows: position (the position of the fruit within the cluster: central versus lateral), weight (fruit weight, a likely indicator of the fruit developmental stage), treatment (untreated versus treated with BA), and destiny (five classes of abscission potential, as described above). The analysis was performed considering as biological replicates the samples with the same predicted abscission potential (AP). The choice of replicates was done in order to reduce the total number of samples and hybridizations, concurrently taking into account a large part of the variation seen at the biological level (i.e. treated/untreated fruits with different dimensions but with the same AP). Four classes were established: naturally abscising fruitlets (L1 fruitlets; 90% < AP < 100%), strongly abscising fruitlets (LB1 and LB3 fruitlets; 90% < AP < 100%), probably persisting fruitlets (CB1 fruitlets; AP < 10%), and strongly persisting fruitlets (L3, C1, C3, and CB3; AP ∼ 0%). Summarizing, three biological replicates for naturally abscising fruits, four for strongly persisting fruits, two for probably persisting fruits, and 11 for the strongly persisting fruits were used (Supplemental Fig. S5).
For hierarchical clustering, raw intensity data were mean centered, normalized, and clustered by means of Cluster 3.0 software (de Hoon et al., 2004), using the uncentered correlation similarity matrix and the centroid linkage clustering method.
All the experimental procedures comply with minimum information about a microarray experiment standards for array data (Brazma et al., 2001). Gene expression data have been submitted to ArrayExpress (accession no. A-MEXP-1852).
Subtractive Analysis
In order to validate the results of the Globaltest analysis, a subtractive approach was carried out on single-slide data by means of the Nudge package of R (Dean and Raftery, 2005). This method can be used also for nonrepeated experiments and was herein applied to estimate genes with significant differential expression according to an all-against-all comparison, using the nudge1 function and posterior P of at least 0.5. Raw intensity data were used as input for the program, since it already implements a normalization step (Loess mean normalization, with default parameters). Negative controls spotted on the microarray were used for false-positive discovery. Accordingly, such control genes were correctly identified as nondifferentially expressed at the end of the analysis. By means of this approach (Supplemental Fig. S5), it was assessed if the differential expression specifically induced by the BA treatment in L3 fruitlets concerned the same functional networks pointed out by the Globaltest analysis carried out above.
The gene sets differentially expressed during the 4-d time course, when abscission is thought to be induced, were considered (each group was labeled as described in Supplemental Fig. S2), and a series of operations was performed separately for up- and down-regulated genes, as follows:
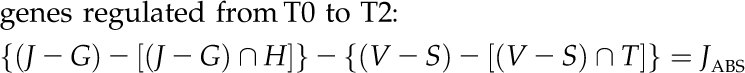 |
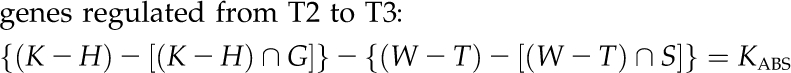 |
In detail, the gene sets G and H were subtracted from J and K, respectively, to remove genes that are naturally regulated during fruitlet development from T0 to T2 (15–17 DAPF) and from T2 to T3 (17–19 DAPF), respectively. Genes with a delayed regulation, given by (J – G) ∩ H, and those regulated in advance, given by (K – H) ∩ G, were further subtracted. At this step, development-independent genes whose regulation was affected by BA from T0 to T2, and from T2 to T3, were obtained. However, among these genes, also those pharmacologically regulated by BA, thus with no specific correlation with abscission, were included. Therefore, development-independent genes regulated by BA in strongly persisting fruitlets (i.e. not directly correlated with abscission) were further subtracted, giving the final sets of development-independent/BA-regulated/abscission-related genes differentially expressed from T0 to T2 (JABS) and from T2 to T3 (KABS).
Quantitative PCR Expression Analyses
cDNA for expression analyses was synthesized from 2 μg of DNA-free total RNA in a final volume of 25 μL containing 200 units of Moloney murine leukemia virus reverse transcriptase (Promega), 1× Moloney murine leukemia virus buffer, 25 units of RNasin (RNase inhibitor; Amersham Biosciences), 1 μg of random hexamers (Invitrogen), and 2 mm deoxyribonucleotide triphosphates. The reaction was carried out for 1 h at 37°C in a Gene Amp PCR System 9700 thermocycler (Applied Biosystems).
Real-time PCR relative quantification was performed in triplicate on two biological replicates in a total volume of 10 μL using the Fast SYBR Green Master Mix (Applied Biosystems) with 3 pmol of every primer and 2 μL of a 1:10 dilution of cDNA. Primers (Supplemental Table S6) were designed with Primer3 software version 0.4.0 (http://frodo.wi.mit.edu/primer3/) according to the instructions given by Applied Biosystems. The specificity of amplification was assessed by subsequent subcloning and sequencing of the PCR products obtained under the same conditions adopted in the real-time experiments. The reaction mixture was amplified in a StepOne Real-Time PCR System (Applied Biosystems) under the following conditions: initial activation step at 95°C for 10 min, followed by 50 cycles including 3 s of denaturation at 95°C and 15 s of annealing/extension at 60°C. After each PCR cycle, a data acquisition step was introduced to record the fluorescent signals at the optimum temperature, previously determined by melting point analysis of every specific amplification product. Data were acquired, elaborated, and exported with the StepOne Software version 2.1 (Applied Biosystems), whereas all the final calculations were carried out with the automated Excel spreadsheet Q-Gene designed by Simon (2003) using the modifications of the delta cycle threshold method suggested by Pfaffl (2001). Besides those found in the literature, additional reference genes were selected among those spotted on the microarray according to the criteria of Vandesompele et al. (2002). The genes were MdUBI, Md18S (Dal Cin et al., 2005a), MdACT (Li and Yuan, 2008), Md_8283:1:a, and Md_4592:1:a for the cortex and Md18S for the seed, the latter being sufficiently stable to be used alone. Gene expression values were normalized to the housekeeping genes identified above and reported as arbitrary units of mean normalized expression, using equation 2 of Q-Gene. The correct size of the amplification products was checked by running each reaction on a 1.5% agarose gel stained with SYBR Safe (Invitrogen) and viewed under UV light.
Quantification of Carbohydrates and Hydrogen Peroxide
For carbohydrate measurements, carried out in three biological replicates, 50 mg of frozen flesh powder was extracted in 1.5 mL of 80% ethanol and 20% water, containing 100 mm HEPES-KOH (pH 7.1) and 10 mm MgCl2, for 45 min at 80°C. After cooling at room temperature, the extract was centrifuged at 15,800g for 5 min. The supernatant, containing soluble sugars (Glc, Fru, Suc, and sorbitol), was either analyzed immediately for the sugar content or stored at −20°C until analysis. The pellet, containing starch, was resuspended and washed, at least four times, with 40 mm acetate buffer (pH 4.5). After washing, the pellet was autoclaved in 1 mL of the washing buffer for 45 min at 120°C to solubilize the starch. After autoclaving, 4 units of α-amylase and 40 units of amyloglucosidase were added to the pellet, and the mixture was incubated for 1 h at 50°C to allow complete starch hydrolysis. After starch hydrolysis, the samples were centrifuged and the supernatant was analyzed immediately for Glc or stored at −20°C until analysis. Soluble sugars, as well as Glc originated from the starch hydrolysis, were analyzed enzymatically as described by Jones et al. (1977) with minor modifications as described by Antognozzi et al. (1996). Suc was analyzed in sequence after Glc and Fru following the addition of 100 units of invertase to the assay mixture. Sorbitol was measured enzymatically following the reduction of NAD+ coupled to sorbitol oxidation to Fru mediated by sorbitol dehydrogenase. The assay was set up to allow the use of a plate reader. The assay contained 100 mm Bicine (pH 9.2), 5 mm MgCl2, 0.01% (w/v) bovine serum albumin, 1 mm NAD+, 2 units of sorbitol dehydrogenase, and the appropriate amount of sample. The carbohydrate extracts were used directly up to 40 μL in the assay without problems. All carbohydrate measurements were performed in dual-wavelength mode (340–405 nm) in an Anthos 2001 plate reader (Anthos Labtec Instruments).
Hydrogen peroxide was quantified by means of the PeroXOquant Quantitative Peroxide Assay kit (Pierce), following the instructions provided by the manufacturer. Briefly, 130 mg of fruit cortex was ground to a fine powder in liquid nitrogen and processed as indicated by the manufacturer in three biological replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The sequence clustering/assembling pipeline specifically set up for apple sequences.
Supplemental Figure S2. Schematic representation of the experimental plan for microarray experiments.
Supplemental Figure S3. GO annotation of the 30,518 probes spotted on the apple microarray.
Supplemental Figure S4. Concentrations of Glc, Fru, sorbitol, and starch in fruitlet samples.
Supplemental Figure S5. Flow chart showing the data analysis procedure.
Supplemental Figure S6. Hierarchical clustering of the significantly variable transcriptomes of cortex and seed.
Supplemental Figure S7. Quantitative real-time PCR validation of selected genes.
Supplemental Table S1. Globaltest statistics summarizing the associations between expression data in the cortex and fruitlet destiny.
Supplemental Table S2. Globaltest statistics summarizing the associations between expression data in the seed and fruitlet destiny.
Supplemental Table S3. Number of genes differentially expressed in all the sample combinations.
Supplemental Table S4. Genes differentially expressed in the cortex of fruitlets that were induced to abscise upon BA treatment.
Supplemental Table S5. Genes differentially expressed in the seed of fruitlets that were induced to abscise upon BA treatment.
Supplemental Table S6. Primers used in quantitative PCR experiments.
Supplemental Materials and Methods S1. The construction of the microarray.
Supplementary Material
Acknowledgments
We thank Alberto Dorigoni and Paolo Lezzer for setting up field trials and treatments. We also thank Andrea Boschetti for ethylene measurements.
References
- Alferez F, Zhong GY, Burns JK. (2007) A citrus abscission agent induces anoxia- and senescence-related gene expression in Arabidopsis. J Exp Bot 58: 2451–2462 [DOI] [PubMed] [Google Scholar]
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390 [DOI] [PubMed] [Google Scholar]
- Angeli D, Comai M, Danesin M, Dorigoni A, Ruperti B, Ramina A. (2002) Interazione tra citochinine ed etilene nel controllo del diradamento del melo. Rivista di Frutticoltura e di Ortofloricoltura 5: 56–59 [Google Scholar]
- Antognozzi E, Battistelli A, Famiani F, Moscatello S, Stanica F, Tombesi A. (1996) Influence of CPPU on carbohydrate accumulation and metabolism in fruits of Actinidia deliciosa (A Chev). Sci Hortic (Amsterdam) 65: 37–47 [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G. (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136: 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer AM, Uhlemann R, Reuter G, Humbeck K. (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58: 333–346 [DOI] [PubMed] [Google Scholar]
- Bangerth F. (2000) Abscission and thinning of young fruit and their regulation by plant hormones and bioregulators. Plant Growth Regul 31: 43–59 [Google Scholar]
- Bangerth F, Li CJ, Gruber J. (2000) Mutual interaction of auxin and cytokinins in regulating correlative dominance. Plant Growth Regul 32: 205–217 [Google Scholar]
- Bassham DC, Brandizzi F, Otegui M, Sanderfoot A. (2008) The secretory system of Arabidopsis. In Somerville CR, Meyerowitz EM, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0116, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botton A, Andreotti C, Costa G, Ramina A. (2009a) Peach (Prunus persica L. Batsch) allergen-encoding genes are developmentally regulated and affected by fruit load and light radiation. J Agric Food Chem 57: 724–734 [DOI] [PubMed] [Google Scholar]
- Botton A, Lezzer P, Dorigoni A, Barcaccia G, Ruperti B, Ramina A. (2008) Genetic and environmental factors affecting allergen-related gene expression in apple fruit (Malus domestica L. Borkh). J Agric Food Chem 56: 6707–6716 [DOI] [PubMed] [Google Scholar]
- Botton A, Lezzer P, Dorigoni A, Ruperti B, Ramina A. (2009b) Environmental factors affecting the expression of apple (Malus domestica L. Borkh) allergen-encoding genes. J Hortic Sci Biotech ISAFRUIT Special Issue: 182–187 [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Bubán T. (2000) The use of benzyladenine in orchard fruit growing: a mini review. Plant Growth Regul 32: 381–390 [Google Scholar]
- Büttner M, Singh KB. (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chen I, Lee S, Pan S, Hsieh H. (2007) GASA4, a GA-stimulated gene, participates in light signaling in Arabidopsis. Plant Sci 172: 1062–1071 [Google Scholar]
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H. (2001) Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet 264: 674–681 [DOI] [PubMed] [Google Scholar]
- Choi JW, Kimi GB, Huh YC, Kwon MR, Mok IG, Kim JW, Lee TS, Kim S, Im KH. (2004) Cloning of genes differentially expressed during the initial stage of fruit development in melon (Cucumis melo cv. Reticulatus). Mol Cells 17: 237–241 [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel-Vedele F. (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC. (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio C, Dunand C. (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60: 391–408 [DOI] [PubMed] [Google Scholar]
- Couée I, Sulmon C, Gouesbet G, El Amrani A. (2006) Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot 57: 449–459 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Barbaro E, Danesin M, Murayama H, Velasco R, Ramina A. (2009a) Fruitlet abscission: a cDNA-AFLP approach to study genes differentially expressed during shedding of immature fruits reveals the involvement of a putative auxin hydrogen symporter in apple (Malus domestica L. Borkh). Gene 442: 26–36 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Boschetti A, Dorigoni A, Ramina A. (2007) Benzylaminopurine application on two different apple cultivars (Malus domestica) displays new and unexpected fruitlet abscission features. Ann Bot (Lond) 99: 1195–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin V, Danesin M, Boschetti A, Dorigoni A, Ramina A. (2005a) Ethylene biosynthesis and perception in apple fruitlet abscission (Malus domestica L. Borkh). J Exp Bot 56: 2995–3005 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Danesin M, Rizzini FM, Ramina A. (2005b) RNA extraction from plant tissues: the use of calcium to precipitate contaminating pectic sugars. Mol Biotechnol 31: 113–119 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Velasco R, Ramina A. (2009b) Dominance induction of fruitlet shedding in Malus × domestica (L. Borkh): molecular changes associated with polar auxin transport. BMC Plant Biol 9: 139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Raftery AE. (2005) Normal uniform mixture differential gene expression detection for cDNA microarrays. BMC Bioinformatics 6: 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- de Jong M, Mariani C, Vriezen WH. (2009) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Faik A, Maclachlan G. (1998) Fucosyltransferase and the biosynthesis of storage and structural xyloglucan in developing nasturtium fruits. Plant Physiol 118: 885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Yao JL, Atkinson RG, Putterill JJ, Morris BA, Gardner RC. (2000) MDH1: an apple homeobox gene belonging to the BEL1 family. Plant Mol Biol 42: 623–633 [DOI] [PubMed] [Google Scholar]
- Du L, Poovaiah BW. (2004) A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol Biol 54: 549–569 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Frébortová J, Fraaije MW, Galuszka P, Sebela M, Pec P, Hrbác J, Novák O, Bilyeu KD, English JT, Frébort I. (2004) Catalytic reaction of cytokinin dehydrogenase: preference for quinones as electron acceptors. Biochem J 380: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K. (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C. (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. (2004) A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20: 93–99 [DOI] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA. (2006) Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84 [DOI] [PubMed] [Google Scholar]
- Greene DW, Autio WR, Erf JA, Mao ZY. (1992) Mode of action of benzyladenine when used as a chemical thinner on apples. J Am Soc Hortic Sci 117: 775–779 [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC. (2007) BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19: 1809–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Olsson ASB, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E. (2005) Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol 139: 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Rodríguez MB, Maldonado JM, Pérez-Vicente R. (2004) Light and metabolic regulation of HAS1, HAS1.1 and HAS2, three asparagine synthetase genes in Helianthus annuus. Plant Physiol Biochem 42: 511–518 [DOI] [PubMed] [Google Scholar]
- Hong-Bo S, Zong-Suo L, Ming-An S. (2005) LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids Surf B Biointerfaces 45: 131–135 [DOI] [PubMed] [Google Scholar]
- Hooks MA, Bode K, Couee I. (1995) Regulation of acyl-CoA oxidases in maize seedlings. Phytochemistry 40: 657–660 [Google Scholar]
- Huang S, Sawaki T, Takahashi A, Mizuno S, Takezawa K, Matsumura A, Yokotsuka M, Hirasawa Y, Sonoda M, Nakagawa H, et al. (2010) Melon EIN3-like transcription factors (CmEIL1 and CmEIL2) are positive regulators of an ethylene- and ripening-induced 1-aminocyclopropane-1-carboxylic acid oxidase gene (CM-ACO1). Plant Sci 178: 251–257 [Google Scholar]
- Jensen P, Makalowska I, Altman N, Fazio G, Praul C, Maximova S, Crassweller R, Travis J, Mcnellis T. (2009) Rootstock-regulated gene expression patterns in apple tree scions. Tree Genet Genomes 6: 57–72 [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MG, Outlaw WH, Lowry OH. (1977) Enzymic assay of 10 to 10 moles of sucrose in plant tissues. Plant Physiol 60: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancien M, Roberts MR. (2006) Regulation of Arabidopsis thaliana 14-3-3 gene expression by gamma-aminobutyric acid. Plant Cell Environ 29: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J. (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. (2001) Dyneins have run their course in plant lineage. Traffic 2: 362–363 [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. (2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117: 9–18 [DOI] [PubMed] [Google Scholar]
- Li J, Yuan R. (2008) NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious‘ apples. J Plant Growth Regul 27: 283–295 [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Iwakawa H, Machida Y, Machida C. (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel A, Levine A. (2002) Induction of glucosyltransferase transcription and activity during superoxide-dependent cell death in Arabidopsis plants. Plant Physiol Biochem 40: 133–140 [Google Scholar]
- Meng C, Cai C, Zhang T, Guo W. (2009) Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci 176: 352–359 [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb RD, Crowhurst RN, Gleave AP, Rikkerink EHA, Allan AC, Beuning LL, Bowen JH, Gera E, Jamieson KR, Janssen BJ, et al. (2006) Analyses of expressed sequence tags from apple. Plant Physiol 141: 147–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch LMC, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH. (2009) Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 229: 1335–1346 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Singh KB. (2002) Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM. (2003) Hormonal interactions in fruit development. J Plant Growth Regul 22: 73–81 [Google Scholar]
- Pandolfini T, Molesini B, Spena A. (2007) Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci 12: 327–329 [DOI] [PubMed] [Google Scholar]
- Park S, Sugimoto N, Larson MD, Beaudry R, van Nocker S. (2006) Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol 141: 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. (2007) Trehalose 6-phosphate. Curr Opin Plant Biol 10: 303–309 [DOI] [PubMed] [Google Scholar]
- Pedersen L, Henriksen A. (2005) Acyl-CoA oxidase 1 from Arabidopsis thaliana: structure of a key enzyme in plant lipid metabolism. J Mol Biol 345: 487–500 [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang XQ, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. (2003) TIGR Gene Indices Clustering Tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45–e50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. (2009) Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol 149: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG. (1998) Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Ray A, Robinson-Beers K, Ray S, Baker SC, Lang JD, Preuss D, Milligan SB, Gasser CS. (1994) Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc Natl Acad Sci USA 91: 5761–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T. (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Baier M, Holman R, May AG, Bevan MW. (2006) Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J 46: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Rorat T. (2006) Plant dehydrins: tissue location, structure and function. Cell Mol Biol Lett 11: 536–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. (2006) Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell 98: 53–67 [DOI] [PubMed] [Google Scholar]
- Rösti J, Barton CJ, Albrecht S, Dupree P, Pauly M, Findlay K, Roberts K, Seifert GJ. (2007) UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19: 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruperti B, Bonghi C, Rasori A, Ramina A, Tonutti P. (2001) Characterization and expression of two members of the peach 1-aminocyclopropane-1-carboxylate oxidase gene family. Physiol Plant 111: 336–344 [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Munemura I, Tomita R, Kobayashi K. (2008) Reactive oxygen species in leaf abscission signaling. Plant Signal Behav 3: 1014–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Mase K, Nakano Y, Nishikubo N, Sugita R, Tsuboi Y, Kajita S, Zhou J, Kitano H, Katayama Y. (2006) 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase is regulated for the accumulation of polysaccharide-linked hydroxycinnamoyl esters in rice (Oryza sativa L.) internode cell walls. Plant Cell Rep 25: 676–688 [DOI] [PubMed] [Google Scholar]
- Schaffer RJ, Friel EN, Souleyre EJF, Bolitho K, Thodey K, Ledger S, Bowen JH, Ma JH, Nain B, Cohen D, et al. (2007) A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiol 144: 1899–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, Salinas J, Mancuso S, Valpuesta V, Baluska F, et al. (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135: 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Walz A, Park S, Cohen JD, Ludwig-Müller J. (2006) Indole-3-acetic acid protein conjugates: novel players in auxin homeostasis. Plant Biol (Stuttg) 8: 340–345 [DOI] [PubMed] [Google Scholar]
- Sepúlveda-Jiménez G, Rueda-Benítez P, Porta H, Rocha-Sosa M. (2005) A red beet (Beta vulgaris) UDP-glucosyltransferase gene induced by wounding, bacterial infiltration and oxidative stress. J Exp Bot 56: 605–611 [DOI] [PubMed] [Google Scholar]
- Sexton R, Roberts J. (1982) Cell biology of abscission. Annu Rev Plant Physiol 33: 133–162 [Google Scholar]
- Simon P. (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440 [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Oñate-Sánchez L. (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Skinner DJ, Hill TA, Gasser CS. (2004) Regulation of ovule development. Plant Cell (Suppl) 16: S32–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen GA, Pawlowski L, Wilensky SE, Bender J. (2002) Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopar M, Resnik M, Pongrac VZ. (2001) Non-structural carbohydrate status and CO2 exchange rate of apple fruitlets at the time of abscission influenced by shade, NAA or BA. Sci Hortic (Amsterdam) 87: 65–76 [Google Scholar]
- Tamaoki M, Nakajima N, Kubo A, Aono M, Matsuyama T, Saji H. (2003) Transcriptome analysis of O3-exposed Arabidopsis reveals that multiple signal pathways act mutually antagonistically to induce gene expression. Plant Mol Biol 53: 443–456 [DOI] [PubMed] [Google Scholar]
- Tani E, Polidoros AN, Flemetakis E, Stedel C, Kalloniati C, Demetriou K, Katinakis P, Tsaftaris AS. (2009) Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol Biochem 47: 690–700 [DOI] [PubMed] [Google Scholar]
- Tatsuki M, Hayama H, Nakamura Y. (2009) Apple ethylene receptor protein concentrations are affected by ethylene, and differ in cultivars that have different storage life. Planta 230: 407–417 [DOI] [PubMed] [Google Scholar]
- Taylor JE, Whitelaw CA. (2001) Signals in abscission. New Phytol 151: 323–339 [Google Scholar]
- Torres MA, Dangl JL, Jones JD. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177: 60–76 [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari J, Kobae Y, Yamaki S, Yamada K, Toyofuku K, Tabuchi T, Shiratake K. (2004) Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant Cell Physiol 45: 1032–1041 [DOI] [PubMed] [Google Scholar]
- Wheeler MCG, Tronconi MA, Drincovich MF, Andreo CS, Flügge UI, Maurino VG. (2005) A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol 139: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Masclaux-Daubresse C, Fischer AM. (2009) Sugars, senescence, and ageing in plants and heterotrophic organisms. J Exp Bot 60: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ. (2009) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150: 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL. (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol 139: 1350–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Yao JL, Dong YH, Kvarnheden A, Morris B. (1999) Seven MADS-box genes in apple are expressed in different parts of the fruit. J Am Soc Hortic Sci 124: 8–13 [Google Scholar]
- Zentgraf U, Laun T, Miao Y. (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zhang X, Hu J. (2010) The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.