Abstract
The heteromeric acetyl-coenzyme A carboxylase catalyzes the first and committed reaction of de novo fatty acid biosynthesis in plastids. This enzyme is composed of four subunits: biotin carboxyl-carrier protein (BCCP), biotin carboxylase, α-carboxyltransferase, and β-carboxyltransferase. With the exception of BCCP, single-copy genes encode these subunits in Arabidopsis (Arabidopsis thaliana). Reverse-genetic approaches were used to individually investigate the physiological significance of the two paralogous BCCP-coding genes, CAC1A (At5g16390, codes for BCCP1) and CAC1B (At5g15530, codes for BCCP2). Transfer DNA insertional alleles that completely eliminate the accumulation of BCCP2 have no perceptible effect on plant growth, development, and fatty acid accumulation. In contrast, transfer DNA insertional null allele of the CAC1A gene is embryo lethal and deleteriously affects pollen development and germination. During seed development the effect of the cac1a null allele first becomes apparent at 3-d after flowering, when the synchronous development of the endosperm and embryo is disrupted. Characterization of CAC1A antisense plants showed that reducing BCCP1 accumulation to 35% of wild-type levels, decreases fatty acid accumulation and severely affects normal vegetative plant growth. Detailed expression analysis by a suite of approaches including in situ RNA hybridization, promoter:reporter transgene expression, and quantitative western blotting reveal that the expression of CAC1B is limited to a subset of the CAC1A-expressing tissues, and CAC1B expression levels are only about one-fifth of CAC1A expression levels. Therefore, a likely explanation for the observed unidirectional redundancy between these two paralogous genes is that whereas the BCCP1 protein can compensate for the lack of BCCP2, the absence of BCCP1 cannot be tolerated as BCCP2 levels are not sufficient to support heteromeric acetyl-coenzyme A carboxylase activity at a level that is required for normal growth and development.
Fatty acids have multiple functions in plant biology (Wallis and Browse, 2002). Most critical of these is probably their universal function as the building blocks of the membranes that physically divide all subcellular and cellular compartments (Bloom et al., 1991). These membranes not only serve as barriers, but they also serve as the matrix upon which important metabolic processes are organized (e.g. photosynthesis and oxidative respiration). Additional functions of fatty acids occur more discretely in more specialized compartments. Examples of these include their use in the assembly of triacylglycerol molecules, which act as carbon and energy reserves in seeds, their use in the assembly of the cuticle (Martin and Juniper, 1970; Post-Beittenmiller, 1996; Kunst and Samuels, 2003), which is the outermost barrier that terrestrial plants present to the abiotic world, and their use in the biosynthesis and assembly of a wide variety of signaling molecules (Shah, 2005), such as jasmonic acid (Liechti and Farmer, 2002; Wasternack and Hause, 2002), sphingolipids (Sperling and Heinz, 2003; Worrall et al., 2003), phosphatidylinositols (Mueller-Roeber and Pical, 2002), and phospholipase D-derived phospholipids (Wang, 2002).
Plants synthesize a wide variety of different fatty acids, which differ in their carbon chain lengths. Two biosynthetic machineries that synthesize these fatty acids are distributed among at least two subcellular compartments, the plastids and the cytosol. Although distinct enzyme systems catalyze these reactions of fatty acid biosynthesis, in both compartments, a similar series of iterative reactions extend the length of the acyl chain by two-carbon atoms per cycle using malonyl-CoA as the donor of the extending carbon atoms. In plastids, a type II fatty acid synthase, primed with acetyl-CoA, produces fatty acids of up to 18-carbon chain length. In these reactions the acyl intermediates are esterified to acyl-carrier protein (Ohlrogge et al., 1993; Ohlrogge and Browse, 1995). In the cytosol, 18-carbon acyl-CoAs are elongated by a partially characterized fatty acid elongase, to chain lengths of up to 36 carbons (Leonard et al., 2004).
The malonyl-CoA that is required for these two fatty acid biosynthetic processes is generated by acetyl-CoA carboxylase (ACCase; EC 6.4.1.2), which catalyzes the ATP-dependent carboxylation of acetyl-CoA. Two subcellular distinct forms of ACCase occur in plants, one in plastids and one in the cytosol (Nikolau et al., 2003). In most plants (with the exception of the Graminae), the plastidic ACCase has a heteromeric quaternary structure, and the cytosolic isoform has a homomeric structure (Konishi and Sasaki, 1994; Konishi et al., 1996). Plant homomeric ACCase (hmACCase) is homologous to the animal and yeast (Saccharomyces cerevisiae) enzymes, and in Arabidopsis (Arabidopsis thaliana), two genes code for this isozyme, ACC1 and ACC2 (Roesler et al., 1994; Yanai et al., 1995).
The heteromeric ACCase (htACCase) is composed of four subunits, and is analogous to the bacterial enzyme (Cronan and Waldrop, 2002; Nikolau et al., 2003). Three of these subunits are nuclear encoded, biotin carboxyl-carrier protein (BCCP), biotin carboxylase (BC), α-carboxyltransferase (α-CT), and the fourth subunit is plastome-encoded, β-carboxyltransferase (β-CT). In most plants, the htACCase subunits are encoded by a small family of genes (Shorrosh et al., 1995, 1996; Elborough et al., 1996; Reverdatto et al., 1999; Mekhedov et al., 2000); in Arabidopsis, this gene redundancy is minimized. Thus, single-copy genes encode BC (CAC2, At5g35360; Shorrosh et al., 1995; Bao et al., 1997; Sun et al., 1997), α-CT (CAC3, At2g38040; Shorrosh et al., 1996; Ke et al., 2000b), and β-CT (accD, AtCg00500; Sasaki et al., 1993; Ke et al., 2000b), but there are two BCCP-coding genes: CAC1A (At5g16390) codes for BCCP1, and CAC1B (At5g15530) codes for BCCP2 (Choi et al., 1995; Ke et al., 1997; Thelen et al., 2001). CAC1A and CAC1B genes are located within 0.32 Mb of each other on chromosome 5.
As may be expected for a multicomponent enzyme system, the htACCase subunit genes are expressed in a spatially and temporally coordinated manner. This has been demonstrated for the analogous enzyme in Escherichia coli (Cronan and Waldrop, 2002), and by detailed in situ hybridization studies of four mRNAs coding for the htACCase subunit genes, CAC1A, CAC2, CAC3, and accD (Ke et al., 2000b). Indeed, similar studies have shown that this pattern of gene expression extends to the metabolic network associated with acetyl-CoA generation and commitment of carbon for de novo fatty acid biosynthesis (Ke et al., 2000a). Specifically, these studies indicate that htACCase shows an expression pattern that is consistent with its role in de novo fatty acid biosynthesis, being expressed at high levels in cells and tissues that hyperaccumulate fatty acids. These tissues include cells of young expanding seedlings that are depositing membrane lipids to accommodate cellular growth and embryonic cells at the time when seed oil reserves are being deposited. These detailed expression studies, coupled with global RNA-profiling studies (Girke et al., 2000; Mentzen et al., 2008), indicate that the expression of the htACCase subunit genes may be part of a regulon that encompasses the fatty acid biosynthesis metabolic network.
The discovery that Arabidopsis expresses two paralogs of the BCCP subunit, which share relatively low sequence similarity (42% amino acid identity) and show distinct patterns of expression (Thelen et al., 2001), opens the possibility of novel mechanisms by which htACCase expression may be regulated, with consequential effects on fatty acid biosynthesis. Indeed, the ectopic alteration of the pattern of BCCP2 expression by antisense and sense RNA technology appears to affect htACCase expression; however, the effect on fatty acid accumulation is more complex (Thelen and Ohlrogge, 2002a; Chen et al., 2009). Specifically, reducing BCCP2 expression with an antisense transgene did not affect normal growth of the plant but slightly reduced the accumulation of oil in the seeds (Thelen and Ohlrogge, 2002a), and Chen et al. (2009) showed that ectopic expression of the BCCP2 subunit with the 35S promoter affected the growth of the vegetative organs of the plant.
In this study, we investigated the biochemical and physiological significance of the two paralogous BCCP-coding genes by identifying and characterizing transfer DNA (T-DNA) knockout alleles of each gene and reducing the accumulation of the BCCP1 protein with antisense RNA technology. These characterizations indicate that the htACCase is essential for plant seed development and that BCCP1 is an essential subunit for htACCase, but BCCP2 appears to be redundant and dispensable to BCCP1. This unidirectional redundancy between these two paralogous subunits appears to be due to the fact that BCCP1 subunit accumulation prevails over BCCP2.
RESULTS
Identification of T-DNA Knockout Mutants for BCCP Subunit Genes
To understand the physiological significance of the two BCCP subunit isoforms, we identified and characterized eight CAC1A and two CAC1B T-DNA-tagged mutant allele stocks (Supplemental Table S1). All but one of these alleles were identified by searching SALK Insertion Sequence Database (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al., 2003), and the one exception was isolated from the T-DNA insertion lines of Arabidopsis Knockout facility at University of Wisconsin-Madison (Sussman et al., 2000). To authenticate the identity of the mutant alleles and deduce their molecular structure and their inheritance, plant genomic DNA sequences flanking the T-DNA borders were amplified from individual progeny plants from each mutant stock, and these were sequenced. The molecular structure of the mutant alleles was deduced by aligning these flanking DNA sequences with the Arabidopsis genome sequence.
Of the eight cac1a alleles that were analyzed, only the cac1a-1 allele, isolated from the Wisconsin T-DNA collection, proved useful in these experiments. The insertion in this allele carries chimeric T-DNAs, in head-to-head arrangement, so that both ends of the insert have outward-facing T-DNA left-border (LB) sequences. The insertion is located in the first intron of CAC1A gene (Fig. 1A), and there is a 4-nucleotide deletion of genomic DNA at the insertion site. Two cac1b alleles were similarly characterized. In the case of the cac1b-1 allele (SALK_056228), the LB end of the T-DNA is located in the third intron of the gene (Fig. 1B). For the cac1b-2 allele (SALK_070569), the T-DNA is inserted at the first exon of the gene, where a 64-nucleotide deletion has occurred in association with the insertion (Fig. 1B). Details of the other mutant alleles that were investigated but proved not to be useful in these experiments are summarized in Supplemental Table S1.
Figure 1.
Molecular structure of the cac1a-1, cac1b-1, and cac1b-2 mutant alleles. Exons are represented by black boxes, introns are represented by white boxes, and untranslated regions are represented by gray boxes. Arrows indicate the position of the PCR primers used to characterize each allele. A, In the cac1a-1 allele, the insertion is located in the first intron of the gene and consists of chimeric T-DNAs, in a head-to-head arrangement, so that both ends of the insert have outward-facing LB sequences. B, T-DNA insertions in the cac1b-1 (SALK_056228) and cac1b-2 (SALK_070569) alleles are located in the third intron and the first exon, respectively.
The cac1a-1 Allele Is Recessive Lethal, But the Two cac1b Alleles Are Viable
As the first step in characterizing loss-of-function mutants, segregation analyses were conducted on progeny from individual heterozygous mutant plants. Seeds were sown on Murashige and Skoog media and the genotype of the progeny seedlings determined by PCR-based assays. For the cac1a-1 allele, 252 progeny from seven heterozygous plants of four sequentially selfed generations were analyzed, and only heterozygous or wild-type progeny were recovered; homozygous mutant plants were never recovered (Table I). Hence, cac1a-1 appears to be recessive lethal. In the siliques of cac1a-1 heterozygous plants approximately 20% of the fertilized seeds aborted during development, whereas only 2% of the seeds aborted in the siliques of the wild-type siblings grown in parallel to the mutant line (Table II; Fig. 2, A–D). Therefore, we deduce that the increased proportion of aborted seeds in the siliques from heterozygous mutant plants is due to homozygous mutant embryos (genotype: cac1a-1/cac1a-1).
Table I. Segregation analysis of T-DNA-tagged alleles.
Genotypes of progeny from heterozygous parents were determined by PCR-based assays.
| Allele | Genotype | No. of Plants | χ22:1 | χ21:2:1 | P Value |
| cac1a-1a | cac1a-1/cac1a-1 | 0 | |||
| cac1a-1/CAC1A | 143 | 11.16 | 0.0008 | ||
| CAC1A/CAC1A | 109 | ||||
| cac1b-1b (SALK_056228) | cac1b-1/cac1b-1 | 48 | |||
| cac1b-1/CAC1B | 95 | 0.01 | 0.995 | ||
| CAC1B/CAC1B | 47 | ||||
| cac1b-2c (SALK_070569) | cac1b-2/cac1b-2 | 56 | |||
| cac1b-2/CAC1B | 79 | 4.40 | 0.11 | ||
| CAC1B/CAC1B | 41 |
Data were collected from seven individual heterozygous parents.
Data were collected from three individual heterozygous parents.
Data were collected from three individual heterozygous parents.
Table II. Number of aborted seeds in siliques from heterozygous cac1a-1 and wild-type sibling plants.
For each genotype, 10 siliques from each of 10 individual sibling plants were examined. Therefore, in total, 100 siliques of each genotype were examined. χ2 test shows that the difference between the two proportions is statistically significant (P value < 0.001).
| Genotype | Aborted | Normal | Total |
| cac1a-1/CAC1A | 551 (18.5%) | 2,429 (81.5%) | 2,980 |
| CAC1A/CAC1A | 70 (2.1%) | 3,245 (97.9%) | 3,315 |
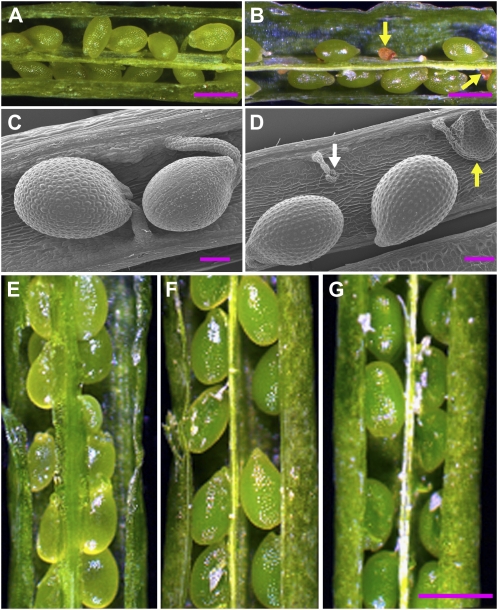
Figure 2.
Mutations in the CAC1A gene, but not in the CAC1B gene, show a seed abortion phenotype. Stereomicrographs (A, B, E–G) and scanning electron micrographs (C and D) of siliques at 9 DAF from sibling wild-type and mutant plants that were grown side by side. Bars = 350 μm (A, B, E–G); 100 μm (C and D). Seeds developing in siliques of wild-type (A and C) or sibling plants that are heterozygous for the cac1a-1 allele (B and D). White arrows identify aborted seeds, and yellow arrows identify nonfertilized ovules. Regardless of the allele carried at the CAC1B locus (homozygous wild-type [E], cac1b-1 [F], or cac1b-2 [G]), seeds develop normally.
In contrast, homozygous mutant plants were recovered for both cac1b-1 and cac1b-2 alleles, and both alleles were inherited in a normal, 1:2:1 Mendelian ratio (Table I). The cac1b-1 and cac1b-2 mutants were grown in soil, side by side with wild-type sibling plants at 22°C with 24-h continuous illumination. In these growth conditions, the two cac1b mutants are indistinguishable in their growth habits from wild-type plants. Siliques from each cac1b mutant as well as wild-type plants were dissected and examined by stereomicroscopy. The seeds in siliques from either cac1b-1 or cac1b-2 mutant are indistinguishable from that of wild-type plants (Fig. 2, E–G). Western-blot analysis of protein extracts from a mixture of flowers and flower buds using BCCP2-specific antibody (see Supplemental Fig. S3) shows that while the BCCP2 protein is readily detectable in the wild-type control, it is undetectable in either the cac1b-1 or cac1b-2 homozygous mutants (Fig. 3). Therefore, these two cac1b alleles are apparently null alleles, but plants that lack the BCCP2 protein are completely viable. It should be noted that although in the stated condition we were unable to detect any morphological difference between cac1b mutants and their wild-type siblings, this does not imply the CAC1B gene has no function. This functionality will become apparent when cac1b mutant plants are placed in conditions where this function becomes limiting.
Figure 3.
Effect of cac1b mutations on the accumulation of the htACCase subunits. Protein extracts from a mixture of flowers and flower buds were prepared from wild-type (WT), cac1b-1 homozygous, or cac1b-2 homozygous plants. Each lane was loaded with extracts containing 200 μg of total protein. Specific antisera were used to detect five htACCase subunits. In addition, streptavidin was used to detect biotinylated BCCP1 and biotinylated BCCP2 subunits. The mitochondrial MCCase subunit A was detected by its specific antibody and served as an internal control.
Genetic Complementation of the cac1a-1 Mutant
Genetic complementation was used to test whether the embryo-lethal phenotype associated with the cac1a-1 allele is due to loss of CAC1A gene function. To conduct this experiment, a 4.5-kb Arabidopsis genomic fragment containing the CAC1A gene was transformed into heterozygous cac1a-1 plants using the plant transformation vector, pCAMBIA3300 (www.cambia.org). Because this transformation vector carries the bar gene that confers glufosinate resistance (Supplemental Fig. S1C), and the T-DNA of the cac1a-1 allele carries an NPTII gene that confers kanamycin resistance (Supplemental Fig. S1B), it was readily possible to simultaneously select for transgenic plants that carried both the ectopic wild-type CAC1A allele and the mutant cac1a-1 allele by resistance to glufosinate and kanamycin, respectively. To molecularly confirm the genotype of these selected transgenic plants, gene-specific primers flanking the T-DNA insertion site of cac1a-1 allele were designed with one primer residing in the genome region beyond the 4.5-kb fragment used for genetic complementation (Supplemental Fig. S1A), so that the endogenous genomic CAC1A allele can be distinguished from the ectopic copy carried by the transformed DNA fragment (Supplemental Fig. S1C). Of 48 T2 generation plants, which were propagated from three independent T1 transgenic lines that were both glufosinate and kanamycin resistant, we were able to recover 14 T2 plants homozygous for cac1a-1 allele, which also carried the ectopic CAC1A transgene allele. The fact that these cac1a-1 homozygous plants successfully developed through embryogenesis established that the ectopic transgenic CAC1A allele genetically complemented the embryo-lethal phenotype associated with the cac1a-1 allele. Therefore, this genetic complementation demonstrates that the cac1a-1 allele is a loss-of-function allele and is responsible for the embryo-lethal phenotype.
Effect of the cac1a-1 Allele on Embryo and Endosperm Development
To establish the ontology of the cac1a-associated embryo-lethal phenotype, developing ovules on cac1a-1 heterozygous plants and wild-type siblings were examined by confocal and scanning electron microscopy (SEM) at different stages after flowering (Fig. 4). Siliques from the middle of the bolt were sampled at flower opening and at 1, 3, 5, and 7 d after flowering (DAF). At each developmental stage, 10 siliques from each genotype were cleared and examined by confocal microscopy (i.e. over 150 ovules of each genotype were examined at each developmental stage). In parallel, at each developmental stage, 75 embryos harvested from 10 siliques of either wild-type or heterozygous cac1a-1 siblings were examined by SEM.
Figure 4.
Effect of the cac1a-1 mutation on embryo and endosperm development. Developing ovules and embryos were examined by CSLM (A–C, J–O, S–U) and SEM (D–I, P–R, V–X), at 3 DAF (A–L), 5 DAF (M–R), and 7 DAF (S–X). em, Embryo; en, endosperm; s, suspensor; oi, outer integument; ii, inner integument; ph, phragmoplasts; cw, cell wall; den, degenerated endosperm. As labeled, the development of the ovules and embryos were compared on sibling plants that were grown side by side, but carried either wild-type alleles at the CAC1A locus (WT) or were heterozygous for the cac1a-1 allele (cac1a). As described in the “Materials and Methods,” for CSLM, flowers and siliques were fixed, cleared, and the enclosed ovules examined in the intact organs. For SEM, collected flowers and siliques were fixed with glutaraldehyde/paraformaldehyde, post fixed in OsO4, and following dehydration and drying, the ovules were fractured and examined. A to L, At 3 DAF, aberrations in ovule development are apparent in plants carrying the cac1a-1 allele. In wild-type plants, the embryos (em) are at the 32-cell stage, whereas in cac1a-1 plants 16-cell (B) and four-cell (C) embryos are present. Formation of cellular connections between sister and nonsister nuclei has begun to take place in the endosperm (en) of wild-type plants (D, G, J), but this process is delayed and less regular in aberrant ovules of cac1a-1 plants (E, F, H, I, K, L). Bars = 50 μm (A–C, J–L); or 10 μm (D–I). M to R, At 5 DAF, aberrations in ovule development become more apparent in plants carrying the cac1a-1 allele. Wild-type embryos are at the heart stage of development (M) and the endosperm has begun to cellularize (P). In cac1a-1 plants, the aberrantly developing ovules have globular embryos with highly vacuolated cells (N and O). In the most severely aberrant ovules, the embryo sac has collapsed (O). Endosperm is uncellularized or aberrantly cellularized and sometimes degenerated (den; Q and R). Bars = 50 μm (M–O); 10 μm (P–R). S to X, At 7 DAF, the aberrant ovules in the cac1a-1 plants have ceased development at the globular or heart stages (T and U), the endosperm is degenerated, and the embryo sacs are often collapsed (W and X). In contrast, in the wild-type siblings the embryos are at the torpedo stage (S) and the endosperm is fully cellularized (V). Bars = 50 μm (S–U); 10 μm (V–X).
During the initial stages of seed development, the ovules and the enclosed embryos of both genotypes are indistinguishable (data not shown). Namely, ovules had a normal-appearing eight-nucleate megagametophyte, composed of an egg cell, two synergids, two polar nuclei, and three chalazal antipodals. Fertilization-associated events, including movement of the sperm nucleus within the egg cell, synergid degeneration, and fusion of the polar nuclei that are positioned near the egg cell at the time of double fertilization all proceeded normally. In post-fertilization ovules, the zygotes and embryos were indistinguishable between the two genotypes through the 16-cell globular stage of development.
However, starting from 3 DAF, about 20% of the fertilized ovules on the cac1a-1 heterozygous parent develop aberrantly and ultimately fail to produce viable seeds. We infer that these nonviable ovules are homozygous for the cac1a-1 mutant allele. The specific differences that develop between these homozygous mutant seeds and the wild-type siblings are illustrated in the images shown in Figure 4.
Starting at 3 DAF, the homozygous cac1a-1 embryos (i.e. about one-fifth of the embryos on heterozygous cac1a-1 plants) begin to show distinct differences from those on wild-type sibling plants. Wild-type embryos are at the 32-cell globular stage (Fig. 4, A and D), whereas mutant embryos are lagging behind and are at the four- or 16-cell stage (Fig. 4, B and C). Despite the slower rate of cell division, the cells of the presumed mutant embryos are indistinguishable from those of the wild type, by confocal (Fig. 4, A–C) or light microscopy (LM; data not shown). In contrast, cells of the endosperm are different. In wild-type ovules, endosperm nuclei are largely coenocytic, but are undergoing nuclei divisions, and forming the cellular connections between sister and nonsister nuclei that ultimately leads to cellularization (Fig. 4, D, G, and J); however, endosperm nuclei of presumed mutant ovules are lagging in nuclear division (Fig. 4, E, F, H, I, K, and L), and there are fewer and less-regular cellular connections between sister and nonsister nuclei (Fig. 4, H and I, as compared with G).
By 5 DAF, wild-type embryos show the polarization associated with formation of the root and shoot apex (i.e. they are heart staged; Fig. 4, M and P). In contrast, the cac1a-1 aberrant embryos are still at the globular stage, and the cells of these embryos have become highly vacuolated (Fig. 4, N, O, Q, and R); in some cases, the embryo sac that contains these aberrant embryos has collapsed (Fig. 4O). The majority of the endosperm of 5-DAF wild-type plants has cellularized (Fig. 4P). In contrast, the endosperm of the aberrant ovules remains predominantly uncellularized (Fig. 4, Q and R). Abnormal nonradial microtubular-like structures may be present, but few phragmoplasts have formed, and cell wall is rarely deposited (Fig. 4Q, left side shows a few cellularized endosperm cells). Cytokinesis is defective as judged by the paucity of endosperm nuclei (Fig. 4N), and of the nuclei that are present, many appear aberrant, degenerated, or multinucleate (Fig. 4, Q and R).
By 7 DAF, whereas normal embryos are at the torpedo stage (Fig. 4S), with cellularized endosperm (Fig. 4, S and V), the aberrant embryos that occur on cac1a-1 siblings lag behind in development, being at the heart or globular stage (Fig. 4, T and U). Moreover, in most ovules, the embryo sac has collapsed, and the embryo cells appear devoid of nuclei (Fig. 4, T and U). These dramatic differences in embryo morphology are associated with abnormal endosperm development (Fig. 4, W and X). The endosperm has not cellularized and appears very abnormal. Unlike the endosperm at 5 DAF, almost no normal-appearing endosperm nuclei or cells are present. Irregular cytoplasmic structures reminiscent of cell wall deposition may exist but appear to be degenerating (Fig. 4W) or have degenerated (Fig. 4X).
cac1a-1 Allele Reduces Male Gamete Transmission
The inheritance of the cac1a-1 allele indicates that the CAC1A function may be required for the normal development of gametophytes. For a recessive embryo-lethal mutant, only heterozygous or wild-type progeny should be recovered from heterozygous plants, at a ratio of 2:1. In the case of the cac1a-1 allele, although only heterozygous (CAC1A/cac1a-1) or wild-type (CAC1A/CAC1A) progeny were recovered, they were recovered at a ratio of 143:109 (Table I). Because the observed deviation from the expected 2:1 ratio is statistically significant (χ2 test, P value < 0.001), these data indicate that the cac1a-1 allele is poorly transmitted through gametogenesis.
To determine whether transmission of the cac1a-1 allele is impaired through the male or female gametes, we crossed heterozygous mutant plants as either males or females (genotype CAC1A/cac1a-1) with wild-type plants (genotype CAC1A/CAC1A), and the genotypes of the progeny from these crosses were scored by PCR analysis (Table III). In the crosses where the cac1a-1 allele was transmitted through the female gamete, it was recovered at a rate that is indistinguishable from that of the CAC1A allele (i.e. heterozygous and wild-type progeny were recovered at a ratio that is statistically indistinguishable from 1:1). However, in the crosses where the cac1a-1 allele was transmitted through the male gamete, transmission of the mutant allele is significantly reduced in comparison with the wild-type allele, resulting in progeny that were 30% heterozygous and 70% wild type. These data establish that transmission of the cac1a-1 allele is reduced through male gametogenesis.
Table III.
Transmission of cac1a-1 gametes
| Crossa | No. of F1 Progeny |
χ21:1 | P Value | ||
| Total | CAC1A/cac1a-1 | CAC1A/CAC1A | |||
| CAC1A/CAC1A$ × CAC1A/cac1a-1# | 86 | 21 | 65 | 22.5 | <0.001 |
| CAC1A/cac1a-1$ × CAC1A/CAC1A# | 64 | 30 | 34 | 0.25 | 0.62 |
Fifteen plants for each genotype were crossed.
Effect of the cac1a-1 Allele on Morphology and Germination of Pollen Grains and on Pollen Tube Growth
The morphology of pollen grains was examined to further characterize the reduced transmission of the cac1a-1 allele through male gametes. Pollen grains were collected from sibling wild-type (CAC1A/CAC1A) and heterozygous mutant plants (CAC1A/cac1a-1) and examined by SEM (Fig. 5). Approximately 20% to 30% of the pollen grains collected from heterozygous mutant plants have an aberrant shape (Fig. 5B, arrows). On these abnormally shaped pollen grains, the exine is abnormally smooth around the aperture area (the aperture is the spot on the pollen grain where the pollen tube will expand from upon germination of the pollen grain). Such aberrant pollen grains are not recovered from the sibling wild-type plants (Fig. 5A).
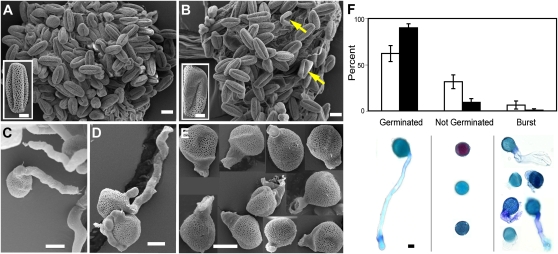
Figure 5.
The CAC1A function is required for normal pollen development and germination. A and B, Scanning electron micrographs of pollen grains collected from sibling plants that are either wild type (A) or carry the cac1a-1 allele (B). Arrows in B point to aberrantly shaped pollen grains that have an altered exine layer; these are recovered from plants carrying the cac1a-1 allele, but not from wild-type siblings. Insets show magnified views of individual pollen grains from wild-type plants and the aberrant pollen grains that were recovered from mutant plants. C to E, Scanning electron micrographs of pollen grains after 20 h on germination medium. Pollen collected from wild-type plants germinate and elongate a pollen tube (C), but those that are collected from plants that carry the cac1a-1 allele have an elevated number of grains that do not germinate or germinate aberrantly (D) or upon germination the elongating pollen tube appears to burst (E). F, The proportion of pollen grains that are classified as germinated, not germinated, or burst after 20 h on germination medium; dark bars are for pollen grains collected from wild-type plants and the white bars are for pollen grains collected from siblings that carry the cac1a-1 allele. Under each category are micrographs of Nile Blue stained pollen grains representative of each category. Bars = 10 μm. [See online article for color version of this figure.]
To investigate whether the cac1a-1 allele affects the functionality of the pollen, pollen grains were collected from heterozygous (CAC1A/cac1a-1) and wild-type (CAC1A/CAC1A) sibling plants and germinated in vitro. Twenty-hours later, the germinated pollen were examined and classified as germinated normally, not germinated, or burst (Fig. 5F). While 90% of the pollen from wild-type plants germinate normally, only 60% of the pollen from cac1a-1 heterozygous sibling plants germinate normally. Concomitantly, there is an increased percentage of pollen grains from these heterozygous plants that either fail to germinate (about 30%), or if germinated, the elongating pollen tube bursts during elongation (about 6%). In contrast, the percentage of nongerminated pollen grains or burst pollen tubes is significantly lower (about 9% and 0.9%, respectively) for pollen collected from wild-type sibling plants.
We expanded the characterization of the cac1a-1-associated pollen phenotype by observing and contrasting the in planta germination of pollen grains and expanding pollen tubes on pistils of heterozygous (CAC1A/cac1a-1) and wild-type (CAC1A/CAC1A) sibling plants (Fig. 6). At pollination, pistils were stained with Aniline Blue and 4′,6-diamidino-2-phenylindole (DAPI) and examined by fluorescence microscopy. In comparison with the wild-type siblings (Fig. 6, A, C, and E), fewer pollen grains germinate on heterozygous plants (Fig. 6, B, D, and F), and of those that do germinate, the migration of the pollen tubes through the style is slower.
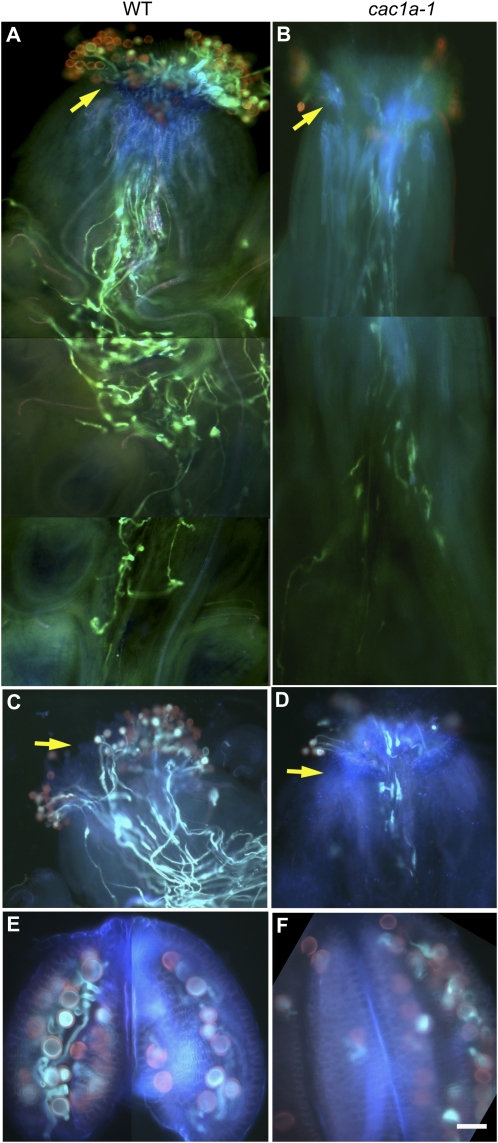
Figure 6.
The CAC1A function is required for normal in planta pollen germination and pollen tube elongation. Larger number of pollen grains germinate on the stigma (arrow), and pollen tubes more fully elongated in the style of flowers of wild-type (WT) plants (A and C) than on plants carrying the cac1a-1 allele (B and D). In addition, during processing for microscopic examination, more pollen grains have germinated in the anthers of wild-type plants (E) than on plants carrying the cac1a-1 allele (F). Pollen was visualized on sibling plants grown side by side that were either wild type (A, C, E) or heterozygous for the cac1a-1 allele (B, D, E). Flowers were stained either with Aniline Blue (A and B) or Aniline Blue and DAPI (C–F) and viewed by fluorescence microscopy with DAPI/fluorescein isothiocyanate/Texas Red filters. Bar = 10 μm.
Reduction of BCCP1 Accumulation by CAC1A Antisense RNA Expression Induces Unique Morphological Changes
Because the cac1a-1 mutant allele shows an embryo-lethal phenotype, we were unable to examine the role of the BCCP1 protein in plant development after embryogenesis. Therefore, transgenic Arabidopsis plants expressing CAC1A antisense RNA, under the transcriptional control of the cauliflower mosaic virus (CaMV) 35S promoter were generated and characterized. As indicated by inheritance of the tightly linked kanamycin-resistance trait, by PCR amplification of the 35S:CAC1A antisense RNA transgene, and Southern-blot hybridization analyses of genomic DNA isolated from individual plants, we were able to confirm that we had recovered 42 independently generated lines that carried the 35S:CAC1A antisense RNA transgene (data not shown).
About half of these transgenic lines show unique morphological alterations as compared with wild-type controls. The altered phenotype includes reduced overall size of the transgenic plants, and altered leaf morphology and pigmentation. These lines can be grouped into five classes (very severe, severe [S], moderate [M], mild, or wild type like) depending upon the degree of morphological alteration and the timing of the appearance of the altered growth phenotype. Figure 7A shows seedlings that are expressing either the M or S phenotype, which were further analyzed in this study.
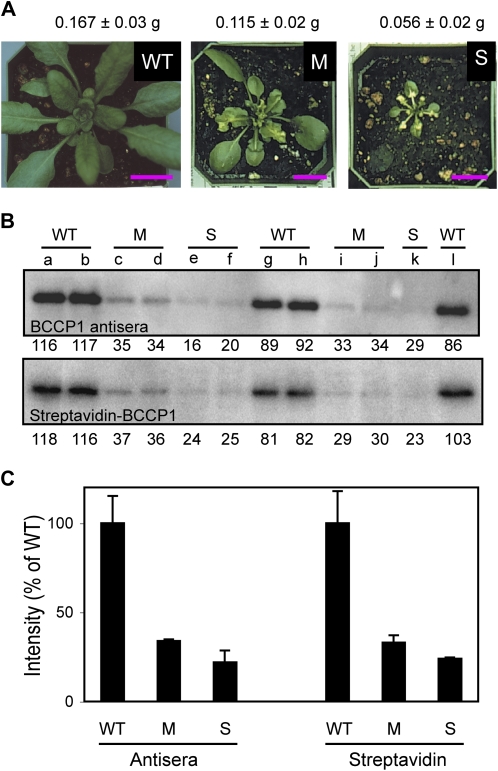
Figure 7.
Reduced BCCP1 abundance in CAC1A antisense plants correlates with the reduced growth phenotype. A, Phenotype of representative plants used to investigate the relationship between BCCP1 abundance and the severity of the resulting reduced growth phenotype. WT, Wild type. Bars = 1 cm. B, Abundance of BCCP1 protein in five independent wild-type plants (WT; lanes a, b, g, h, l), four independent CAC1A antisense plants expressing the M phenotype (lanes c, d, i, j), and three independent CAC1A antisense plants expressing the S phenotype (lanes e, f, k). Protein extracts were prepared from young rosette leaves of 30-d-old plants, and 50 μg of protein from each plant extract was subjected to western-blot analysis, using either BCCP1 antisera followed by 125I-labeled Protein A or 125I-labeled streptavidin. Numbers below each blot are signal intensities relative to the average intensity of the WT (100). C, Comparison of BCCP1 abundance in wild-type and CAC1A antisense plants analyzed with either subunit-specific antisera or streptavidin as in B. BCCP1 signal intensities from wild type (WT; n = 5), and CAC1A antisense plants expressing a M (n = 4) and S (n = 3) phenotype were averaged. Three repetitions of this experiment gave similar results. [See online article for color version of this figure.]
Plants with an S phenotype germinate normally and have normal-appearing cotyledons, but beginning at 1 to 2 weeks after germination, the growth rate of these seedlings is noticeably decreased. These plants may have up to three wild-type-like true leaves, but subsequent emerging rosette and cauline leaves are significantly smaller, extremely undulated, and mostly yellow variegated with small green areas. By 30 d after imbibition (DAI), these plants are approximately one-fifth the size of wild-type plants.
In plants with an M phenotype, the switch from wild-type-like leaves to aberrant leaves occurs sometime between the emergence of the fourth and seventh leaf. Those aberrant leaves are slightly crinkly and have distinct yellow patches. By 30 DAI, these plants are approximately two-thirds the size of wild-type plants.
To determine whether CAC1A expression is reduced in the CAC1A antisense plants, the accumulation of the BCCP1 protein was assessed by western-blot analyses (Fig. 7, B and C). In these analyses, the immunoreactive BCCP1 polypeptide was detected with specific antisera (Choi et al., 1995), and the biotin prosthetic group associated with this protein was detected with streptavidin (Nikolau et al., 1985).
The accumulation of the BCCP1 protein in shoots of 30-d-old antisense plants with an M phenotype is significantly lower than that in wild-type plants (about 35% of wild-type levels; P value < 0.0001, for both antisera and streptavidin detection); in plants with an S phenotype BCCP1 level is even lower (about 20% of wild-type levels; P value = 0.001 and 0.0002 for antisera and streptavidin detection, respectively). In transgenic plants with a mild or wild-type-like phenotype, BCCP1 polypeptide and holoprotein levels are indistinguishable from wild-type plants (data not shown). The ratios of the band detected by BCCP1 antisera to that detected by streptavidin are near identical regardless of the genotypes tested (compare with Fig. 7, B and C). These comparisons indicate that the biotinylation status of BCCP1 is similar in wild-type and CAC1A antisense plants. Moreover, these analyses indicate that there is a direct relationship between the reduction in BCCP1 accumulation and the severity of the CAC1A antisense phenotype.
Reduction of BCCP1, But Not of BCCP2 Levels, Affects Fatty Acid Accumulation
To investigate the individual effect of BCCP1 and BCCP2 on fatty acid accumulation, lipids extracted from wild-type and CAC1A antisense plants or cac1b mutant plants were compared. In leaves of CAC1A antisense plants the concentration of fatty acids (on the basis of fresh weight) is increased by about 20% and 30% in plants showing an M and an S phenotype, respectively (Fig. 8B). However, because these antisense RNA plants are considerably smaller (rosettes of plants with the S phenotype weigh between 10% and 20% of wild-type rosettes), the total fatty acid content of each individual plant is considerably less than that of a wild-type plant (Fig. 8C). Because CAC1B is primarily expressed in developing siliques, and not in leaves (Thelen et al., 2001), fatty acids were analyzed in the seeds of the cac1b knockout mutants. The amount of seed fatty acids in either cac1b-1 or cac1b-2 homozygous mutant plants are indistinguishable from those of wild-type plants (Fig. 8A).
Figure 8.
Reduction in the abundance of BCCP1, but not BCCP2, affects fatty acid accumulation without affecting fatty acid concentration or composition. Each value in the chart represents the averaged value of (n) biological replications. Error bars indicate the sds. A, Total fatty acid concentration in seeds harvested from wild type (WT; n = 6), cac1b-1 homozygous (cac1b-1; n = 6), and cac1b-2 homozygous (cac1b-2; n = 6) mutant plants. B, Total fatty acid concentration in leaves of wild-type (WT; n = 3) plants, CAC1A antisense plants with M (n = 7) phenotype, and CAC1A antisense plants with S (n = 3) phenotype. C, Total fatty acid content (per plant) in leaves from wild-type plants (WT; n = 5), CAC1A antisense plants with M phenotype (n = 4), and CAC1A antisense plants with S phenotype (n = 3). D, Fatty acid composition of seeds harvested from homozygous plants that carried wild-type CAC1B (white bars; n = 6), and mutant cac1b-1 (gray bars; n = 6) or cac1b-2 (black bars; n = 6) alleles. E, Fatty acid composition of leaves from wild-type plants (white bars; n = 3), and of leaves from CAC1A antisense plants with M phenotype (gray bars; n = 7), or S phenotype (black bars; n = 3).
The fatty acid composition of the leaf lipids of CAC1A antisense plants with an M or an S phenotype was compared with that of wild-type leaves (Fig. 8D). In addition, the fatty acid composition of lipids from mature seeds of cac1b-1 and cac1b-2 alleles was compared with the wild type (Fig. 8E). There is no detectable difference in the relative amounts of each fatty acid irrespective of the CAC1A or CAC1B allele in any of the organs that we examined. Hence, reduction in the expression of the BCCP1 protein, but not of the BCCP2 protein, quantitatively affects fatty acid accumulation; but reduced expression of either paralogous protein does not affect fatty acid composition.
The Expression of the Other htACCase Subunits Is Unaffected by the Absence or Reduction of Either BCCP1 or BCCP2
To investigate the effect of losing either CAC1A or CAC1B function on the expression of the other htACCase subunit genes, the accumulation of each subunit was determined by western-blot analysis with subunit-specific antibodies. The generation of antibodies specific to BC, α-CT, or β-CT has been described previously (Sun et al., 1997; Ke et al., 2000b). To generate antibodies specific for each BCCP subunit, the low similarity region of each subunit was identified from the alignment of the amino acid sequences of BCCP1 and BCCP2 (Supplemental Fig. S3A). The cDNA sequence coding this region of each BCCP subunit was PCR amplified and cloned into an E. coli expression vector (pET-30; Novagen), and recombinant proteins produced from these two constructs were used to immunize rabbits to generate polyclonal antibodies. Protein extracts from Arabidopsis flowers were subjected to western-blot analyses with these two antibodies as well as with streptavidin. While streptavidin reveals both BCCP1 and BCCP2 as two individual protein bands of about 35 and 25 kD, BCCP1 antibody detects only the 35-kD band, and BCCP2 antibody detects only the 25-kD band (Supplemental Fig. S3, B and C). These experiments demonstrate that monoreactive antibodies for the BCCP1 and BCCP2 proteins were generated.
Western-blot analyses with htACCase subunit-specific antibodies on protein extracts prepared from a mixture of flowers and flower buds of wild-type plants and the two homozygous CAC1B knockout mutants show that the elimination of BCCP2 subunit has no effect on the accumulation of the other four subunits (Fig. 3). Because tissue and organs were not recoverable from plants that lacked CAC1A functionality (i.e. cac1a homozygous plants), we assessed the consequence of BCCP1 reduction on the accumulation of the other htACCase subunits in the CAC1A antisense plants. Western-blot analyses of leaf extracts from plants that showed an M or S growth phenotype demonstrated that apart from the expected reduction in BCCP1 accumulation, none of the other htACCase subunits showed any significant change in accumulation relative to the wild-type plants (Fig. 9A). Because prior studies had shown that BCCP2 accumulates at a low level (Thelen et al., 2001) in leaves, and in the study reported herein it was undetectable with our BCCP2-specific antisera and barely detectable with streptavidin upon very long exposures (Fig. 9A), we extended the western analysis to developing siliques where BCCP2 is readily detectable in wild-type plants. As with the observations made in leaf extracts, the abundance of BCCP2, BC, α-CT, and β-CT was indistinguishable between wild-type and CAC1A antisense plants (Fig. 9, B and C). Taken together, these results demonstrate that a decrease in the accumulation of either BCCP1 or BCCP2 does not affect the accumulation of the other htACCase subunits.
Figure 9.
Reduced accumulation of BCCP1 subunit in CAC1A antisense RNA plants does not affect the accumulation of the other subunits of htACCase. A, Protein extracts were prepared from rosette leaves of three wild-type plants (WT) and five CAC1A antisense RNA plants expressing the M or S phenotypes. Aliquots of extracts containing 50 μg of protein were fractionated by SDS-PAGE and subjected to western-blot analyses. The blots were incubated with streptavidin or antisera against each individual subunit of htACCase. B, Protein extracts prepared from excised siliques isolated at between 1 and 3 DAF from wild-type (WT) and CAC1A antisense RNA plants with a S phenotype were subjected to western-blot analysis using either streptavidin or an anti-BCCP antibody that reacts with both BCCP1 and BCCP2 (Choi et al., 1995). C, The same protein samples used in B were subjected to western-blot analysis using antisera specific for BC, α-CT, or β-CT subunit.
CAC1A and CAC1B mRNAs Have Similar Accumulation Patterns during Embryo Development
Because BCCP1 and BCCP2 proteins have potentially redundant functions, to decipher the differential metabolic and morphological effects arising from the loss of each protein, we compared the expression of the CAC1A and CAC1B genes via a combination of different approaches (promoter:GUS transgenes, in situ hybridization of mRNAs, and quantitative western-blot analyses). These characterizations focused on the expression of these genes during the early stages of silique and seed development, when the unidirectional redundancy between the two genes was most readily observed. In addition, we also compared the expression of these two genes in other organs to ensure that our data are consistent with prior such characterizations (Ke et al., 1997, 2000b; Thelen et al., 2001; Ruuska et al., 2002; Thelen and Ohlrogge, 2002b).
The cellular distribution of the CAC1A and CAC1B mRNAs was compared within embryos and siliques, particularly at the early stages of development when the differential effects of the cac1a-1 and cac1b-1 mutations were initially detected. The relative accumulation of these mRNAs was determined by in situ hybridization at five distinct developmental stages starting from 1 DAF (Fig. 10A). In rapidly enlarging siliques (1 DAF), the sporophyte within the ovule is a zygote or several-celled embryo, and CAC1A mRNA is evenly distributed throughout the silique tissues, including the silique walls and developing ovules. By 3 DAF, siliques have ceased enlargement. CAC1A mRNA is concentrated in the rapidly growing globular embryos and is considerably reduced in silique walls and ovule integuments. At 5 to 7 DAF, the time of maximal seed oil accumulation, CAC1A mRNA is highly concentrated in the embryos. By 12 DAF, embryos have ceased growing and are approaching desiccation; at this stage, CAC1A mRNA is no longer detected in the embryos or siliques.
Figure 10.
The expression pattern of CAC1A and CAC1B genes as determined by in situ hybridization and histological staining of GUS activity expressed from promoter:GUS transgenes. A, The spatial and temporal patterns of CAC1A and CAC1B mRNAs are near identical during embryo development. The CAC1A and CAC1B mRNAs were detected by in situ hybridization procedure described in the “Materials and Methods,” using gene-specific DIG-labeled antisense RNAs as probes; control hybridizations were conducted with DIG-labeled sense RNAs. Bar = 50 μm. B, Histological staining of GUS activity expressed in different Arabidopsis organs by CAC1A- and CAC1B-GUS transgenes. These data reveal that CAC1B is only expressed in a subset of the organs and tissues, whereas CAC1A is expressed at a higher level. Organs that are shown are as follows, and these are taken from plants that are developmentally staged as defined by Boyes et al. (2001). B1: rosette of plants between principal growth stages 1.08 and 1.12 ; B2: roots of same plants as shown in B1; B3: florets of plants at principal growth stage 6.30, green horizontal arrows point to flowers at 1 DAF and red vertical arrows point to flowers at 2 DAF; B4: flowers at 2 DAF from plants at principal growth stage 6.30; B5: silique at 5 DAF; B6: silique at 8 and 9 DAF, for CAC1A:GUS and CAC1B:GUS, respectively; B7: silique at 12 DAF. Red bars = 1 mm; blue bars = 0.1 mm.
Like CAC1A, CAC1B mRNA is distributed evenly throughout the silique tissues at 1 DAF. By 3 DAF, CAC1B mRNA is concentrated in the embryos and is reduced in silique walls and integuments of the ovules. At 5 and 7 DAF, CAC1B mRNA level decreases from that of 3-DAF siliques. CAC1B mRNA is undetectable in the silique walls or ovule integuments from 5 DAF and onwards. In 12-DAF siliques, CAC1B mRNA is no longer detected in the embryos. These data establish that at the early stages of silique and seed development (at 1–5 DAF), these two transcripts have a similar spatial pattern of expression, the one difference being that CAC1B expression is at lower levels (as judged by the fact that the digoxigenin (DIG)-labeling reactions to visualize the CAC1B mRNA was conducted for 2- to 3-fold longer than those for the CAC1A mRNA), and the level of CAC1B mRNA reduced somewhat earlier in silique development than CAC1A accumulation.
Additional insights into the expression patterns and expression levels of the CAC1A and CAC1B genes were explored with transgenic plants that carried a GUS reporter gene under the control of the promoter of each BCCP-encoding gene. These analyses revealed that while CAC1A is expressed in all the examined organs including the cotyledons, leaves, and roots of seedlings, flowers, and siliques, CAC1B is only expressed in a subset of the tissues where CAC1A is expressed, and at lower levels (Fig. 10B). Consistent with prior studies and with the western-blot analyses of BCCP2 protein, we find that the CAC1B:GUS expression is considerably limited in the aerial organs of young seedlings, but is expressed in roots, although at a lower level than CAC1A:GUS. Another significant difference in the expression patterns between the two genes occurs in young developing siliques, at 1 to 3 DAF. Specifically, whereas CAC1A-driven GUS activity is strongly detected in these young siliques, CAC1B-driven GUS activity is barely detectable and is focused at the senescing stigma. Interestingly, and in parallel with the in situ mRNA hybridization studies, CAC1B:GUS activity rises later in silique development but decays earlier than CAC1A:GUS activity as seeds mature and the siliques begin to senesce.
Accumulation of BCCP1 Is 5-Fold Higher Than That of BCCP2 in Developing Siliques
BCCP1 and BCCP2 expression was also evaluated at the protein level using a quantitative western-blot procedure. The absolute concentration of each BCCP subunit was determined by using recombinantly produced S-tag fusion proteins as standards. The absolute concentration of these standards was initially determined by measuring the RNase activity reconstituted by the binding of either BCCP1 S·tag or BCCP2 S·tag with S protein (Richards and Wyckoff, 1971). Subsequently, parallel western-blot analyses were performed with antiserum for each subunit with plant extracts and a serial dilution of each recombinant standard protein (Fig. 11A). The standard curve constructed from each recombinant protein provides a means of calculating the molar concentration of BCCP1 and BCCP2 in identical protein extracts (Fig. 11B). Using this method, we found that in developing siliques the BCCP1 subunit accumulates to levels that are about 5-fold higher than that of the BCCP2 subunit (Fig. 11C). Consistent with both the need for de novo fatty acid biosynthesis and the pattern of CAC1A mRNA accumulation, maximal levels of BCCP1 protein occur in siliques of between 6 and 12 DAF.
Figure 11.
BCCP1 is the major paralog that accumulates during silique development. Using recombinantly produced authentic proteins as standards and BCCP1- or BCCP-2-specific antisera the absolute amount of each paralog was determined in protein extracts prepared from siliques at the indicated stage of development. A, The indicated amounts of recombinant BCCP1 or BCCP2 proteins, and aliquots of silique extracts (μg of protein loading indicated under each lane) were subjected to electrophoresis on the same gel. Gels were then subjected to western-blot analysis with subunit-specific antisera. B, The standard curve for BCCP-1 protein. C, The standard curve for BCCP-2 protein. D, BCCP1 and BCCP2 concentrations in developing siliques. Average of three determinations of three individual harvests of siliques at each indicated developmental stage; error bars indicate sds.
DISCUSSION
A common biochemical feature of fatty acid biosynthesis is the carboxylation of acetyl-CoA to form malonyl-CoA. This is a chemical requirement of the Claisen condensation-based mechanism that is common to the biosynthesis of all polyketides, including fatty acid biosynthesis. Specifically this carboxylation reaction chemically activates the methylene group of the resulting malonyl moiety, facilitating the subsequent chemical condensation that drives alkyl-chain elongation in these processes. The biotinylated enzyme, ACCase, catalyzes this carboxylation reaction. Unique to the plant kingdom, two distinct malonyl-CoA-generating enzymes occur in separate subcellular compartments of these organisms, one located in the plastids and the other in the cytosol (Nikolau et al., 2003). This apparent redundancy is probably the consequence of the evolutionary origin of the metabolic networks within these two subcellular compartments, which is a remnant of the endosymbiotic origin of the plastid organelle (Reyes-Prieto et al., 2007). Thus, the two separated pools of malonyl-CoA are thought to be metabolically distinct because membranes that separate these pools are physical barriers that do not allow the free movement of CoA derivatives, and there are no known transporters to facilitate the movement of malonyl-CoA across the plastidic membrane. Indeed, in vivo isotope-labeling experiments indicate that these two malonyl-CoA pools are metabolically distinct (Schwender and Ohlrogge, 2002). The plastidic malonyl-CoA serves as the precursor for de novo fatty acid biosynthesis, and the cytosolic pool is used for the biosynthesis of a number of different classes of specialized metabolites, including the elongation of preexisting fatty acids, the biosynthesis of flavonoids and other polyketides, and the malonylation of specialized metabolites. In most plants, with the exception of the Graminae, these two malonyl-CoA pools are generated by structurally distinct isozymes of ACCase (Nikolau et al., 2003), with the plastid-localized isozyme being a heteromeric enzyme (htACCase), composed of four separate subunits, whereas the cytosolic isozyme is homomeric (hmACCase) composed of two identical subunits.
Unidirectional Redundancy between CAC1A and CAC1B Paralogs
In Arabidopsis, single-copy genes encode all but one of the four subunits that comprise the plastidic htACCase (Choi et al., 1995; Shorrosh et al., 1995, 1996; Roesler et al., 1996; Sun et al., 1997; Ke et al., 2000b). The exception is the BCCP subunit, which is encoded by two genes, CAC1A and CAC1B (Ke et al., 1997; Thelen et al., 2001), located within 0.32 Mb of each other on chromosome 5. The presence of two paralogous BCCP-coding genes raises the question about their individual significance to the physiological function of htACCase and their consequential role in de novo fatty acid biosynthesis. Based on the observation that BCCP2 primarily accumulates in developing seeds rather than other organs and tissues, it has been suggested that this paralog is specialized for generating malonyl-CoA that is used for the biosynthesis of fatty acids destined for seed oil deposition (Thelen et al., 2001).
To directly test this, we identified and characterized T-DNA-tagged mutant alleles for each BCCP-coding gene and characterized the effect of down-regulating the expression of BCCP1 via antisense RNA technology. These characterizations indicate that the BCCP2 subunit is redundant relative to BCCP1, in that plants devoid of the BCCP2 subunit are indistinguishable from wild-type plants, and fatty acid accumulation appears to be unaffected by the loss of this subunit (at least under the growth conditions that we explored). This conclusion is based on three sets of observations: (1) plants that are homozygous for either one of the two T-DNA disrupted cac1b mutant alleles are recovered at a normal Mendelian inheritance rate from heterozygous parents, (2) the growth and developmental program of the cac1b homozygous mutants is indistinguishable from the wild-type sibling plants, and (3) the amount and composition of fatty acids that accumulate in the cac1b mutants are indistinguishable from the wild-type siblings.
In contrast, the lack of the BCCP1 subunit has drastic effects on growth and development. Specifically, homozygous cac1a mutant plants are not recoverable among the progeny of parents that are heterozygous for this mutant allele, and the transmission of this allele is retarded through male gametogenesis due to abnormal pollen development and/or germination. The inability to recover homozygous cac1a mutant seeds indicates that the CAC1A gene is essential for the development of seeds. Thus, CAC1 is one of 358 gene loci whose functionality is required for normal seed development (McElver et al., 2001; Tzafrir et al., 2003, 2004). Additional characterizations of CAC1A antisense plants establish that BCCP1 plays a significant role in vegetative growth, as evidenced by the fact that reducing the accumulation of this subunit to one-third of wild-type levels generates a dramatically altered growth phenotype. In toto, these findings indicate that BCCP2 cannot compensate for the loss of BCCP1 for maintaining sufficient ACCase activity to sustain a supply of malonyl-CoA for fatty acid biosynthesis required for normal growth and development, and that this becomes critical at two developmental stages, seed and pollen development.
Unidirectional Redundancy Is Probably Due to Expression Difference between CAC1A and CAC1B
To understand the unidirectional redundancy between the two BCCP-coding genes, one needs to ascertain the relative expression patterns of the two genes during the specific developmental stage when the critical phenotype initially demonstrates its expression. Because the lethality of the cac1a allele is uniformly expressed in all seeds that are homozygous for this mutant allele (in contrast to pollen where the penentrance of the mutant phenotype is not near 100%) and because developing seeds (as compared with pollen) are larger and more accessible for characterization, we focused our expression analysis on developing seeds.
Previous studies (Ke et al., 2000b) have shown that the accumulation of the mRNAs coding for four of the htACCase subunits (i.e. those coding for BCCP1, BC, α-CT, and β-CT) is coordinated and is in a spatial and temporal pattern consistent with the burst of fatty acid biosynthesis associated with seed oil deposition, which occurs as embryos develop through the heart and torpedo stages of embryogenesis (O’Hara et al., 2002; Ruuska et al., 2002). Furthermore, parallel studies have suggested that BCCP2 may be specifically required for oil deposition because it is specifically expressed during seed development (Thelen et al., 2001), whereas BCCP1 accumulates in most of the organs of the Arabidopsis plant (Ke et al., 1997; Thelen et al., 2001). In this study, we have extended these observations by assessing the expression of each BCCP-coding gene with promoter:GUS transgenes, in situ hybridization studies of mRNA accumulation patterns, and by quantitative western-blot analyses of BCCP1 and BCCP2 accumulation. Consistent with the prior results, these studies showed that the CAC1A gene is expressed in many organs and tissues of Arabidopsis, but CAC1B expression is constrained to a smaller subset of these organs and tissues. Moreover, at the early stages of embryo development (between 1 and 3 DAF) when the cac1a-associated embryo defects are first detected, by all measures of gene expression that are presented herein, the level of CAC1B expression is lower than that of CAC1A. Most relevant in terms of possibly explaining the unidirectional redundancy between the two BCCP-coding genes is the finding that in developing siliques BCCP2 accumulates at only about 20% of the level of BCCP1. Namely, total BCCP levels in the developing siliques of the cac1b mutants would remain at more than 80% of wild-type levels, but in the cac1a mutant, total BCCP levels would be lowered to less than 20% of wild-type levels. Considering that in the CAC1A antisense plants a significant growth phenotype is not observed in aerial portions of the Arabidopsis (where BCCP2 does not accumulate to significant levels) until BCCP1 levels are lowered to less than 35% of wild-type levels, we surmise therefore that BCCP2 level in the cac1a mutant embryos is not sufficient to rescue the embryo-lethal phenotype. A similar argument can be made for the haploid pollen grains. Here, however, the effect is manifested as decreased penentrance of the phenotype, with only 50% of the haploid cac1a mutant pollen failing to be transmitted.
Given the fact that biochemically BCCP1 and BCCP2 function as the biotin-containing subunits of htACCase (Choi et al., 1995; Thelen et al., 2001), the above quantitative model seems to be the most reasonable explanation for the unidirectional genetic redundancy presented herein. This is not unlike a well-known example of unequal genetic redundancy between APETALA1 (AP1) and CAULIFLOWER (CAL), two homologous genes involved in controlling floral meristem identity. In this latter case, AP1 is essential for normal sepal and petal development, but CAL is dispensable, and this phenomenon has been attributed to the fact that in these organs AP1 shows much higher expression levels than CAL (Kempin et al., 1995). However, we cannot rule out the possibility that BCCP1 and BCCP2 subunits may each confer distinct catalytic characteristics to the htACCase complex.
Distinct and Essential Function for the Plastidic htACCase Isoforms
The htACCase is considered to be a key enzyme for de novo fatty acid biosynthesis in that it generates the committing substrate for this process, malonyl-CoA. Because fatty acids are required for the formation of many important molecules, including membrane lipids, waxes, and signaling molecules, it would be expected that disruption of fatty acid biosynthesis should have severe physiological effects on growth and development. Specific studies (Reverdatto et al., 1999) and sequence-based queries of plant genomic data (i.e. EST sequences and completed genome sequences) indicate that most plant species contain multiple htACCase-subunit coding genes. Therefore, plants have the potential of expressing multiple isoforms of htACCase. However as a model, Arabidopsis probably represents the simplest system to explore the individual contributions of these isoforms to overall htACCase function and therefore to fatty acid biosynthesis, because there are only two apparent gene redundancies that encode this complex enzyme. Thus barring any posttranscriptional and/or posttranslational processing, Arabidopsis has the genetic potential to express at minimum BCCP1-containing and BCCP2-containing htACCase isoforms. Our characterization of Arabidopsis T-DNA insertional null-mutant alleles, which individually eliminates each of these isoforms revealed an unexpected unidirectional redundancy between these two paralogous genes, which could be interpreted as an indication of distinct metabolic functionalities for each of htACCase isoforms.
Consistent with the essential nature of fatty acid biosynthesis, mutations in the CAC1A gene encoding the BCCP1 subunit of htACCase generates an embryo-lethal phenotype, as is the case with mutations in the gene coding for the E2 subunit of the plastidic pyruvate dehydrogenase, which provides the acetyl-CoA substrate for the htACCase (Lin et al., 2003). Moreover, mutations in the enoyl-acyl carrier protein reductase gene, which reduce de novo fatty acid biosynthesis flux, also dramatically alters the growth morphology of plants (Mou et al., 2000).
The embryo lethality of the cac1a mutation is also consistent with characterizations of mutations that affect biotin biosynthesis and utilization. As a biotin-containing enzyme, ACCase is absolutely dependent on the availability of this covalently bound cofactor for catalysis (Nikolau et al., 2003). Thus, mutations in every known biotin biosynthesis gene of Arabidopsis (i.e. bio1, bio2, bio4, and the bifunctional bio3::bio1 locus) and in the hcs1 gene that catalyzes the biotinylation of such proteins as BCCP1, all show an embryo-lethal phenotype (Shellhammer and Meinke, 1990; Patton et al., 1998; Pinon et al., 2005; Muralla et al., 2008; Puyaubert et al., 2008). Moreover, this study helps refine the explanation as to the essentiality of biotin in plants. Specifically, deficiencies in biotin status could affect embryo development through a number of different metabolic lesions associated with the fact that biotin occurs on five Arabidopsis gene products in addition to BCCP1 and BCCP2. These are the hmACCase (products of the ACC1 and ACC2 loci) that generates malonyl-CoA for fatty acid elongation and cytosolic derived secondary metabolites (Yanai et al., 1995; Baud et al., 2003, 2004), mitochondrial methylcrotonyl-CoA carboxylase (MCCase) that is involved in Leu catabolism (Song et al., 1994; Wang et al., 1994; Anderson et al., 1998; McKean et al., 2000), and a seed biotin-storage protein. Of these, the embryo-lethal phenotype is only associated with acc1 alleles (Baud et al., 2003, 2004), and now with the cac1a allele. Unpublished data indicates that mutations associated with seed biotin-storage protein and MCCase do not affect embryo development (G. Ding, E.S. Wurtele, and B.J. Nikolau, unpublished data), which therefore argues that the essentiality of biotin is to support ACCase, and thus fatty acid biosynthesis and elongation, and/or the biosynthesis of malonyl-CoA derived secondary metabolites. However, the fact that many of the cytosolic-malonyl-CoA derived metabolites are classified as secondary metabolites, indicates their dispensable nature (e.g. flavonoids, polyketides), and the fact that fatty acid elongation genes are themselves essential (Millar et al., 2000; Dietrich et al., 2005; Zheng et al., 2005; Beaudoin et al., 2009) further narrows the essentiality of biotin to support ACCase metabolic functionalities associated with the biosynthesis and elongation of fatty acids. Consistent with this generalized concept is the fact that there are no known organisms that do not express ACCase, and the corollary that the biosphere contains organisms in which the only biotinylated protein is ACCase (e.g. E. coli; Cronan and Waldrop, 2002).
The unexpected finding however was the lack of a morphological phenotype associated with the cac1b null alleles. A similar observation has also been reported in earlier studies in which the abundance of the BCCP2 protein was either reduced with antisense RNA technology, or increased by expression of a sense gene construct, using the constitutive 35S CaMV promoter to control the expression of the transgenes (Thelen and Ohlrogge, 2002a). Although relatively subtle changes in fatty acid accumulation were achieved by these transgenic modifications of BCCP2 expression, appearance of the resulting plant morphology was largely unchanged. However, upon the targeted overexpression of BCCP2 in developing seeds (transcriptionally controlled by the napin promoter), ACCase expression was reduced due to the accumulation of nonbiotinylated apo-BCCP2 protein, and this transgenic modification altered the developmental program of the developing seeds (Thelen and Ohlrogge, 2002a). It would appear therefore, that this BCCP2-sense transgene is acting as a dominant negative allele, and a biochemical explanation may be that overexpression of a nonfunctional apo-BCCP2 protein is poisoning the htACCase complex and thus inhibiting flux to fatty acid biosynthesis. More recent proteomics analysis of these seeds further substantiates these alterations by cataloging a large number of pleotropic effects of the dominant negative napin:CAC1B allele (Chen et al., 2009).
Essential Nature of the Plastidic Malonyl-CoA Pool
The finding that htACCase is essential for embryogenesis establishes that the plastidic malonyl-CoA pool generated by this enzyme cannot be supplemented by other sources of this metabolite. For example, plant cells generate a second pool of malonyl-CoA via the action of hmACCase. In Arabidopsis, there are two hmACCase-coding genes, ACC1 and ACC2 (Yanai et al., 1995). The ACC1 protein is located in the cytosol, but the ACC2 protein has a putative transit peptide and may be targeted to plastids (Schulte et al., 1997). The finding that the cac1a mutation is embryo lethal indicates that the malonyl-CoA produced by the ACC1- and ACC2-coding hmACCase is unable to supplement the deficiency in plastidic malonyl-CoA generation. This suggests that cytosolic malonyl-CoA cannot enter the plastids and that ACC2-coding hmACCase is either not located in plastids, or that even if it is located in plastids, its ability to generate malonyl-CoA is insufficient to meet the requirement for this metabolite in this organelle. Given that the only known metabolic fate of plastidic malonyl-CoA is for the biosynthesis of fatty acids, these findings demonstrate that htACCase is essential in generating malonyl-CoA for normal de novo fatty acid biosynthesis.
It is noteworthy that although fatty acids are required to support membrane synthesis in the growth and development of embryos and endosperm, such development appears normal at the very early stages of seed development (0–2 DAF), even in the absence of the BCCP1 protein. This is probably enabled by the ability of the BCCP2 protein to support htACCase function, by the expression of the plastid-targeted ACC2-encoded hmACCase, or by a small amount of malonyl-CoA (or downstream metabolites) that may be donated by maternal cells to the developing seed. Regardless of which mechanism supports growth of the embryo and endosperm at the early stages of seed development, it is insufficient to continue to support the generation of malonyl-CoA needed for fatty acid biosynthesis as membrane lipids and seed oil deposition demands increase as the seed progresses past the globular stage of embryo development.
Cytosolic malonyl-CoA has a more versatile metabolic fate than the plastidic malonyl-CoA. It is an intermediate in a variety of pathways including fatty acid elongation and flavonoid biosynthesis. Mutations in the ACC1 gene also generate an embryo-lethal phenotype, however, the acc1 mutant embryos undergo a distinctly different abnormal embryo developmental program from that of the cac1a mutant (Baud et al., 2003, 2004). In the acc1 mutants, the defects in embryo morphogenesis include the lack of cotyledons and the disruption of cellular organization of the apical region of the embryo. In spite of these defects, the abnormal embryo still matures. In contrast, the cac1a mutant shows delayed development and premature abortion of the embryo, without any specific alteration of embryo morphology until the cells become vacuolated and the embryo collapses. These morphogenetic differences between acc1 and cac1a mutants suggest that the role of BCCP1 (and hence htACCase) during embryo development is related to a fundamental metabolic process providing structural components such as membrane lipids, rather than a signaling process, which is most likely the case for hmACCase. Indeed, the defects associated with the cytosolic malonyl-CoA generation appear to be associated with fatty acid elongation and biosynthesis of sphingolipid, a molecule with many signaling properties (Smith and Merrill, 2002; Sperling and Heinz, 2003; Worrall et al., 2003; Dietrich et al., 2005; Zheng et al., 2005).
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the wild-type and mutant Arabidopsis (Arabidopsis thaliana) genetic stocks were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The cac1a-1 mutant line is in the Wassilewskija background. The cac1b-1 and cac1b-2 alleles are in the Columbia-0 background.
Seeds were sown either in LC1 Sunshine mix (Sun Gro Horticulture) or on Murashige and Skoog agar medium (Invitrogen). To break dormancy, sown seeds were placed at 4°C for 2 d, and then moved into a growth room maintained at 22°C with continuous illumination (100 μmol m−2 s−1). When sown on Murashige and Skoog media, seeds were first sterilized by a short wash with 95% ethanol and a 10-min incubation in 50% bleach solution containing 0.1% Tween 20. The seeds were then extensively washed with sterile water before sowing. If needed, the seedlings were transferred from Murashige and Skoog media to soil at 7 to 10 d after germination.
CAC1A antisense plants were grown under a controlled photoperiod of 16 h of illumination followed by 8 h of darkness. All studies of rosette leaves were from plants between 28 and 30 DAI, and all plant materials were harvested between 3 to 5 h after the start of the illumination period.
To developmentally stage siliques, color threads were used to tag flowers on the day of flowering (day 0) when petals just appeared. The siliques that developed from those tagged flowers were collected at specific time points thereafter.
PCR Genotyping and DNA Gel Blotting
Plant DNA extraction and PCR reactions were conducted as described by Sussman et al. (2000). Two PCR reactions were performed to genotype individual plants carrying T-DNA-tagged alleles. One reaction used a T-DNA border primer and a gene-specific primer to detect the T-DNA-tagged allele, the other reaction used a pair of gene-specific primers that flank the T-DNA insertion site to detect the wild-type allele. Supplemental Table S2 lists the primers used in this study for genotyping of the T-DNA-tagged mutants.
CAC1A cDNA-specific primers (pC-F and pC-R) were designed to track the CAC1A antisense RNA transgene (Supplemental Table S2). This PCR reaction amplified a 933-bp fragment from the endogenous CAC1A genomic gene and a 462-bp fragment from the CAC1A antisense RNA transgene. Thus, a PCR reaction using these primers yields a 933-bp product from wild-type plants and 933- and 462-bp products from transgenic plants.
For DNA gel-blot analysis, DNA was isolated from plant leaves based on the method of Rogers and Bendich (1994). To confirm the integration of the CAC1A antisense RNA transgene into the genome, DNA was digested with restriction enzymes EcoRI and HindIII and fractionated by electrophoresis in 0.8% agarose gels. The DNA was then transferred onto MAGNA nylon membrane (Osmonics) and hybridized with 32P-labeled CAC1A cDNA. Blots were prehybridized and hybridized under standard conditions (Sambrook et al., 1989) and were washed at 65°C for 10 min in 2× SSC, 0.1% SDS, and 10 min in 1× SSC, 0.1% SDS.
In Situ Hybridization
Arabidopsis siliques were harvested at 1, 3, 5, 7, and 12 DAF, cut into 4-mm pieces, fixed, embedded, and sectioned as previously described (Ke et al., 1997). After Proteinase K digestion, hybridization, and washing at 65°C, slides were treated with RNaseA to remove the RNA probe that had not hybridized with mRNA. The Boehringer Mannheim DIG nucleic acid detection kit (Boehringer Mannheim) was used for immunological detection (Canas et al., 1994). In these experiments, about 30 silique pieces were used in total. For each developmental stage, about eight ovules per silique piece were examined, and the entire experiment was repeated three times.
DIG-labeled, gene-specific RNA probes (antisense and sense) were transcribed from the following insert/vector combinations: 676 nucleotides at 3′-end (positions 451–1,114) of CAC1A cDNA in pBluescript SK (±), and 789 nucleotides at 3′-end (positions 451–1,217) of CAC1B cDNA in pSPORT 1 (GIBCO). To ensure that these are CAC1A- and CAC1B-specific probes, each was tested on reconstituted northern-blot analyses, which directly demonstrated that the CAC1A probe does not detect the CAC1B RNA, and that the CAC1B probe does not detect the CAC1A RNA (Supplemental Fig. S4).
Binary Vector Construction
To express the CAC1A antisense RNA, a plant transformation vector was constructed. The binary vector pBI121 (CLONTECH) was modified by removing the GUS gene and replacing it with an oligonucleotide-containing XhoI and SstI restriction enzyme sites. A reverse-orientated full-length CAC1A cDNA (Choi et al., 1995) was then inserted into the modified pBI121 vector using XhoI/SstI restriction enzymes such that the antisense CAC1A cDNA was under the control of the CaMV 35S promoter.
The construct used for complementation of cac1a-1 mutant was made by subcloning the 4.5-kb genomic BamHI fragment containing CAC1A gene (Ke et al., 1997) into pCAMBIA3300 (see Supplemental Fig. S1).
Plant Transformation and Screening for Transgenic Plants
The binary vectors were transformed into Agrobacterium tumefaciens (strains C58C1 or GV3101) by electroporation. Plant transformation was performed with a simplified Arabidopsis transformation protocol (Clough and Bent, 1998). Kanamycin and glufosinate ammonium were used to screen for transformants of the antisense CAC1A cDNA construct and genomic CAC1A construct, respectively. To screen for kanamycin resistance, the T1 seeds were sown on Murashige and Skoog medium containing 50 μg/mL kanamycin. After about 12 d the kanamycin-resistant seedlings were transferred into soil. To screen with glufosinate ammonium, T1 seeds were sown in soil and 14-d seedlings were sprayed with 1:1,500 dilution of Finale Concentrate (Farnam Companies).
Complementation experiment was performed by transforming heterozygous cac1a-1 plants with the pCAMBIA-CAC1A construct. Seeds were collected from the individual T1 transformants. To test whether the construct complements the cac1a-1 mutant, the T2 plants were PCR genotyped by using the primers listed in Supplemental Table S2 and Supplemental Figure S1.
Generation of BCCP1- and BCCP2-Specific Antibodies
Primers A-F and A-R were used to amplify a CAC1A-specific cDNA region. Primers B-F and B-R were used to amplify a CAC1B-specific cDNA region (Supplemental Table S2). The resulting two PCR products were cloned into pET30 vector (Novagen). These two constructs were named CAC1A-pET and CAC1B-pET. Expression of these constructs in Escherichia coli strain BL21 (DE3) produced recombinant proteins with both an S tag and a His tag located at the N terminus. BCCP2-specific recombinant protein was purified with a His·bind column, as described by the manufacturer (Novagen). Affinity purification of BCCP1-specific recombinant protein with either S tag or His tag was not successful in either nondenaturing or denaturing conditions. Thus, this protein was purified by SDS-PAGE and the band that contained the BCCP1-specific recombinant protein was excised for antibody generation. The antibodies were generated in New Zealand white female rabbits, following the procedure previously described (Ke et al., 2000b).
Immunoblot Analysis
Proteins were extracted from plant tissues as described previously (Che et al., 2002). The protein concentration in the extracts was determined with the Bradford method (Bradford, 1976). Proteins were separated by SDS-PAGE (Laemmli, 1970) and transferred to a nitrocellulose membrane (Towbin et al., 1979) using mini-PROTEAN II electrophoresis and mini trans-blot system (Bio-Rad Laboratories). The BCCP1, BCCP2, BC, α-CT, and β-CT subunits of htACCase were immunologically detected with antisera that had been generated and characterized previously (Sun et al., 1997; Ke et al., 2000b) or during the course of this work. The antigen-antibody complexes were detected by either autoradiography with 125I-Protein A or ECL chemiluminescent detection system with horseradish peroxidase-linked anti-rabbit IgG (Amersham Biosciences). The biotin-containing proteins were detected using 125I-labeled or horseradish peroxidase-linked streptavidin (Nikolau et al., 1985). For quantitative analysis, a Storm 840 scanner (Amersham Biosciences) was used to scan the membrane. The signal intensities were quantified by ImageQuant software (Molecular Dynamics).
Promoter:GUS Transgenic Analysis
Promoter:GUS fusion constructs were made with both the CAC1A and CAC1B promoter regions, which were PCR amplified from isolated Arabidopsis genomic DNA using two sets of primers: (1) for the CAC1A promoter, the primer combination was CAC1Ap1F (5′-AAGCTTGATTGTAACCAAGACA-3′) and CAC1ApR (5′-TTCGTCTTCTTATTGTTATTGG-3′); and (2) for the CAC1B promoter, the primer combination was CAC1Bp1F (5′-CAGGGTCATATGGAGCAAGC-3′) and CAC1Bp2R (5′-ATTGTTGAGACAGTGGACGATG-3′). The resulting 1,124- and 1,231-bp fragments upstream of the −1 position, starting from the translational start codons of each gene, were initially cloned upstream of promoterless GFP-GUS reporter genes in the Entry vector, pDONR221 (Gateway Cloning Technology, Invitrogen,), and then moved into the binary vector pBGWFS7 (Karimi et al., 2002). Gateway vectors were transformed into A. tumefaciens strain GV3101, and transgenic plants were generated as described above.
Histochemical staining of GUS activity was conducted on transgenic, T2-generation seedlings that had been germinated in soil. Plants were harvested at various stages of development, and a minimum of five plants was assayed for each independently transformed line; all plants or organs from the same line were stained in the same tube using the same staining solution. GUS staining solution was a mixture of the following five solutions mixed in the ratio of 5:470:25:2:2. (1) Triton X-100:ethanol:water (1:4:5); (2) 0.5 m potassium phosphate buffer, pH 7.0; (3) 10 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcUA dissolved in dimethyl sulfoxide; (4) 0.1 m potassium ferricyanide, pH 7.0; and (5) 0.1 m potassium ferrocyanide, pH 7.0. Staining incubation was either for 3 h or an overnight period, at room temperature (Jefferson et al., 1987). Staining patterns were observed and documented using an Olympus stereomicroscope at the Bessey Microscopy Facility (Iowa State University).
Quantification of BCCP1 and BCCP2 Subunits
Standard molecular cloning procedures (Sambrook et al., 1989) were used to make constructs for generation of S-tag fusion proteins of each BCCP paralog. The 1.1-kb CAC1A cDNA (Choi et al., 1995) was released from pBlueScript vector with EcoRI and Sau3AI and cloned into EcoRI and BamHI sites of pET30a vector (Novagen). The CAC1B cDNA clone 180M1T7 (GenBank accession no. H37386) was obtained from the Arabidopsis Biological Research Center (Columbus, OH). SalI and NotI were used to subclone the CAC1B cDNA into pET30a vector. The resulting plasmids were named pET-CAC1A and pET-CAC1B, respectively. The recombinant proteins resulting from the expression of these two plasmids were designated as S-tag BCCP1 and S-tag BCCP2, respectively. The presence of S-tag fusion proteins in the total protein extract was detected by SDS-PAGE and western blotting with S-protein conjugates (Novagen). The molar concentration of each S-tag fusion protein was determined by using FRETWorks S-tag assay kit (Novagen).
To quantify the BCCP1 subunit in plants, plant protein extracts and a serial dilution of the recombinant BCCP1 standard of known concentration were loaded in a single SDS polyacrylamide gel. After electrophoresis, the proteins were transferred onto a nitrocellulose membrane. Antiserum of BCCP1 was used together with ECL chemiluminescent detection system (Amersham Biosciences) to detect the recombinant protein as well as native BCCP1 in plant samples. The membrane was exposed to Kodak X-OMAT film. Several exposures of different length of time were made to generate a signal intensity within the linear range of the film. The signal intensities of the target bands were quantified by using ImageJ software (National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2005). Standard curves were constructed by plotting the intensity of recombinant protein standard against its concentration. The BCCP1 concentration in each plant sample was determined from this standard curve. For each plant sample, the entire procedure was repeated at least twice. The average of the replicate measurements for a single sample was used as the BCCP1 concentration in that sample. An identical procedure, but using recombinant BCCP2 protein derived from pET-CAC1B, and BCCP2 antiserum was used to quantify the BCCP2 subunit.
Fatty Acid Analysis
Extraction and methylation of fatty acids were conducted using the direct transesterification method (Lewis et al., 2000). Triheptadecanoate was used as internal standard. Analysis of the fatty acid methyl esters was performed with a gas chromatograph (model 6890 series, Agilent Technologies), equipped with a mass detector model 5973 (Agilent Technologies). Chromatography was conducted with a HP-1 column, using helium as the carrier gas. The injection temperature was at 300°C. The oven temperature was initially at 80°C for 2 min and then increased to 150°C at a rate of 40°C min−1. Then the temperature was ramped to 200°C at a rate of 2°C min−1. After holding this temperature for 2 min, it was ramped to 300°C at a rate of 5°C min−1 and held there for 2 min. Peak identification was facilitated by using the HP enhanced chemical analysis software G1701BA (version B.01.00) with Windows NT operating system.
Cytological Preparations of Pollen
Pollen was collected from freshly dehiscent anthers of flowers that were opening or fully open. These flowers (and enclosed pollen) were fixed in Farmer’s fixative (ethanol:acetic acid, 3:1 [v/v]) and stored in 70% ethanol at 4°C. To collect the pollen, fixed or freshly collected anthers were disrupted on a microscope slide using dissecting needles, and the released pollen grains were gently placed under a coverslip for staining. Alternatively, pollen was collected from three to four open flowers, which were placed in a microcentrifuge tube containing 300 μL of stain solution. After briefly vortexing the mixture, the flowers were removed, and the released pollen grains were collected by centrifugation. The pollen pellet was transferred to a microscope slide for viewing by LM and by UV epiillumination.
Five different staining assays were used to compare pollen viability and germination (Alexander’s stain, iodine-potassium iodide [IKI], DAPI, Aniline Blue, and Nile Blue). Fixed pollen was stained with Alexander’s stain for 30 min (Alexander, 1969). Alexander’s stain differentially stains viable pollen grains as dark purple and nonviable pollen grains as either pale green or splotchy dark purple and pale green. Viable pollen is stained by IKI as dark red/brownish, while aborted pollen are slightly red. With Nile Blue stain, aborted pollen appear greenish blue. Aniline Blue (0.1% [w/v] in 0.1 m phosphate buffer, pH 8.5) was used for viewing pollen tube walls under epifluorescence. The stain was applied at least 30 min prior to microscopic examination. To visualize nuclei by UV epiillumination, mature pollen grains were stained for 1 to 2 h in the dark with DAPI (0.4 μg mL−1 DAPI in 0.1 m sodium phosphate, pH 7, 1 mm EDTA, 0.1% Triton X-100). Stained samples were examined with bright-field microscopy with a Zeiss Axiovert microscope (Alexander’s stain, IKI, Nile Blue stain), and UV epiillumination was with a UV filter set for observing the DAPI and Aniline Blue fluorescent stains. Digital images were acquired using AxioVision software.
In vitro germination of isolated pollen was performed in a medium containing 30 μg L−1 boric acid and 25% Suc. Glass slides were dipped into this warm medium, and then cooled to form a thin layer on the slide. Pollen was placed on these slides, and the slides were placed in a sealed box and incubated overnight at 22°C at 100% humidity under continuous illumination.
To determine in vivo germination efficiency, stigmas were cut at the base and inserted vertically into germination medium (30 μg L−1 boric acid, 25% Suc) in a 9-cm petri dish. Plates were sealed and incubated overnight at 22°C, at 100% humidity, under continuous illumination. For each genotype about 300 to 400 pollen grains deposited from 20 stigmas were analyzed. Pollen grains were scored and length of pollen tubes measured by direct observation of plates using a Zeiss Axiovert inverted microscope.
Anthers and the path of pollen tubes inside the pistils were visualized by fixing whole pistils and staining with Aniline Blue and DAPI. The tissue was then cleared for 24 h at room temperature with a drop of clearing solution (240 g of chloral hydrate and 30 g of glycerol in 90 mL deionized water). Pollen was examined with Zeiss Axioplan 2 light microscope and images were captured with a Zeiss AxioCamHRc digital camera (Carl Zeiss, Inc.) using AxioVision 4.3 software. The microscope was equipped with a DAPI filter set comprised of an excitation filter (BP 365/12 nm), a beam splitter (395 nm), and an emission filter (LP 397 nm). The objectives used for imaging were the Neofluar 40× oil, the Apochromat 63× oil, and the Neofluar 100× oil.
Cytological Preparations of Siliques and Seeds
For differential interference contrast microscopy (Nomarski optics), siliques at different stages of development were collected and fixed at room temperature, under vacuum (15 psi) for 48 h with formalin-acetic acid-alcohol (prepared by mixing 50 mL of 50% [v/v] ethanol, 10 mL of 37% [v/v] formalin, 5 mL of acetic acid, and 35 mL of deionized water; Sass, 1958). Siliques were taken through a series of 50% and 70% ethanol, and stored in 70% ethanol (v/v) at 4°C. Fixed siliques were cleared for 1 to 20 h with Herr’s fluid (85% lactic acid:chloral hydrate:phenol:clove oil:xylene; 2:2:2:2:1, w/w; Herr, 1971), and then viewed with a Zeiss Axioplan 2 light microscope equipped with Nomarski optics and a Zeiss AxioCamHRc digital camera (Carl Zeiss, Inc.).
Developing seed were fixed in ethanol:acetic acid (3:1, v/v) for 1 d, followed by a partial rehydration series in ethanol. Clearing was obtained after 24 h in a derivative of Hoyer’s medium (7.5 g gum Arabic, 100 g chloral hydrate, 5 mL glycerol in 30 mL deionized water). The preparations could then be conserved at 4°C for more than 24 h.
To prepare siliques for confocal scanning laser microscopy (CSLM), siliques at different stage of development were collected and fixed under vacuum with formalin-acetic acid-alcohol, then dehydrated, and stored in 70% ethanol. Siliques were then hydrated in a descending series of ethanol (50%, 30% ethanol, and deionized water; 20 min per step), and then thoroughly washed with deionized water. Modified Kasten’s fluorescent Feulgen reaction and Kasten’s fluorescent periodic acid-Schiff reaction (Kasten, 1981) were used to stain nuclei and walls, respectively. For the Feulgen reaction, siliques were subjected to hydrolysis for 20 min in 6 n HCl at room temperature, and then rinsed with deionized water. For the periodic acid-Schiff reaction, siliques were hydrolyzed with 0.5% periodic acid for 25 min at room temperature, thoroughly rinsed with deionized water, and stained for 20 to 30 min in a 0.1% to 0.25% fluorescent acriflavine-Schiff-type reagent (Kasten, 1981). The stained siliques were rinsed with deionized water until very little acriflavine leached out (1–4 h). The siliques were dehydrated in a graded ethanol series (25% to 100% ethanol, 1 h per step), and cleared in an ethanol-methyl salicylate series (3:1, 1:1, 1:3), followed by two changes of 100% methyl salicylate; at least 1 h per step. The siliques were mounted in methyl salicylate on slides, a coverslip was gently added, and siliques were sealed with nail polish or liquid transparent glue. Slides were stored at 4°C to decrease evaporation prior to viewing. Ovules at each developmental stage were viewed using a Leica TCS-NT laser-scanning microscope with an Argon/Krypton laser (the excitation wavelength was 488 nm and 518; Omnichrome). Images (40–60 1-μm optical images per ovule, the serial 1-μm optical-section images) were collected digitally.
SEM of Flowers/Pollen and Seeds/Ovules
Flowers and siliques were fixed under vacuum (18 psi Hg) for 7 h at room temperature, and then overnight at 4°C with 2% glutaraldehyde, 2% paraformaldehyde in 0.1 m sodium cacodylate buffer, pH 7.2. Fixed samples were washed three times in the same buffer, post fixed in buffered 1% OsO4 for 2 h. Following two washes with buffer followed by deionized water, the samples were dehydrated in an ethanol series to absolute ethanol. Samples were subsequently critical point dried in a DCP-1 Denton critical point-drying apparatus (Denton Vacuum Inc.), and mounted on aluminum stubs with double-sided sticky tape and silver cement. For viewing inside the seeds/ovules, seeds/ovules were fractured using a fine surgery razor blade. Flowers and seeds/ovules were sputter coated with gold and palladium in a Denton vacuum LLC desk II cold sputter unit (Denton Vacuum Inc.) for 120 s, and viewed with JEOL 5800LV SEM (Japan Electron Optics Laboratory) at 10 kV. For SEM of pollen, released pollen grains were directly mounted on stubs and sputter coated with gold particles before specimens were examined with an SEM. All digitally collected images including the LM, CSLM, and SEM images were processed in Adobe PhotoShop 7.0 and made into plates using Adobe Illustrator 10.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transgenic complementation of the cac1a-1 mutant allele.
Supplemental Figure S2. Representative PCR genotyping results for transgenic complementation of the cac1a-1 mutant allele.
Supplemental Figure S3. Generation of BCCP1- and BCCP2-specific antibodies.
Supplemental Figure S4. The CAC1A- and CAC1B-specific probes used in RNA hybridization experiments.
Supplemental Table S1. Characterization of T-DNA-tagged alleles of BCCP subunit genes.
Supplemental Table S2. PCR primers used to characterize CAC1A antisense plants and T-DNA-tagged alleles for htACCase subunit genes.
Supplementary Material
Acknowledgments
We are grateful to Dr. Ann Perera for helpful discussion on fatty acid analysis and for guidance during the use of equipment at the W.M. Keck Metabolomics Research Facility and to Dr. Harry Horner and Tracey Pepper for help with using equipment at the Iowa State University Microscopy and NanoImaging Facility. We are greatly thankful to Dr. Carol Foster for helpful advice and assistance on in situ hybridization analysis. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson MD, Che P, Song JP, Nikolau BJ, Wurtele ES. (1998) 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol 118: 1127–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Shorrosh BS, Ohlrogge JB. (1997) Isolation and characterization of an Arabidopsis biotin carboxylase gene and its promoter. Plant Mol Biol 35: 539–550 [DOI] [PubMed] [Google Scholar]
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C. (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep 5: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C. (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L. (2009) Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom M, Evans E, Mouritsen OG. (1991) Physical properties of the fluid lipid-bilayer component of cell membranes: a perspective. Q Rev Biophys 24: 293–397 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Canas LA, Busscher M, Angenent GC, Beltran JP, van Tunen AJ. (1994) Nuclear localization of the petunia MADS box protein FBP1. Plant J 6: 597–604 [Google Scholar]
- Che P, Wurtele ES, Nikolau BJ. (2002) Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. Plant Physiol 129: 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Mooney BP, Hajduch M, Joshi T, Zhou M, Xu D, Thelen JJ. (2009) System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Yu F, Wurtele ES, Nikolau BJ. (1995) Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol 109: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cronan JE, Jr, Waldrop GL. (2002) Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res 41: 407–435 [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Perera MA, D Yandeau-Nelson M, Meeley RB, Nikolau BJ, Schnable PS. (2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42: 844–861 [DOI] [PubMed] [Google Scholar]
- Elborough KM, Winz R, Deka RK, Markham JE, White AJ, Rawsthorne S, Slabas AR. (1996) Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J 315: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr JM., Jr (1971) A new clearing-squash technique for the study of ovule development in angiosperms. Am J Bot 58: 785–790 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kasten FH. (1981) Methods for fluorescence microscopy. Clark G, Biological Stain Commission, , Staining Procedures, Ed 4 Published for the Biological Stain Commission by Williams & Wilkins, Baltimore, pp 39–103 [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. (2000a) The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol 123: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Choi JK, Smith M, Horner HT, Nikolau BJ, Wurtele ES. (1997) Structure of the CAC1 gene and in situ characterization of its expression: the Arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol 113: 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Wen TN, Nikolau BJ, Wurtele ES. (2000b) Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol 122: 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y. (1994) Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA 91: 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y. (1996) Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37: 117–122 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL. (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leonard AE, Pereira SL, Sprecher H, Huang YS. (2004) Elongation of long-chain fatty acids. Prog Lipid Res 43: 36–54 [DOI] [PubMed] [Google Scholar]
- Lewis T, Nichols PD, McMeekin TA. (2000) Evaluation of extraction methods for recovery of fatty acids from lipid-producing microheterotrophs. J Microbiol Methods 43: 107–116 [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE. (2002) The jasmonate pathway. Science 296: 1649–1650 [DOI] [PubMed] [Google Scholar]
- Lin M, Behal R, Oliver DJ. (2003) Disruption of plE2, the gene for the E2 subunit of the plastid pyruvate dehydrogenase complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Mol Biol 52: 865–872 [DOI] [PubMed] [Google Scholar]
- Martin JT, Juniper BE. (1970) The Cuticle of Plants. Edward Arnold Ltd., Edinburgh [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean AL, Ke J, Song J, Che P, Achenbach S, Nikolau BJ, Wurtele ES. (2000) Molecular characterization of the non-biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase. J Biol Chem 275: 5582–5590 [DOI] [PubMed] [Google Scholar]
- Mekhedov S, de Ilárduya OM, Ohlrogge J. (2000) Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol 122: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzen WI, Peng J, Ransom N, Nikolau BJ, Wurtele ES. (2008) Articulation of three core metabolic processes in Arabidopsis: fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol 8: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kunst L. (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C. (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Chen E, Sweeney C, Gray JA, Dickerman A, Nikolau BJ, Meinke D. (2008) A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in Arabidopsis. Plant Physiol 146: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES. (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414: 211–222 [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Wurtele ES, Stumpf PK. (1985) Use of streptavidin to detect biotin-containing proteins in plants. Anal Biochem 149: 448–453 [DOI] [PubMed] [Google Scholar]
- O’Hara P, Slabas AR, Fawcett T. (2002) Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol 129: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG, Post-Beittenmiller D. (1993) De novo fatty acid biosynthesis. Moore TS, Jr, , Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 3–32 [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Ravanel S, Douce R, Alban C. (2005) Biotin synthesis in plants: the first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiol 139: 1666–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D. (1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 405–430 [DOI] [PubMed] [Google Scholar]
- Puyaubert J, Denis L, Alban C. (2008) Dual targeting of Arabidopsis holocarboxylase synthetase1: a small upstream open reading frame regulates translation initiation and protein targeting. Plant Physiol 146: 478–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto S, Beilinson V, Nielsen NC. (1999) A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol 119: 961–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. (2007) The origin and establishment of the plastid in algae and plants. Annu Rev Genet 41: 147–168 [DOI] [PubMed] [Google Scholar]
- Richards FM, Wyckoff HW. (1971) Bovine pancreatic ribonuclease. Boyer PD, , The Enzymes, Ed 3, Vol 4 Academic Press, New York, pp 647–806 [Google Scholar]
- Roesler KR, Savage LJ, Shintani DK, Shorrosh BS, Ohlrogge JB. (1996) Co-purification, co-immunoprecipitation, and coordinate expression of acetyl-coenzyme A carboxylase activity, biotin carboxylase, and biotin carboxyl carrier protein of higher plants. Planta 198: 517–525 [DOI] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB. (1994) Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiol 105: 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. (1994) Extraction of total cellular DNA from plants, algae and fungi. Gelvin SB, Schilperoort RA, , Plant Molecular Biology Manual, Ed 2 Kluwer Academic Press, Dordrecht, The Netherlands, pp D1: 1–8 [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. (1993) Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem 268: 25118–25123 [PubMed] [Google Scholar]
- Sass JE. (1958) Botanical Microtechnique, Ed 3 Iowa State College Press, Ames, IA [Google Scholar]
- Schulte W, Topfer R, Stracke R, Schell J, Martini N. (1997) Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA 94: 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB. (2002) Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol 130: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. (2005) Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol 43: 229–260 [DOI] [PubMed] [Google Scholar]
- Shellhammer J, Meinke D. (1990) Arrested embryos from the bio1 auxotroph of Arabidopsis thaliana contain reduced levels of biotin. Plant Physiol 93: 1162–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Roesler KR, Shintani D, van de Loo FJ, Ohlrogge JB. (1995) Structural analysis, plastid localization, and expression of the biotin carboxylase subunit of acetyl-coenzyme A carboxylase from tobacco. Plant Physiol 108: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Savage LJ, Soll J, Ohlrogge JB. (1996) The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J 10: 261–268 [DOI] [PubMed] [Google Scholar]
- Smith WL, Merrill AH., Jr (2002) Sphingolipid metabolism and signaling minireview series. J Biol Chem 277: 25841–25842 [DOI] [PubMed] [Google Scholar]
- Song J, Wurtele ES, Nikolau BJ. (1994) Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonoyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc Natl Acad Sci USA 91: 5779–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Heinz E. (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632: 1–15 [DOI] [PubMed] [Google Scholar]
- Sun J, Ke J, Johnson JL, Nikolau BJ, Wurtele ES. (1997) Biochemical and molecular biological characterization of CAC2, the Arabidopsis thaliana gene coding for the biotin carboxylase subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol 115: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Mekhedov S, Ohlrogge JB. (2001) Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol 125: 2016–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. (2002a) Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana. Plant J 32: 419–431 [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. (2002b) The multisubunit acetyl-CoA carboxylase is strongly associated with the chloroplast envelope through non-ionic interactions to the carboxyltransferase subunits. Arch Biochem Biophys 400: 245–257 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. (2003) The Arabidopsis SeedGenes project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JG, Browse J. (2002) Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res 41: 254–278 [DOI] [PubMed] [Google Scholar]
- Wang X. (2002) Phospholipase D in hormonal and stress signaling. Curr Opin Plant Biol 5: 408–414 [DOI] [PubMed] [Google Scholar]
- Wang X, Wurtele ES, Keller G, McKean AL, Nikolau BJ. (1994) Molecular cloning of cDNAs and genes coding for beta-methylcrotonyl-CoA carboxylase of tomato. J Biol Chem 269: 11760–11768 [PubMed] [Google Scholar]
- Wasternack C, Hause B. (2002) Jasmonates and octadecanoids: signals in plant stress responses and development. Prog Nucleic Acid Res Mol Biol 72: 165–221 [DOI] [PubMed] [Google Scholar]
- Worrall D, Ng CK, Hetherington AM. (2003) Sphingolipids, new players in plant signaling. Trends Plant Sci 8: 317–320 [DOI] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.