Abstract
Phytophthora sojae encodes hundreds of putative host cytoplasmic effectors with conserved FLAK motifs following signal peptides, termed crinkling- and necrosis-inducing proteins (CRN) or Crinkler. Their functions and mechanisms in pathogenesis are mostly unknown. Here, we identify a group of five P. sojae-specific CRN-like genes with high levels of sequence similarity, of which three are putative pseudogenes. Functional analysis shows that the two functional genes encode proteins with predicted nuclear localization signals that induce contrasting responses when expressed in Nicotiana benthamiana and soybean (Glycine max). PsCRN63 induces cell death, while PsCRN115 suppresses cell death elicited by the P. sojae necrosis-inducing protein (PsojNIP) or PsCRN63. Expression of CRN fragments with deleted signal peptides and FLAK motifs demonstrates that the carboxyl-terminal portions of PsCRN63 or PsCRN115 are sufficient for their activities. However, the predicted nuclear localization signal is required for PsCRN63 to induce cell death but not for PsCRN115 to suppress cell death. Furthermore, silencing of the PsCRN63 and PsCRN115 genes in P. sojae stable transformants leads to a reduction of virulence on soybean. Intriguingly, the silenced transformants lose the ability to suppress host cell death and callose deposition on inoculated plants. These results suggest a role for CRN effectors in the suppression of host defense responses.
Many plant pathogens, including bacteria, fungi, oomycetes, and nematodes, secrete distinct proteins into different cellular compartments of their hosts to modulate host defense circuitry and benefit parasite colonization (Bhavsar et al., 2007; Hogenhout et al., 2009; Tyler, 2009). These pathogen-secreted proteins are named effectors (Hogenhout et al., 2009). Through coevolution, plants have developed effective surveillance systems to recognize particular effectors and induce defense pathways. This host response is called effector-triggered immunity (ETI; Chisholm et al., 2006; Jones and Dangl, 2006; Cui et al., 2009). A major function of pathogen effectors is believed to be the suppression of host defense responses through their interaction with critical host targets, including signal transduction pathways involved in pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and ETI (Chisholm et al., 2006; Jones and Dangl, 2006). For example, many characterized effectors of bacterial or oomycete plant pathogens, such as Pseudomonas syringae AvrPtoB (Abramovitch et al., 2003), Phytophthora sojae Avr1b (Dou et al., 2008a), and Phytophthora infestans Avr3a (Bos et al., 2006), can suppress host cell death and/or the hypersensitive response (HR), which are involved in PTI and/or ETI.
P. sojae, causing soybean (Glycine max) root and stem rot, is a major threat to soybean cultivation and leads to annual losses of $1 to $2 billion worldwide (Tyler, 2007). Besides P. sojae, the genus Phytophthora contains over 90 species, almost all of which are destructive pathogens of a huge range of agriculturally and ornamentally important plants (Erwin and Ribiero, 1996). Phytophthora belongs to the fungus-like oomycetes, which are evolutionarily related to algae in the kingdom Stramenopila (Kamoun, 2003; Tyler et al., 2006). Since oomycetes are distinct from fungi in many aspects, including biochemical pathways and infection strategies, management approaches used for fungal pathogens are often not effective for controlling oomycete diseases (Erwin and Ribiero, 1996; Kamoun, 2003). The study of effectors and relevant functional analyses is important for understanding oomycete pathogenesis and for developing novel and effective strategies to manage disease (Hogenhout et al., 2009; Tyler, 2009). For instance, several potato (Solanum tuberosum) late-blight resistance (R) genes specific for particular P. infestans strains have been identified based upon advanced knowledge of oomycete effectors (Vleeshouwers et al., 2008; Lokossou et al., 2009).
A major group of oomycete effectors targeted to the host cytoplasm are the RXLR effectors, which were discovered based on a common sequence pattern identified in oomycete avirulence proteins and their homologs in the genome of P. sojae and Phytophthora ramorum (Tyler et al., 2006; Jiang et al., 2008). The common pattern, containing a signal peptide and a conserved RXLR-dEER motif, was shown to be responsible for delivering proteins into host plant cells colonized by P. infestans (Whisson et al., 2007) or P. sojae (Dou et al., 2008b) and is presumed to have the same function in most oomycetes. The oomycete RXLR-dEER motif and Plasmodium host target signals are interchangeable (Bhattacharjee et al., 2006; Dou et al., 2008b). Also, effectors of fungal pathogens contain functional variants of the RXLR motif (Kale et al., 2010). The translocation process is independent of pathogen machinery (Dou et al., 2008b) and mediated by binding to phosphatidylinositol 3-phosphate on the outer surface of host plasma membranes (Kale et al., 2010), suggesting a potential durable disease management strategy by blocking the entry of effectors. Once delivered inside host cells, RXLR effectors are involved in the suppression of host basal resistance (Sohn et al., 2007) and cell death (Bos et al., 2006; Dou et al., 2008a) when the cognate R genes are absent.
Oomycete CRN effectors are another group of pathogen proteins presumed to enter host cytoplasm (Torto et al., 2003; Win et al., 2007; Haas et al., 2009). This group of effectors was first identified based on their ability to elicit plant cell death and defense responses (Torto et al., 2003). Similar to RXLR effectors, CRN effectors contain a conserved motif, FLAK (F, Phe; L, Leu; A, Ala; and K, Lys), following the signal peptide (Win et al., 2007; Haas et al., 2009). Each species of Phytophthora sequenced to date encodes 61 to 451 CRN genes in their genomes (Tyler et al., 2006; Haas et al., 2009). Interestingly, Hyaloperonospora arabidopsidis contains many proteins with overlapping RXLR and FLAK motifs (RXLRLFLAK; Win et al., 2007), suggesting that these two motifs share a similar delivery function, although further validation is required. Apart from this, however, the functions and mechanisms of the CRN effectors in pathogenesis are mostly unknown.
Here, two P. sojae CRN effectors with predicted nuclear localization signals (NLS) were studied using a combination of computational and functional genomic approaches. Surprisingly, although the two proteins share close sequence similarity, they induce contrasting and apparently opposite responses when expressed in Nicotiana benthamiana and the host soybean: PsCRN63 induces cell death, while PsCRN115 blocks cell death. Cosilencing of both genes in stable transformants leads to a significant reduction of virulence on host plants. Furthermore, strong host cell death and callose deposition were only observed in plants inoculated with the CRN-silenced transformants. Therefore, we propose a role for CRN effectors in the suppression of host defense responses.
RESULTS
Identification of a Group of P. sojae-Specific Effectors Containing the FLAK Motif and NLS
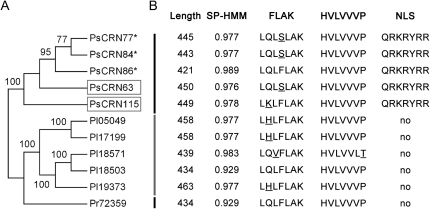
The P. sojae CRN family, containing 202 genes, has been extensively analyzed (Tyler et al., 2006; Haas et al., 2009). In this study, we selected one group of genes for functional analysis for three reasons: first, the predicted gene models encode proteins with a predicted NLS (Fig. 1; Cokol et al., 2000); second, the group shows high sequence similarity with P. infestans CRN2 (PiCRN2), which has the ability to elicit cell death in N. benthamiana and induce the expression of defense responses in tomato (Solanum lycopersicum; Torto et al., 2003); and third, selected genes within the group are the most highly expressed among all P. sojae RXLR and CRN effectors based upon transcriptional profiling of various life cycle and infection stages (W. Ye, X. Wang, D. Dou, and Y. Wang, unpublished data). Five genes in this group are PsCRN63, PsCRN77, PsCRN84, PsCRN86, and PsCRN115 (Haas et al., 2009). Among these, PsCRN77, PsCRN84, and PsCRN86 are apparently pseudogenes, because they possess one or more stop codons in their predicted open reading frames that would result in truncated proteins (Supplemental Fig. S1).
Figure 1.
Identification of a group of P. sojae-specific effectors containing FLAK motifs and NLS. A, Phylogenetic relationships of a group of CRN effectors in three Phytophthora species. The phylogenetic tree was constructed with amino acid sequences using MEGA 4.1 with the neighbor-joining method, 1,000 replicates, and pairwise-deletion option. Putative pseudogenes are indicated with asterisks, and the two functional analyzed genes are denoted by boxes. CRN genes, designated Ps, Pi, and Pr, correspond to P. sojae, P. infestans, and P. ramorum, respectively. The names and gene models of each gene follow published data (Haas et al., 2009). The unmasked sequence alignments used to reconstruct the phylogenic tree are available as Supplemental Figure S1. B, Predicted gene structures. Length, SP-HMM, and NLS represent the length of the predicted protein, signal peptide prediction based on hidden Markov models, and nuclear localization signals, respectively. The signal peptide probabilities and NLS were predicted from the SignalP3.0 server (Bendtsen et al., 2004) and the NLS server (Cokol et al., 2000), respectively.
We determined the actual transcription units for PsCRN63 and PsCRN115 by reverse transcription (RT)-PCR from total RNA of the cultured mycelium of P. sojae isolate P6497 (Förster et al., 1994). Primers were designed to amplify the predicted open reading frame regions, excluding the putative signal peptide, for expression in planta. The cDNA sequence of PsCRN63 is identical to the gene model from the reference genome, while PsCRN115 differs from the reference genome at two sites, having one nucleotide substitution at the 973th site, from A to G, and an addition of nine nucleotides at the 826th site. Both genes lack introns and encode proteins of 450 amino acids (PsCRN63) and 499 amino acids (PsCRN115; Fig. 1; Supplemental Fig. S1).
After removing the stop codons or revising the frame shifts by deleting one or two nucleotides, the theoretical protein sequences encoded by PsCRN77, PsCRN84, and PsCRN86 were obtained and analyzed together with PsCRN63 and PsCRN115. They share a high level of sequence similarity and contain identical sequence segments corresponding to a NLS motif (Fig. 1; Supplemental Fig. S1). In total, the sequences of PsCRN63 and PsCRN115 are 95.7% identical at the amino acid level. The C-terminal portion of PsCRN63, which is sufficient to trigger cell death in N. benthamiana (described below; Fig. 1; Supplemental Fig. S1), differs by only four amino acids from that of PsCRN115. To identify potentially orthologous genes in other Phytophthora species, we searched the P. infestans and P. ramorum genomes using BLASTN, with a cutoff of E < 10−120. As a result, five genes were obtained from P. infestans and one gene was obtained from P. ramorum. All of the predicted protein sequences contain the conserved signal peptides and FLAK motif but lack the NLS (Fig. 1; Supplemental Fig. S1).
Phylogenetic relationships among the 11 CRN proteins from three species of Phytophthora were modeled by using MEGA 4.1 with the neighbor-joining method (Tamura et al., 2007). On the phylogenetic tree, the 11 CRNs were sorted into three distinct groups according to species of origin (Fig. 1). The recent expansion of this class of CRN genes in the genomes of P. sojae and P. infestans, and the presence of pseudogenes in P. sojae, suggest that the genes are subject to high selective pressure and are consistent with a “rapid birth-and-death” mechanism (Tyler, 2009). Among this group of 11 closely related proteins, the P. sojae CRNs are exceptional because they the only ones with clearly predicted NLS motifs (Fig. 1).
Expressional Analysis of Genes in the P. sojae Group
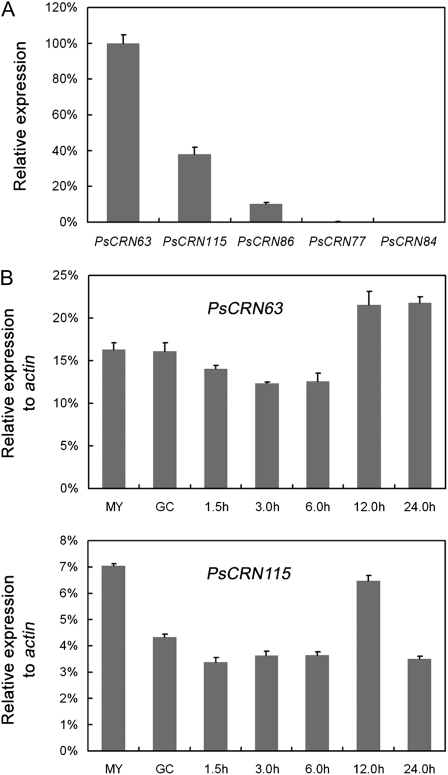
To investigate the relative expression level of each gene in the P. sojae group, we designed specific primers and performed quantitative RT-PCR using RNA from growing hyphae. Figure 2A shows that PsCRN63 expression is the highest among the five genes tested. Expression levels of PsCRN115 and PsCRN86 were 38.1% and 10.4% of that of PsCRN63, respectively. Transcripts of pseudogenes PsCRN77 and PsCRN84 were nearly undetectable in this assay. We also determined the expression patterns of PsCRN63 and PsCRN115 in different stages of development, including mycelium, germinated cysts, and infection stages at 1.5, 3.0, 6.0, 12.0, and 24.0 h post infection (hpi). PsCRN63 transcripts are slightly (approximately 1.5-fold) induced during the late infection stages (12.0 and 24.0 hpi), whereas the highest levels of PsCRN115 RNA are found at the mycelium stage. Overall, these results show that transcripts of PsCRN63 and PsCRN115 are detectable in all the tested stages (Fig. 2B). In addition, relative transcript levels of PsCRN63 and PsCRN115 were 12.4% to 21.8% and 3.4% to 7.1% of that of the P. sojae actin gene, respectively.
Figure 2.
Expression analysis of the group of CRN effectors in P. sojae. A, Relative expression of PsCRN63, PsCRN77, PsCRN84, PsCRN86, and PsCRN115 in vegetative hyphae. Expression levels relative to PsCRN63 were calculated as described in “Materials and Methods.” B, Expression of PsCRN63 and PsCRN115 during stages of asexual development and infection. Mycelium was inoculated on leaves of soybean cv Williams during the indicated time course (i.e. 1.5, 3.0, 6.0, 12.0, and 24.0 hpi). The cultured hyphae (MY) and germinated cysts (GC) were used as controls. Expression levels relative to the P. sojae actin gene were calculated as described in “Materials and Methods.” In A and B, PCR was replicated three times (i.e. technical replicates). Error bars indicate se.
Contrasting Effects of PsCRN63 and PsCRN115 on Cell Death
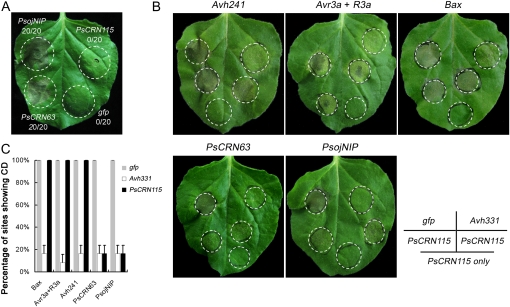
We tested whether PsCRN63 or PsCRN115 proteins could induce necrosis in N. benthamiana using infiltration of Agrobacterium tumefaciens cells with a potato virus X (PVX) vector carrying each of the mature genes (the predicted signal peptides were removed). These methods have been widely used for transient expression of genes in N. benthamiana and for determining whether particular genes can induce or suppress cell death (Torto et al., 2003; Bos et al., 2006; Dou et al., 2008a; Haas et al., 2009). A known elicitor of cell death, the P. sojae necrosis-inducing protein (PsojNIP; Qutob et al., 2002), was used as positive control. Figure 3A shows that PsCRN63 triggered cell death in N. benthamiana. In contrast, only mosaic symptoms due to PVX were observed following infiltration with PsCRN115 or gfp under the same conditions. The necrotic symptoms trigged by PsCRN63 developed about 5 d after infiltration and showed light-colored lesions, whereas PsojNIP triggered dark necrotic lesions around the site of infiltration and the symptoms developed about 4 d after infiltration (Fig. 3A). Considering that the cell death-inducing activity of PsCRN63 might possibly be caused by gene overexpression in the PVX system, we performed additional plant transformation experiments by transient expression of PsCRN63 in N. benthamiana leaves by agroinfiltration. The leaves infiltrated with PsCRN63 exhibited cell death at 5 d after infiltration. In contrast, no obvious cell death was found when a control gene, gfp, was expressed under the same conditions (Supplemental Fig. S2).
Figure 3.
Functional analysis of PsCRN63 and PsCRN115. A, PsCRN63 triggers cell death in N. benthamiana. The plant leaves were infiltrated with A. tumefaciens cells containing a PVX vector carrying the genes PsojNIP (positive control), PsCRN63, PsCRN115, or gfp (negative control). Photographs were taken 5 d after infiltration. B and C, PsCRN115 suppresses cell death in N. benthamiana. B, Agroinfiltration sites in each N. benthamiana leaf expressing gfp (top left), Avh331 (top right), or PsCRN115 (bottom three sites) were challenged with A. tumefaciens expressing Avh241, Avr3a + R3a, Bax, PsCRN63, and PsojNIP at the four top-most sites (the bottom-most site was left as a control). Photographs were taken 5 d after cell death inducer infiltration. C, Quantification of the percentage of cell death (CD) sites infiltrated with cell death inducers. The mean percentages of sites showing cell death were scored from 24 infiltration sites based on three independent experiments.
To further explore the functions of PsCRN115, we tested whether it could suppress cell death like other bacterial and oomycete effectors (Abramovitch et al., 2003; Bos et al., 2006; Dou et al., 2008a). Several cell death inducers were used in this assay: BAX (triggering HR-mimicking cell death in plants; Lacomme and Santa Cruz, 1999), PsojNIP (Qutob et al., 2002), a combination of P. infestans Avr3a and potato R3a as an HR inducer (Armstrong et al., 2005), a P. sojae RXLR effector identified as an elicitor of cell death (Avh241; Q. Wang, C. Han, and Y. Wang, unpublished data), and PsCRN63. Cell death symptoms were observed when A. tumefaciens cells carrying the above genes were infiltrated into N. benthamiana leaves (Fig. 3, B and C). However, when PsCRN115 was infiltrated in N. benthamiana leaves 12 h prior to infiltration of each of the inducers, this pretreatment blocked cell death normally triggered by PsojNIP and PsCRN63. In contrast, PsCRN115 could not protect N. benthamiana tissue from cell death triggered by BAX, Avh241, or Avr3a/R3a (Fig. 3, B and C). Furthermore, prior infiltration with A. tumefaciens cells containing the gfp gene, a negative control, did not protect against cell death. A previously identified cell death suppressor, Avh331 (Dou et al., 2008a), was used as a positive control and could block cell death triggered by all the tested elicitors (Fig. 3, B and C).
To independently confirm the activities of PsCRN63 and PsCRN115 in the host soybean, we performed cobombardment assays using a double-barreled attachment for the Bio-Rad Gene Gun, which enables us to shoot two different DNA samples side by side into a leaf simultaneously. This method improves the reproducibility of the results (Dou et al., 2008a, 2008b; Kale et al., 2010). The GUS gene was used as a reporter with the indicated combination of tested genes (Table I). Cell death induced or suppressed by the test gene results in elimination or restoration of GUS reporter gene expression, respectively. Table I shows that expression of the PsCRN63 gene reduced the number of GUS-positive blue patches by 52% (compared with a gfp control gene), while expression of the PsCRN115 gene did not significantly alter reporter gene expression. These results indicate that PsCRN63 is a cell death inducer in soybean cells and that PsCRN115 is not. When PsCRN115 was coexpressed with PsCRN63, the number of GUS-positive blue patches was doubled, indicating that PsCRN115 could partially suppress PsCRN63-induced cell death. This result was obtained when the cobombardment of PsCRN63 and PsCRN115 was compared directly with cobombardment of PsCRN63 and GFP using the double-barreled bombardment (direct assay) and when each mixture was separately compared with a reference consisting of GUS plus empty vector (indirect assay; Table I). Furthermore, the PsojNIP-induced cell death inhibition activity of PsCRN115 was also tested by the direct and indirect assays described above. All of the results indicate that PsCRN115 can inhibit PsojNIP-induced cell death in soybean and thus confirm the observations from the N. benthamiana assays (Table I).
Table I. Cell death manipulation activities of PsCRN63 and PsCRN115 measured by double-barreled particle bombardment.
| Experiment | Barrel 1a | Barrel 2a | Direct Ratiob | Indirect Ratioc | Pd |
| a | GFP | GFP | 0.99 ± 0.01 | ||
| b | GFP | PsCRN63 | 0.48 ± 0.02 | b/a = 0.48 | <0.001 |
| c | GFP | PsCRN115 | 1.10 ± 0.04 | c/a = 1.11 | >0.1 |
| d | GFP | PsCRN63 + GFP | 0.54 ± 0.03 | ||
| e | GFP | PsCRN63 + PsCRN115 | 1.01 ± 0.02 | e/d = 1.87 | <0.001 |
| f | PsCRN63 + GFP | PsCRN63 + PsCRN115 | 2.06 ± 0.16 | ||
| g | GFP | PsojNIP + GFP | 0.04 ± 0.00 | ||
| h | GFP | PsojNIP + PsCRN115 | 0.38 ± 0.03 | h/g = 9.50 | <0.001 |
| i | PsojNIP + GFP | PsojNIP + PsCRN115 | 1.84 ± 0.40 |
Barrels 1 and 2 are physically identical. Half of all the replicates were conducted using the configuration of DNA samples indicated plus GUS, and half were conducted with the samples reversed between barrels 1 and 2. In all cases, the mass of DNA for each barrel was identical.
Ratios between the numbers of spots produced by each barrel. Geometric averages and se were calculated from log ratios obtained from 10 to 16 pairs of shots.
Comparison of the two averaged ratios from the experiments indicated by the lowercase letters.
P values for the indirect comparisons were calculated from the log ratios using the Wilcoxon rank sum test.
Deletion Analysis of PsCRN63 and PsCRN115
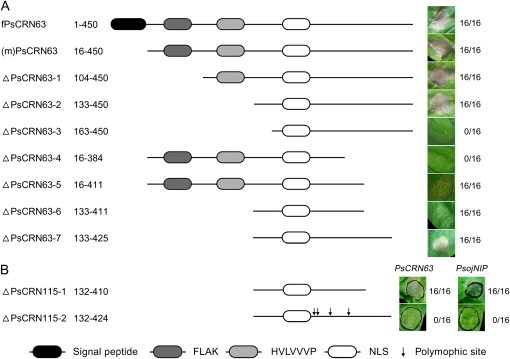
To determine the functional regions in PsCRN63 and PsCRN115, we created seven deletion mutants for PsCRN63 and tested their ability to induce cell death using the above-introduced methods. Figure 4 demonstrates that positions from 133 to 411 of PsCRN63 can trigger a weak cell death, while positions from 133 to 425 are required for full cell death activity. We also tested the corresponding positions of PsCRN115 for cell death suppression ability. The region from 132 to 424 was sufficient to inhibit cell death induced by PsCRN63 or PsojNIP, but the region from 132 to 410 was insufficient for the suppressive activity of PsCRN115. The characterized regions do not contain the conserved FLAK motif, indicating that this motif is not required for the activity when the protein is expressed inside plant cells. Thus, the FLAK motif, as well as RXLR-dEER motifs in oomycete RXLR effectors (Whisson et al., 2007; Dou et al., 2008b) and RXLXE(Q) motifs in Plasmodium (Hiller et al., 2004; Marti et al., 2004), might serve as host translocation signals and deliver proteins into host cells. The full-length PsCRN63 protein including the putative signal peptide also triggered cell death (Fig. 4). This result supports the hypothesis that the FLAK motif performs a host-targeting function similar to the RXLR-dEER motif. However, further experiments are required to validate this hypothesis.
Figure 4.
Deletion analysis of PsCRN63 and PsCRN115 defines their functional fragments. Deletion mutants of PsCRN63 (A; induction of cell death) and PsCRN115 (B; suppression of cell death) were expressed by agroinfiltration in N. benthamiana. A schematic view of the different deletion mutant constructs is shown on the left. The typical symptoms for each infiltration site are shown on the right. Photographs of symptoms were taken 5 d after infiltration. The numbers show the ratio of necrotic responses and the total number of infiltrated sites. In B, cell death inducers (PsCRN63 and PsojNIP) are indicated.
The Contradictory Roles of the Predicted NLS Motif in PsCRN63 and PsCRN115
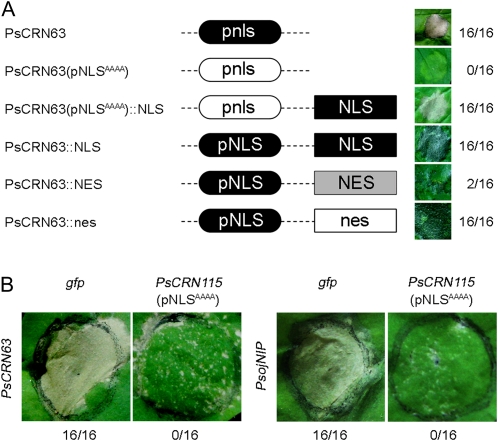
Although orthologous genes to this group of P. sojae CRN effectors are present in other Phytophthora species, the NLS was only found in the P. sojae orthologs (Fig. 1). To test whether this signal is required to induce cell death, we first made mutations of this NLS [PsCRN63(pNLSAAAA)] and then expressed it in N. benthamiana leaves by infiltration methods. Figure 5 shows that mutations of the NLS abolished the ability to trigger cell death. Furthermore, when we added a synthetic NLS to the C terminus of the mutant PsCRN63 gene [PsCRN63(pNLSAAAA)], we found that this complemented the mutation and led to restoration of cell death-triggering activity. These results indicate that a functional NLS is required for the activity. To further confirm this, we sequestered PsCRN63 either in the nucleus or the cytoplasm by attachment of a NLS or a nuclear exclusion signal (NES; Shen et al., 2007) to the C terminus, respectively. Cell death symptoms were not observed when the NES was attached (Fig. 5). To exclude the possibility that the NES could interfere with activity, a nonfunctional NES (nes) was fused to the C terminus of PsCRN63. In the nonfunctional nes, the second and third Leu residues and the first Ile residue were all replaced with Ala residues (Shen et al., 2007). This construct triggered the same symptoms as wild-type PsCRN63. Considering these findings together, we infer that PsCRN63-mediated cell death is triggered in the plant nucleus.
Figure 5.
Functional characterization of the predicted NLS in PsCRN63 and PsCRN115. The predicted NLS is necessary for PsCRN63 to trigger cell death (A) but is not required for cell death suppression of PsCRN115 (B). N. benthamiana was transformed with the mutants of PsCRN63 or PsCRN115 by agroinfiltration. In A, a schematic view of the different mutant constructs is shown on the left. Symptoms of infiltration sites are shown on the right. The numbers show the ratio of necrotic responses and the total number of agroinfiltration sites. Photographs of symptoms were taken 5 d after infiltration. In B, cell death inducers (PsCRN63 and PsojNIP) are indicated.
To test whether the predicted NLS of PsCRN115 is required to suppress cell death, we made mutations of this NLS [PsCRN115(pNLSAAAA)] and then expressed it in N. benthamiana to test its activity. Figure 5 shows that the mutations did not abolish the ability of PsCRN115 to suppress PsCRN63- or PsojNIP-derived cell death, indicating that the predicted NLS is not required for its function. Significantly, in the region of PsCRN63 (positions 133–411) that is required for triggering cell death, the PsCRN115 sequence differs by only four residues from PsCRN63 (Supplemental Fig. S1). Despite the fact that PsCRN63 and PsCRN115 are nearly identical in sequence, the two proteins have contrasting activities and differential requirements for the NLS. We propose that PsCRN115 and PsCRN63 may share the same molecular host targets that are involved in the cell death signal transduction pathway and that their differential activities are dependent on plant nuclear localization or not.
Generation of Transformants with Silenced PsCRN63 and PsCRN115
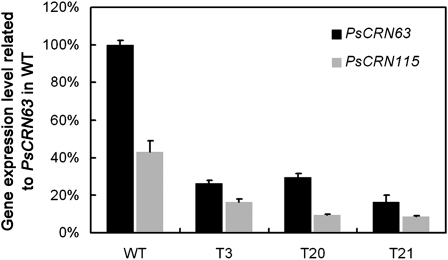
To obtain transformants that do not express PsCRN63 and PsCRN115, we used a gene-silencing strategy based on polyethylene glycol-mediated protoplast stable transformation of P. sojae (Judelson et al., 1991; Dou et al., 2008a; McLeod et al., 2008). The construct pTH209 that carries the selectable marker gene for geneticin resistance was used for cotransformation with the construct containing the open reading frame of PsCRN63 driven by the constitutive Ham34 promoter (Judelson et al., 1991). Using established procedures (Dou et al., 2008a, 2008b), three independent silenced transformants were identified from 25 putative transformants that could grow on selection medium containing 50 μg mL−1 geneticin (Shanghai Sangon BS723). In the three transformants, T3, T20, and T21, expression levels of PsCRN63 were 26.49%, 29.35%, and 16.32% relative to the wild type, respectively (Fig. 6). Meanwhile, we assessed the expression levels of PsCRN115 because it shares high sequence similarity with PsCRN63. Figure 6 shows that the expression level of PsCRN115 was also impaired, and the silencing of this gene was strongly correlated to that of PsCRN63. These results indicate that the transformants T3, T20, and T21 are deficient in mRNA accumulation for both PsCRN63 and PsCRN115.
Figure 6.
Generation of transformants with silenced PsCRN63 and PsCRN115 genes. Relative expression of PsCRN63 and PsCRN115 in silenced lines T3, T20, and T21 and the wild type (WT) is shown. The expression of PsCRN63 in the wild type was set as 100%. Each bar represents the mean of three independent experiments with se.
Silenced Lines Exhibit Reduced Virulence on Soybean
To examine the pathogenicity of the silenced transformants, we inoculated etiolated soybean seedlings (Dong et al., 2009) of cv Williams, which is susceptible to most strains of P. sojae, including P6497. The expression deficiency of these two CRN effectors resulted in a significant reduction of virulence (Fig. 7). Slow-spreading necrotic lesions (0.5 ± 0.08 to 0.9 ± 0.29 cm) developed on plant hypocotyls inoculated with the CRN gene-silenced transformants, whereas rapidly spreading water-soaked lesions (3.45 ± 0.42 cm) were observed on plants inoculated with the wild type (Fig. 7).
Figure 7.
Reduced virulence in silenced lines on soybean. A, Etiolated seedlings of the susceptible soybean cv Williams were inoculated with the wild type (WT) and the silenced line (SL) T21. The inoculated seedlings were photographed at 48 hpi. The experiments were repeated four times in all mutants with similar results, and only one silenced line (T21) is shown as an example. B, Lesion lengths of inoculated sites measured 48 h after inoculation.
Reduction of the Ability in Silenced Lines to Suppress Host Cell Death and Callose Deposition
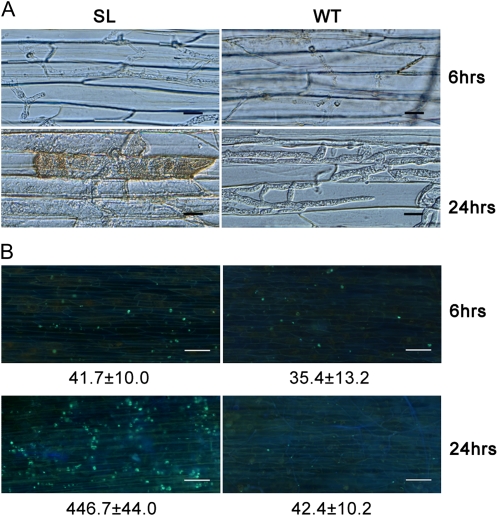
To analyze which stage of pathogen development is affected, we used an inverted microscope to visualize hyphae in infected tissue. At 6 hpi, hyphae of silenced line T21 and of the wild type could be equally well detected in epidermal cells (Fig. 8A). Infectious hyphae of the wild type grew actively and occupied neighboring primary infected cells by 24 hpi. However, infectious hyphae of the silenced lines were mostly restricted to primary infected cells, and only a few infectious hyphae extended into neighboring cells, where cell death was observed (Fig. 8A). Callose deposition was also observed under the same conditions. At 6 hpi, soybean epidermal cells infected by T21 and the wild type showed similar low or undetectable levels of callose deposition. However, a strong signal of callose deposition was only found at 24 hpi in soybean epidermal cells inoculated with T21 (Fig. 8B).
Figure 8.
Silenced lines could not suppress host cell death and callose deposition. A, Microscopic observations of invasive hyphae (top panels) and cell death (bottom panels) in soybean root epidermal cells. Bars = 20 μm. B, Callose deposition in soybean root epidermal cells detected with aniline blue staining. Bars = 100 μm. In A and B, the experiments were repeated four times in all mutants with similar results; only one silenced line (T21) is shown as an example, and the average number of callose deposits per microscopic field of 1 mm2 was calculated using the ImageJ software. SL, Silenced line; WT, wild type.
Since the silenced lines apparently lost the ability to suppress host cell death and defense responses, we further determined whether the silencing of the two CRN effectors interfered with ETI. The wild-type P. sojae strain (P6497) is virulent on soybean cultivars without any known resistance genes (rps) or those carrying Rps1b, but it is avirulent on soybean cultivars carrying Rps1a, Rps1c, Rps1d, Rps1k, Rps2, Rps3a, Rps3b, Rps3c, Rps4, Rps5, or Rps6 (Table II). Two silenced lines (T3 and T21) were determined to be virulent on Rps1b- or rps-containing soybean seedlings, although development of the susceptible phenotype was delayed compared with plants inoculated with the wild type. In other incompatible reactions, the silenced transformants and wild-type strains remained avirulent (Table II). These results indicate that a reduction in expression of the genes of PsCRN63 and PsCRN115 does not interfere with ETI, at least for the interactions of the tested Avr-Rps genes.
Table II. The phenotypes of different soybean cultivars inoculated with the silenced lines (T3 and T21) and the wild type.
| Soybean Cultivara | T3 | T21 | Wild Type | |||
| Harlon (Rps1a) | 1/8b | Rc | 0/10 | R | 2/10 | R |
| L77-1863 (Rps1b) | 15/20 | Sd | 13/20 | S | 19/20 | S |
| Williams79 (Rps1c) | 0/10 | R | 0/9 | R | 0/11 | R |
| PI103091 (Rps1d) | 1/10 | R | 0/10 | R | 1/9 | R |
| Williams82 (Rps1k) | 0/8 | R | 0/10 | R | 0/10 | R |
| L76-1988 (Rps2) | 1/8 | R | 1/9 | R | 1/10 | R |
| Chapman (Rps3a) | 1/10 | R | 0/9 | R | 1/9 | R |
| PRX146-36 (Rps3b) | 0/9 | R | 0/10 | R | 0/9 | R |
| PRX145-48 (Rps3c) | 1/8 | R | 0/9 | R | 1/8 | R |
| L85-2352 (Rps4) | 3/18 | R | 1/16 | R | 4/17 | R |
| L85-3059 (Rps5) | 0/10 | R | 0/10 | R | 0/9 | R |
| Harosoy62xx (Rps6) | 1/10 | R | 0/10 | R | 1/10 | R |
| Williams (rps) | 16/20 | S | 11/17 | S | 21/21 | S |
Soybean cultivars contain 14 soybean differentials of P. sojae physiological races; in parentheses are the genes carrying resistance.
Values shown refer to numbers of plants killed/numbers of all inoculated plants.
R (resistance) ratio below 40%.
S (susceptible) ratio more than 60%.
DISCUSSION
We have examined two CRN effectors of P. sojae with high sequence similarity and high expression levels. We demonstrate that these two proteins cause opposite effects when expressed in plant cells: PsCRN63 induces cell death, while PsCRN115 suppresses cell death induced by PsojNIP or PsCRN63. Next, we showed that the C-terminal fragment of each protein was necessary and sufficient for their activities when the proteins were transiently expressed in N. benthamiana, supporting the hypothesis that oomycete CRN effectors are translocated into host cells. We further showed that the predicted NLS was required for PsCRN63 to trigger cell death but not for PsCRN115 to suppress cell death. We obtained P. sojae transformants with expression deficiencies in both genes and showed that the silenced lines had significantly reduced infection ability. The silenced lines could penetrate host cells but had lost the abilities to suppress cell death and callose deposition. From these results, we inferred that the two effectors were critical to pathogenesis by modulating host defenses.
P. sojae, as well as other oomycete pathogens, contains a vast repertoire of effectors predicted to act in the host cytoplasm (Tyler et al., 2006; Haas et al., 2009). RXLR and CRN effectors are two major classes of such effectors and are both identified by conserved motifs following the signal peptide (Kamoun, 2007). So far, three Phytophthora genome sequences have been reported, P. sojae, P. ramorum, and P. infestans (Tyler et al., 2006; Haas et al., 2009). Each encodes over 350 RXLR effectors (Jiang et al., 2008; Haas et al., 2009). There are 202, 61, and 451 CRN effectors in P. sojae, P. ramorum, and P. infestans, respectively (Tyler et al., 2006; Haas et al., 2009). The RXLR and CRN effector families show extensive expansion and diversification subsequent to speciation (Jiang et al., 2008; Haas et al., 2009). At least two lines of evidence suggest that these genes may have overlapping or redundant functions: first, many avirulence genes are absent in various strains of P. sojae and P. infestans that nonetheless remain fully virulent on their host plants (Tyler, 2009); second, less than 25% of the RXLR effector family has close homologs in other genomes (Win et al., 2007; Jiang et al., 2008). Nevertheless, we showed that the two effectors were critical for pathogenesis. This finding, together with that of Bos et al. (2010) that the RXLR effector Avr3a is required for P. infestans infection, suggests that despite these huge repertoires, several effectors may have indispensable functions.
Plants have evolved two layers of defense against infection by pathogenic microbes. Recognition of PAMPs initiates PTI, while specific pathogen effector molecules initiate ETI (Chisholm et al., 2006; Jones and Dangl, 2006). Although ETI is usually an accelerated and magnified defense response compared with PTI, both may be accompanied by cell death of infected cells, referred to as the HR. The HR is an effective and ultimate defense mechanism against obligate biotrophic and hemibiotrophic pathogens (Clem, 2007). However, delayed HR could be counterproductive and benefit hemibiotrophic pathogens in necrotrophic growth stages. P. sojae is a hemibiotroph and typically switches from biotrophic to necrotrophic growth 16 to 24 h following the invasion of host tissue (Enkerli et al., 1997). Therefore, the ability of P. sojae to suppress or delay the HR of soybean tissue is likely a major component of its pathogenic strategy (Tyler, 2009). Here, we show that two CRN effectors are important factors for P. sojae to manipulate host HR.
CRN family effectors are named such because they can induce necrosis and leaf crinkling in plants (Torto et al., 2003). Surprisingly, we demonstrated that PsCRN115 could block cell death induced by PsojNIP and PsCRN63, indicating that this family of effectors also has similar abilities to RXLR effectors in suppressing plant defense (Bos et al., 2006; Dou et al., 2008a). Wild-type P. sojae can successfully invade compatible soybean cultivars, and only weak or no cell death symptoms are observed in the early infection stage (Chen et al., 2008). Since infection of PsCRN63/115-deficient transformants was hindered by premature host cell death under the same conditions, it appears that cell death suppression was severely weakened in the silenced lines. Considering that PsCRN63 is a cell death inducer and that PsCRN115 is a cell death suppressor, and that both genes are constitutively expressed, we hypothesize that these two effectors serve as essential suppressors of cell death mediated by PAMPs such as PsojNIP (Qutob et al., 2002). However, we cannot exclude other possibilities that may cause the altered host responses we observed in plants infected with the CRN-silenced transformants.
We show that PsCRN63 is a cell death inducer. Our results, together with deletion analysis of PiCRN2 mutants (Haas et al., 2009), indicate that the N termini of CRN proteins, including the signal peptide, are not required to trigger cell death. This result provides indirect evidence to support the hypothesis that CRNs are cytoplasmic effectors (Kamoun, 2007). We also demonstrate that an intact NLS is required for cell death-inducing processes of PsCRN63, suggesting that this protein is not only translocated inside the host cell but also localized to the nucleoplasm during infection. The PsCRN63 protein shares 54% sequence identity with PiCRN2 at the amino acid sequence level, and no obvious NLS has been found in PiCRN2 (Torto et al., 2003). It is not known whether PiCRN2 contains a cryptic signal for nuclear localization or whether the cell death triggered by PiCRN2 and PsCRN63 occurs in different host cellular compartments. Considering that several R gene-mediated cell death responses, such as barley (Hordeum vulgare) MLA (Shen et al., 2007), Nicotiana tabacum N (Burch-Smith et al., 2007), and Arabidopsis (Arabidopsis thaliana) RPS4 (Gassmann et al., 1999), have been traced to the nucleus, we suggest that PsCRN63 might be recognized by plant R genes, resulting in ETI, albeit weaker or slower than the ETI mediated by pathogen avirulence genes and their cognate plant R genes. The signal involved in this process and the role of PsCRN115 are currently under investigation.
Many secreted proteins from Phytophthora have been identified as modulators of host cell death. Proteins secreted into the host apoplast, such as elicitins (Huitema et al., 2005) and PsojNIP (Qutob et al., 2002), can trigger cell death and defense, while selected RXLR effectors (Bos et al., 2006; Dou et al., 2008a) and the P. infestans SNE1 protein (Kelley et al., 2010) have the ability to suppress cell death and defense. The P. infestans RXLR effector appears to play both roles (Bos et al., 2010), as is required for full virulence. In analogous fashion, PsCRN63 and PsCRN115 have contrasting roles in manipulating plant cell death. Expression of PsCRN63 and PsCRN115 is jointly required for full pathogenesis. Our functional investigations and discoveries contribute to the understanding of how Phytophthora and other oomycete pathogens invade plants and defeat their immune systems.
In addition, the concept of accessibility was proposed to describe host cellular conditioning toward a symbiotic relationship induced by compatible pathogens (Ouchi, 1983, 2006), based on the observations that the susceptibility of a plant to an incompatible pathogen and nonpathogen could be induced by prior inoculation with a compatible pathogen (Ouchi et al., 1974). Thus, it has been assumed that compatible pathogens have evolved a variety of strategies to establish a symbiotic relationship with the host, such as suppressing defense responses in the host cell (Ouchi, 1983, 2006). Our findings that CRN effectors, as well as RXLR effectors (Bos et al., 2006; Dou et al., 2008a; Sohn et al., 2007; Kelley et al., 2010), can modulate host defense circuitry and benefit parasite colonization fit the concept of accessibility.
MATERIALS AND METHODS
Sequence Search, Structure Prediction, and Phylogenetic and Molecular Evolution Analyses
The 202 Phytophthora sojae CRN effectors (Tyler et al., 2006; Haas et al., 2009) were renamed by the order of localization within the different scaffolds (Tyler et al., 2006). The PsCRN63, PsCRN77, PsCRN84, PsCRN86, and PsCRN115 genes were obtained by a TBLASTN search (Altschul et al., 1997) using the query sequence PiCRN2 (Torto et al., 2003) against the gene models of P. sojae. Then, PsCRN63 was used in TBLASTN searches against the gene models of Phytophthora ramorum and Phytophthora infestans (Tyler et al., 2006; Haas et al., 2009) with an E value cutoff of E < 10−120. To obtain the theoretical protein sequences encoded by their nonpseudogene progenitors, the sequences of three pseudogenes, PsCRN77, PsCRN84, and PsCRN86, were adjusted by removal of one or more stop codons or by correcting frame shifts by deleting one or two nucleotides.
Signal peptide scores for each gene were predicted by the SignalP3.0 server (Bendtsen et al., 2004), and NLS was analyzed by the Predict NLS server (Cokol et al., 2000). Two conserved motifs following the signal peptide, FLAK and HVLVVVP (Haas et al., 2009), were searched by sequence similarity. We reconstructed the phylogenetic tree of 11 CRN genes using MEGA 4.1 (Tamura et al., 2007) using neighbor joining, 1,000 replicates, and the pairwise-deletion option. The sequences were aligned by MUSCLE 3.6 (Edgar, 2004).
Plasmids and Strain Construction
The oligonucleotides used for the following plasmid constructions are documented in Supplemental Table S1. For the PVX assay, fPsCRN63 (full length of PsCRN63), PsCRN63, PsojNIP and PsCRN115 were amplified using combinations of oligonucleotide primers PsCRN63-F1 and PsCRN63-R, PsCRN63-F2 and PsCRN63-R, PsojNIP-F and PsojNIP-R, and PsCRN115-F2 and PsCRN63-R, respectively. Then, the amplicons were cloned using appropriate restriction enzymes (Supplemental Table S2) into the PVX vector pGR107 (Lu et al., 2003). For the PsCRN63 deletions, the mutants were amplified using combinations of oligonucleotide primers (Supplemental Tables S1 and S2).
To make the sense construct of pHamPsCRN63 (driven by the Ham34 promoter; Judelson et al., 1991), we used fPsCRN63 in the vector of pGR107 as template to amplify PsCRN63 using PrimeSTAR HS DNA Polymerase (Takara code DR010A) with the primers PsCRN63-F1 and PsCRN63-R. The PCR product was inserted into SmaI-digested pTH210 (Wang et al., 2009; Judelson et al., 1991).The constructs were screened and confirmed by sequencing. The other constructs are shown in Supplemental Table S2. All the above plasmids were validated by sequencing by GenScript.
SYBR Green Real-Time RT-PCR Assay
Total RNA was isolated from the hyphae and the germinating cysts using NucleoSpin RNA II (Macherey-Nagel) following the manufacturer’s protocol. The integrity of total RNA was confirmed by agarose gel electrophoresis. The RNA was quantified using a spectrophotometer (Nanodrop ND-1000). To remove contaminating genomic DNA in RNA preparations, 10 μg of total RNA was treated with 4 units of RNase-free DNase I (Takara) at 37°C for 30 min. The removal of DNA was verified under the same conditions as those used for the RT-PCR, except that the 30-min cDNA synthesis step at 37°C was omitted. First-strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (RNase free) and oligo(dT)18 primer (Invitrogen). Quantitative RT-PCR was performed in 20-μL reactions including 20 ng of cDNA, 0.2 μm gene-specific primer or reference actin gene (Supplemental Table S1), 10 μL of SYBR Premix ExTaq (Takara), and 6.8 μL of deionized water. PCR was performed on a ABI PRISM 7300 Fast Real-Time PCR System (Applied Biosystems) under the following conditions: 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 31 s to calculate cycle threshold values, followed by a dissociation program of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s to obtain melt curves. The 7300 System Sequence Detection Software (version 1.4; SDS) was used to obtain relative expression levels of each sample.
P. sojae Strains, Manipulation, and Inoculation Assays
P. sojae reference strain P6497 (race 2; Förster et al., 1994) was routinely grown and maintained on V8 agar (Erwin and Ribiero, 1996). P. sojae transformation was carried out as described (Dou et al., 2008a). Putative P. sojae transformants were screened for PsCRN63 transgenes by amplifying genomic DNA from each line as described previously (Dou et al., 2008a). For screening transformants for silencing of the PsCRN63 gene, total RNA was extracted from mycelia with the above methods. The silenced transformants were identified by quantitative real-time RT-PCR assay following the same steps as above.
The pathogenicity phenotypes of selected transformants were determined by inoculation of etiolated soybean (Glycine max) seedlings (Dong et al., 2009) and by hypocotyl inoculation (Tyler et al., 1995). For inoculation of etiolated soybean seedlings, cv Williams (rps) was used. The lesion diameter was measured as lesion length for quantitative virulence of P. sojae transformants. There were three replications of five to 10 plants each within each experiment. For light microscopy, after inoculation of P. sojae, the epidermis of hypocotyls of etiolated soybean seedlings was detached using an Olympus 1X71 inverted microscope to visualize hyphae in infected tissue. For callose staining, after the inoculation, the hypocotyls of etiolated soybean seedlings was stained with aniline blue for 30 min, and then the epidermal cells were observed with an Olympus 1X71 inverted fluorescence microscope. The average number of callose deposits per microscopic field of 1 mm2 was calculated from all the tested hypocotyls using the ImageJ software (http://www.uhnresearch.ca/wcif).
For hypocotyl inoculation, the following differential cultivars were used: Harlon (Rps1a), L77-1863 (Rps1b), Williams79 (Rps1c), PI103091(Rps1d), Williams82 (Rps1k), L76-1988 (Rps2), Chapman (Rps3a), PRX146-36 (Rps3b), PRX145-48 (Rps3c), L85-2352 (Rps4), L85-3059 (Rps5), Harosoy62xx (Rps6), and Williams (rps). Seedlings of each soybean cultivar were grown in the greenhouse as described (Dou et al., 2008b) for 7 d. Each virulence determination was repeated at least three times. A strain was considered avirulent if more than 60% of the inoculated seedlings survived.
Agrobacterium tumefaciens Infiltration Assays
The A. tumefaciens infiltration assays were performed as described by Dou et al. (2008a), except that A. tumefaciens strain GV3101 (Hellens et al., 2000) was used. For infiltration, recombinant strains were cultured in Luria-Bertani medium supplemented with 50 mg mL−1 kanamycin in a test tube at 28°C to 30°C and 220 rpm for 48 h. The cells were collected by centrifugation (3,000g, 5 min), washed three times in 10 mm MgCl2, and then resuspended in 10 mm MgCl2 to an optical density at 600 nm of 0.4 to 0.6. Infiltration experiments were performed on 7- to 8-week-old Nicotiana benthamiana plants. Plants were grown and maintained throughout the experiments in a greenhouse with an ambient temperature of 22°C to 25°C and high light intensity under a 16-h/8-h light/dark photoperiod. For cell death induction experiments, A. tumefaciens cell solutions carrying the respective constructs were infiltrated into N. benthamiana leaves by pressure infiltration: a small nick was placed in each leaf with a needle, and then 30 to 50 μL of cell suspension was infiltrated through the nick using a syringe without a needle. For cell death suppression experiments, A. tumefaciens cell solutions carrying the PsCRN115, Avh331 (positive control), and gfp (negative control) constructs were infiltrated following the above method. A. tumefaciens cells carrying the cell death-inducing genes (PsCRN63, PsojNIP, Bax, and the combination of Avr3a and R3a) were infiltrated into the same site 12 h later. Symptom development was monitored from 4 to 8 d after infiltration, and photographs were taken after 5 d. The experiments were repeated at least three times. Although it is likely that PVX replication occurred in the transformed plant cells, resulting in amplified expression of the genes in the PVX vector, no attempt was made to quantitate PVX replication (Dou et al., 2008a).
Particle Bombardment Assays
Particle bombardment assays were performed using a double-barreled extension of the Bio-Rad He/1000 particle delivery system (Dou et al., 2008a, 2008b; Kale et al., 2010). Analyzing the bombardment data as a ratio between the test and control shots improves the reproducibility of the measurements greatly (Dou et al., 2008a, 2008b; Kale et al., 2010).
The cell death-inducing activity of PsCRN63 and PsCRN115 constructs was measured as the reduction in the number of blue spots comparing the PsCRN63/115 + GUS bombardment with GUS + GFP bombardment. The cell death suppression activity of PsCRN115 was measured in two ways: (1) the number of blue spots comparing the PsCRN115 + cell death inducer (PsojNIP or PsCRN63) + GUS bombardment with the cell death inducer (PsojNIP or PsCRN63) + GUS bombardment (direct assay); and (2) the number of blue spots comparing the ratio of PsCRN115 + PsojNIP/PsCRN63 + GUS bombardment with the GFP + GUS bombardment control with the ratio of the PsojNIP/PsCRN63 + GUS bombardment with the GFP + GUS bombardment control (indirect assay). For each paired shot, the logarithm of the ratio of the spot numbers of PsCRN63 or PsCRN115 to that of the control was calculated. P values was calculated using the Wilcoxon rank sum test for the indirect assay results and the Wilcoxon signed rank test for the direct assay results (Dou et al., 2008a, 2008b; Kale et al., 2010).
The nucleotide sequences of PsCRN63 and PsCRN115 have been submitted to GenBank with accession numbers HQ231783 and HQ231784, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Text file of unmasked alignment corresponding to the phylogenetic tree in Figure 1.
Supplemental Figure S2. PsCRN63 triggers cell death when expressed from plant expression vector pCHF3.
Supplemental Table S1. Oligonucleotides used for PCR and plasmid construction.
Supplemental Table S2. Description of plasmids used.
Supplementary Material
Acknowledgments
We gratefully acknowledge Mark Gijzen (Agriculture and Agri-Food Canada) and Brett Tyler (Virginia Bioinformatics Institute) for editing and comments on the manuscript. We thank Sophien Kamoun (The Sainsbury Laboratory) for the constructs of R3a/Avr3a.
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Böhme U, Brooks K, Cherevach I, et al. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA 102: 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Hiller NL, Liolios K, Win J, Kanneganti TD, Young C, Kamoun S, Haldar K. (2006) The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog 2: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Guttman JA, Finlay BB. (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449: 827–834 [DOI] [PubMed] [Google Scholar]
- Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JI, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PR, Kamoun S. (2006) The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J 48: 165–176 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XR, Wang XL, Zhang ZG, Wang YC. (2008) Differences in the induction of the oxidative burst in compatible and incompatible interactions of soybean and Phytophthora sojae. Physiol Mol Plant Pathol 73: 16–24 [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clem RJ. (2007) Baculoviruses and apoptosis: a diversity of genes and responses. Curr Drug Targets 8: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B. (2000) Finding nuclear localization signals. EMBO Rep 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol 11: 1453–1461 [DOI] [PubMed] [Google Scholar]
- Dong S, Qutob D, Tedman-Jones J, Kuflu K, Wang Y, Tyler BM, Gijzen M. (2009) The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. PLoS ONE 4: e5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X, Jiang RHY, Arredondo FD, Anderson R, Thakur P, et al. (2008a) Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20: 1118–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, Zhang X, Tyler BM. (2008b) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20: 1930–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkerli K, Hahn MG, Mims CW. (1997) Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae. Can J Bot 75: 1494–1508 [Google Scholar]
- Erwin DC, Ribiero OK. (1996) Phytophthora Diseases Worldwide. APS Press, St. Paul [Google Scholar]
- Förster H, Tyler BM, Coffey MD. (1994) Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol Plant Microbe Interact 7: 780–791 [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. (2000) Technical focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estraño C, Haldar K. (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306: 1934–1937 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Huitema E, Vleeshouwers VGAA, Cakir C, Kamoun S, Govers F. (2005) Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol Plant Microbe Interact 18: 183–193 [DOI] [PubMed] [Google Scholar]
- Jiang RH, Tripathy S, Govers F, Tyler BM. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci USA 105: 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Judelson HS, Tyler BM, Michelmore RW. (1991) Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant Microbe Interact 4: 602–607 [DOI] [PubMed] [Google Scholar]
- Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, Rumore A, Arredondo FD, Hanlon R, Fudal I, Rouxel T, et al. (2010) External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2003) Molecular genetics of pathogenic oomycetes. Eukaryot Cell 2: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. (2007) Groovy times: filamentous pathogen effectors revealed. Curr Opin Plant Biol 10: 358–365 [DOI] [PubMed] [Google Scholar]
- Kelley BS, Lee SJ, Damasceno CM, Chakravarthy S, Kim BD, Martin GB, Rose JK. (2010) A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J 62: 357–366 [DOI] [PubMed] [Google Scholar]
- Lacomme C, Santa Cruz S. (1999) Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci USA 96: 7956–7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PR, Visser RG, Jacobsen E, et al. (2009) Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact 22: 630–641 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC. (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306: 1930–1933 [DOI] [PubMed] [Google Scholar]
- McLeod A, Fry BA, Zuluaga AP, Myers KL, Fry WE. (2008) Toward improvements of oomycete transformation protocols. J Eukaryot Microbiol 55: 103–109 [DOI] [PubMed] [Google Scholar]
- Ouchi S. (1983) Induction of resistance or susceptibility. Annu Rev Phytopathol 21: 289–315 [Google Scholar]
- Ouchi S. (2006) A retrospective of an unconventionally trained plant pathologist: plant diseases to molecular plant pathology. Annu Rev Phytopathol 44: 1–17 [DOI] [PubMed] [Google Scholar]
- Ouchi S, Oku H, Hibino C, Akiyama I. (1974) Induction of accessibility to a nonpathogen by preliminary inoculation with a pathogen. J Phytopathol 79: 142–154 [Google Scholar]
- Qutob D, Kamoun S, Gijzen M. (2002) Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J 32: 361–373 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Sohn KH, Lei R, Nemri A, Jones JD. (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19: 4077–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, van West P, Kamoun S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res 13: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM. (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 8: 1–8 [DOI] [PubMed] [Google Scholar]
- Tyler BM. (2009) Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell Microbiol 11: 13–20 [DOI] [PubMed] [Google Scholar]
- Tyler BM, Forster H, Coffey MD. (1995) Inheritance of avirulence factors and restriction-fragment-length-polymorphism markers in outcrosses of the oomycete Phytophthora sojae. Mol Plant Microbe Interact 8: 515–523 [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang M, Bouwmeester K, Vosman B, Visser RG, et al. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3: e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dou D, Wang X, Li A, Sheng Y, Hua C, Cheng B, Chen X, Zheng X, Wang Y. (2009) The PsCZF1 gene encoding a C2H2 zinc finger protein is required for growth, development and pathogenesis in Phytophthora sojae. Microb Pathog 47: 78–86 [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Win J, Morgan W, Bos J, Krasileva KV, Cano LM, Chaparro-Garcia A, Ammar R, Staskawicz BJ, Kamoun S. (2007) Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19: 2349–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.