Abstract
Auxin signaling is central to plant growth and development, yet hardly anything is known about its evolutionary origin. While the presence of key players in auxin signaling has been analyzed in various land plant species, similar analyses in the green algal lineages are lacking. Here, we survey the key players in auxin biology in the available genomes of Chlorophyta species. We found that the genetic potential for auxin biosynthesis and AUXIN1 (AUX1)/LIKE AUX1- and P-GLYCOPROTEIN/ATP-BINDING CASSETTE subfamily B-dependent transport is already present in several single-celled and colony-forming Chlorophyta species. In addition, our analysis of expressed sequence tag libraries from Coleochaete orbicularis and Spirogyra pratensis, green algae of the Streptophyta clade that are evolutionarily closer to the land plants than those of the Chlorophyta clade, revealed the presence of partial AUXIN RESPONSE FACTORs and/or AUXIN/INDOLE-3-ACETIC ACID proteins (the key factors in auxin signaling) and PIN-FORMED-like proteins (the best-characterized auxin-efflux carriers). While the identification of these possible AUXIN RESPONSE FACTOR- and AUXIN/INDOLE-3-ACETIC ACID precursors and putative PIN-FORMED orthologs calls for a deeper investigation of their evolution after sequencing more intermediate genomes, it emphasizes that the canonical auxin response machinery and auxin transport mechanisms were, at least in part, already present before plants “moved” to land habitats.
In plants, the conquest of terrestrial habitats was associated with an expansion of gene families involved in signaling pathways (Lang et al., 2008; Rensing et al., 2008). This includes signaling mediated by the phytohormone auxin, which is a key regulator of growth and development in land plants (Paponov et al., 2009; Vanneste and Friml, 2009), and we concentrated on auxin signaling to address the question of how and when signaling pathways developed in the green plant lineage. Auxin signaling can be split into three aspects: biosynthesis and metabolism (Woodward and Bartel, 2005; Normanly, 2010); directional transport mediated by specific proteins like PIN-FORMED (PIN) efflux carriers, AUXIN1 (AUX1)/LIKE AUX1 (LAX) influx carriers, and ATP-BINDING CASSETTE subfamily B (ABCB)/P-GLYCOPROTEIN (PGP)/MULTIDRUG RESISTANCE (MDR) transporters (Vieten et al., 2007); and, finally, cell- or tissue-specific responses (Lau et al., 2008). Briefly, the auxin response takes place as follows (Chapman and Estelle, 2009). Intracellular auxin is perceived by the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN1-3 (TIR1/AFB1-3) receptors. TIR1 is an integral component of the SKP1/CULLIN/F-BOX PROTEINTIR1 complex that mediates the auxin-dependent ubiquitination of AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressor proteins and thereby destines them for 26S proteasome-dependent degradation. AUX/IAAs, at low auxin concentrations, form dimers with AUXIN RESPONSE FACTOR (ARF) transcription factors, thereby blocking the activity of at least the activating ARFs. When freed from the AUX/IAAs, these ARFs regulate the expression of auxin-responsive genes.
Although the auxin signaling pathways in land plants are nowadays relatively well understood and the respective signaling mechanisms are also present in basal lineages such as bryophytes (Rensing et al., 2008; Paponov et al., 2009), their evolution is much less clear. The auxin signaling pathway has undergone substantial functional diversification and specialization within vascular plants since they diverged from bryophytes, but it is still not clear when certain aspects of auxin biology emerged in evolution. It has been suggested that auxin signaling emerged to coordinate multicellular growth in land plants (Rensing et al., 2008), but particular aspects of auxin biology could of course have an earlier origin. This is suggested by (1) the presence of auxin in various green, red, and brown algae (Sztein et al., 2000; Basu et al., 2002; Lau et al., 2009; Le Bail et al., 2010; Ross and Reid, 2010), (2) the circumstance that auxin seems to play a role in establishing polarity, regulating rhizoid growth, and/or controlling overall growth (Lau et al., 2009), and (3) possible alternative ways to convey an auxin response in single-celled green algae, such as AUXIN BINDING PROTEIN1 (ABP1)- or INDOLE-3-BUTYRIC ACID RESPONSE5 (IBR5)-mediated auxin responses (Lau et al., 2009; Tromas et al., 2010).
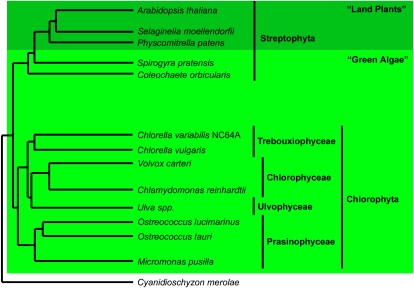
Algae contribute substantially to biological diversity on Earth. They consist of various separate clades, namely green, red, and brown algae, and comprise unicellular and multicellular organisms, which can be colony forming, filamentous, or thalloid (Bhattacharya and Medlin, 1998). The green plants or Viridiplantae evolved in two major lineages, the Chlorophyta and the Streptophyta (Lewis and McCourt, 2004). The Streptophyta comprise several lineages of freshwater green algae and the land plants, and the highly diverse Chlorophyta include all other green algae (McCourt et al., 2004; Fig. 1). In addition to this distinction, making the green algae interesting from an evolutionary point of view (Lewis and McCourt, 2004; Lang et al., 2008), detailed analyses are also appealing in view of the emerging significance of algae for the production of biofuels (Beer et al., 2009; Wijffels and Barbosa, 2010).
Figure 1.
Schematic tree indicating the relationships within the supergroup Plantae. The included species are used in our analyses, except for Ulva species and Cyanidioschyzon merolae (Rhodophyta outgroup). [See online article for color version of this figure.]
Therefore, we investigated all of the above described aspects of auxin biology in the newly available green algal Chlorophyta genomes. Furthermore, we probed EST libraries from two green Streptophyta algae for the presence of the auxin transport and response machineries. Here, we present, to our knowledge for the first time, a comprehensive overview of auxin-related genes in green algae, and we shed some light on the evolution of the genetic information underlying auxin biology.
RESULTS AND DISCUSSION
Algal Species Selected for Analysis
The genomes of the “basal” land plants, such as the moss Physcomitrella patens, already contain members of gene families involved in auxin biosynthesis, metabolism, transport, and signaling (Rensing et al., 2008; Paponov et al., 2009). However, since very few nuclear green algal genomes have been described so far, and with the additional caveat that those fully sequenced and annotated genomes are almost exclusively from single-celled species, it is not clear when these gene families emerged. Here, we largely focused our comprehensive analysis on the genomes of single-celled or colony-forming Chlorophyta from three different classes (Fig. 1): Ostreococcus tauri (Prasinophyceae; Derelle et al., 2006), Ostreococcus lucimarinus (Prasinophyceae; Palenik et al., 2007), Micromonas pusilla strain CCMP1545 and strain RCC299 (Prasinophyceae; Worden et al., 2009), Chlamydomonas reinhardtii (Chlorophyceae; Merchant et al., 2007), Volvox carteri (Chlorophyceae), Chlorella vulgaris (Trebouxiophyceae), and Chlorella variabilis NC64A (Trebouxiophyceae; Blanc et al., 2010). While most of these species are unicellular, V. carteri is multicellular (colony forming) and serves as a model for the evolution of multicellularity and differentiation (Kirk, 1999; Kirk and Nishii, 2001; Schmitt, 2003). Moreover, for the analyses of auxin transport and response, we included EST libraries for green algae members of the Streptophyta (Fig. 1): the multicellular, unbranched filamentous Spirogyra pratensis (Zygnematophyceae) and the complex circular thalloid or disc-shaped Coleochaete orbicularis (Coleochaetophyceae; Timme and Delwiche, 2010). In our analyses, we also included P. patens (Bryophyta; Rensing et al., 2008) and Selaginella moellendorffii (Lycopodiophyta), representing those land plants with fully sequenced genomes closest to the green algae (Lang et al., 2008).
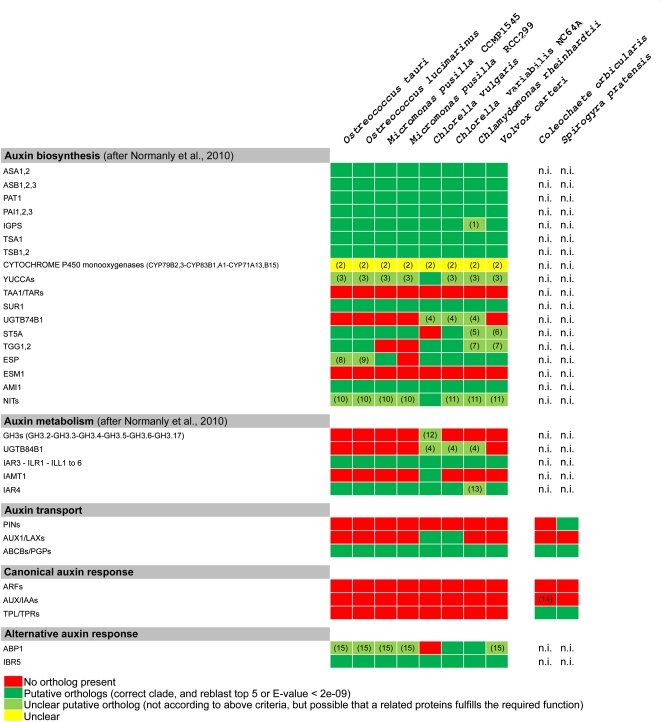
To detect putative orthologs for proteins involved in auxin signaling in Chlorophyta and Streptophyta, we used a combination of reciprocal BLAST and phylogenetic tree analyses. We blasted the respective protein sequences from Arabidopsis (Arabidopsis thaliana) against a database containing all predicted proteins of the respective green algae, identified the potential orthologs using Unweighted Pair Group Method with Arithmetic mean (UPGMA) tree analyses, and then compared the sequences of the closest putative orthologs with a database of Arabidopsis proteins (The Arabidopsis Information Resource 9; http://www.arabidopsis.org/Blast/index.jsp; for details, see “Materials and Methods”). The results of these analyses are discussed below and summarized in Figure 2.
Figure 2.
Scheme indicating the presence or absence of putative orthologs of various auxin-related genes that were investigated in Chlorophyta species. The color code is explained in the legend. Notes in squares are as follows: (1) related based on tree, but unrelated based on ReBlast; (2) difficult to pinpoint precise orthologs, but clear orthologs very likely absent; (3) ReBlast with good E-value, but not in correct clade or outgrouped; (4) difficult to assess, but very likely a putative ortholog present; (5) wrong clade, but ReBlast position 2, E-value = 1.7; (6) related to distant orthologs from C. reinhardtii; (7) good ReBlast with E-value < 2e-09; (8) outgrouped, but with ReBlast E-value = 3e-05; (9) outgrouped, but with ReBlast E-value = 6e-07; (10) closely related to PUTATIVE NITRILASE (AT4G08790); (11) closely related to NITRILASE-LIKE PROTEIN1 (NLP1; AT2G27450); (12) outgrouped, ReBlast only hit, but E-value = 2.2; (13) outgrouped, but with ReBlast E-value = 1e-30 and first hit; (14) putative ancestral, incomplete AUX/IAA similar to AtIAA33; (15) fall within clade, in ReBlast top five but very poor E-value. n.i., Presence of putative orthologs not investigated.
Auxin Biosynthesis and Metabolism in Chlorophyta
As summarized recently, various measurements showed that auxin is present in several kingdoms and in a wide range of organisms, including several single-celled and multicellular green algae (Cooke et al., 2002; Lau et al., 2009; Ross and Reid, 2010). Recently, an in silico survey of components of auxin signaling, such as biosynthesis, conjugation, response, and transport, showed that IAA biosynthesis genes from land plants have orthologs in the brown alga Ectocarpus siliculosus (Cock et al., 2010; Le Bail et al., 2010), and even several bacteria are able to produce auxin (Costacurta and Vanderleyden, 1995). However, so far, there has not been genetic support for the biosynthesis and metabolism of auxin in green algae. Therefore, we probed the available Chlorophyta genomes for the presence of an auxin biosynthesis pathway. While identifying putative orthologs for various enzymes involved in the auxin biosynthesis and metabolism pathway (Figs. 2 and 3), one has to keep in mind that such proteins, although biochemically and evolutionarily related to the respective Arabidopsis homologs, could have a very different function in Chlorophyta.
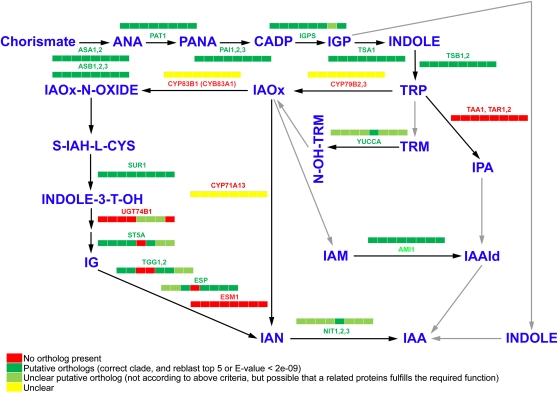
Figure 3.
Auxin biosynthesis as described by Normanly (2010) with absence/presence of key components in Chlorophyta. From left to right on the color coded bars: O. tauri, O. lucimarinus, M. pusilla CCMP1545, M. pusilla RCC299, C. vulgaris, C. variabilis NC64A, C. reinhardtii, and V. carteri. The color code is explained in the legend.
iaaM and iaaH
We first investigated if the Chlorophyta contained the two important enzymes from bacteria that convert Trp into the auxin IAA. The iaaM gene codes for a Trp-2-monoxygenase enzyme that converts Trp to indole-3-acetamide, which is then hydrolyzed to the auxin IAA by iaaH (Lehmann et al., 2010). However, we could not identify any orthologs of iaaM and iaaH in the investigated Chlorophyta (data not shown).
Chlorophyta Orthologs of the Auxin Biosynthesis Genes
Using up-to-date overviews of the molecular components of auxin biosynthesis and metabolism as a template (Normanly, 2010; Zhao, 2010), we investigated the presence of orthologs of the known auxin biosynthesis proteins from land plants in Chlorophyta (Figs. 2 and 3; Supplemental Figs. S1–S17). First of all, the key enzymes that convert the important precursor chorismate into indole and Trp in land plants had clear orthologs in the investigated Chlorophyta (Figs. 2 and 3; Supplemental Fig. S1–S7). Recently, members of a small family of aminotransferases with strong sequence similarity to C-S lyases were shown to be essential in the indole-3-pyruvic acid branch of the Trp-dependent auxin biosynthetic pathway (Stepanova et al., 2008). Orthologs of these proteins are present in P. patens and S. moellendorffii but were not identified in the investigated Chlorophyta (Figs. 2 and 3; Supplemental Fig. S8). Notwithstanding that a number of algal proteins display similarities to TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and TRYPTOPHAN AMINOTRANSFERASE RELATED1 to -4 (TAR1–TAR4), this is most likely based on their common aminotransferase nature. The latter appear to be more closely related to, for example, the ALANINE AMINOTRANSFERASE1 (Supplemental Fig. S8).
The SUPERROOT1 (SUR1) protein, which converts S-IAH-l-Cys into indole-3-T-OH (Mikkelsen et al., 2004), was retrieved in the investigated Chlorophyta (Figs. 2 and 3; Supplemental Fig. S9). Following the activity of SUR1, proteins like UDP-glucosyl transferase 74B1 (UGT74B1), SULFOTRANSFERASE5A (ST5A), THIOGLUCOSIDE GLUCOHYDROLASE1 (TGG1), TGG2, epithiospecifier (ESP), and epithiospecifier modifier 1 (ESM1) are involved in the conversion of indole-3-T-OH to indole-3-acetonitrile (IAN; Normanly, 2010). No clear orthologs of UGT74B1 (Jackson et al., 2001, 2002) were identified in most of the investigated Chlorophyta, but in C. reinhardtii and Chlorella species, putative orthologs might be present (Figs. 2 and 3; Supplemental Fig. S10). However, the enormous diversification of this family (Ross et al., 2001) hampers the identification of true orthologs. For ST5A, putative orthologs are present in most investigated Chlorophyta species, except C. vulgaris (Figs. 2 and 3; Supplemental Fig. S11). Putative orthologs for TGG1 and TGG2 were present in all the investigated Chlorophyta, except in Micromonas species (Figs. 2 and 3; Supplemental Fig. S12). While putative ESP orthologs could be identified in all Chlorophyta, apart from Micromonas strain RCC299, no ESM1 orthologs were identified in the various Chlorophyta (Figs. 2 and 3; Supplemental Figs. S13 and S14).
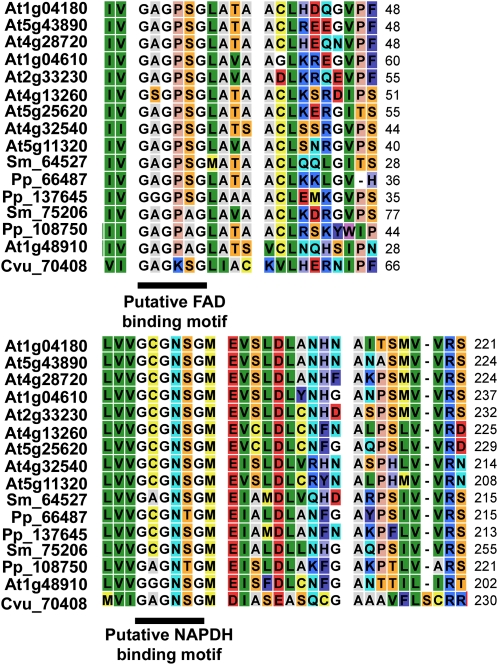
Another key group of proteins in auxin biosynthesis are the YUCCAs (YUCs), which are widespread in plants and which are thought to encode the enzyme for converting tryptamine to N-hydroxytryptamine (Zhao, 2008, 2010). Only one of the investigated green algal species, namely C. vulgaris, encodes a putative functional YUC, since the putative FAD-binding (GAGPSG) and NADPH-binding (GCGNSG) motifs near the N terminus and in the middle of the protein, respectively (Zhao et al., 2001), are largely conserved (Figs. 2–4; Supplemental Fig. S15). In the other Chlorophyta species, we found several proteins that are related to proteins from the flavin-containing monooxygenase family (Hao et al., 2009), for which a YUC family member is retrieved in the reciprocal blast and for which at least one of the above-mentioned critical motifs is partially present; however, these appear to clearly group outside the YUC clade (Figs. 2 and 3; Supplemental Fig. S15).
Figure 4.
Conserved motifs and residues in YUC proteins. Alignment of the FAD-binding and NADPH-binding motifs in the putative YUC in C. vulgaris is shown. Residue colors are according to the rasmol color scheme, which is consistent with traditional amino acid properties. At, Arabidopsis; Cvu, C. vulgaris; Pp, P. patens; Sm, S. moellendorffii. [See online article for color version of this figure.]
Also for the amidohydrolase that is responsible for the in vitro conversion of indole-3-acetamide into IAA, namely AMIDASE1 (AMI1; Lehmann et al., 2010), putative orthologs could be identified in Chlorophyta (Figs. 2 and 3; Supplemental Fig. S16).
Several members of the CYTOCHROME P450 family play important roles in auxin biosynthesis (Mizutani and Ohta, 2010; Normanly, 2010). However, no clear orthologs of the CYTOCHROME P450 MONOOXYGENASES that play a role in auxin biosynthesis in Arabidopsis could be identified (Figs. 2 and 3; data not shown), which is further supported by the absence of these proteins in P. patens and S. moellendorffii (Mizutani and Ohta, 2010).
NITRILASES (NITs) are thought to convert IAN into IAA (Normanly, 2010). Although several NIT-like proteins are present in Chlorophyta, only C. vulgaris appears to contain a clear ortholog of NIT1, NIT2, and NIT3 (Figs. 2 and 3; Supplemental Fig. S17).
In conclusion, it appears that although not all genetic pathways known from land plants for auxin biosynthesis are represented in the investigated Chlorophyta, based on the genome evidence, they very likely can produce auxin from IAN and possibly even from AMI1-dependent production of indole-3-acetaldehyde.
Orthologs for Auxin Metabolism Genes from Land Plants
Various pathways have a (potential) role in auxin metabolism, the formation of conjugates, and the release of IAA from these conjugates (Normanly, 2010). In addition, the formation and hydrolysis of IBA conjugates may also contribute to IAA homeostasis (Normanly, 2010).
Our extended survey showed that orthologs of the IAA-amino acid amidohydrolases IAA-LEUCINE RESISTANT1 (ILR1), IAA-ALANINE RESISTANT3 (IAR3), and ILR-LIKE1 (ILL1) to ILL6 are represented in the investigated Chlorophyta (Fig. 2; Supplemental Fig. S18).
On the other hand, for GH3s, a family of enzymes involved in IAA conjugate formation (Staswick et al., 2005; Woodward and Bartel, 2005; Normanly, 2010), no obvious orthologs appear to be present in the investigated Chlorophyta, except for one unlikely candidate in C. vulgaris (Fig. 2; Supplemental Fig. S19).
For the putative pyruvate dehydrogenase E1 α-subunit encoded by IAR4, which was proposed to be involved in some aspect of auxin homeostasis (Quint et al., 2009), orthologs are present in the investigated Chlorophyta (Fig. 2; Supplemental Fig. S20). As described above, only C. reinhardtii and Chlorella species might have putative orthologs for UGT84B1 (Fig. 2; Supplemental Fig. S10; Jackson et al., 2001, 2002). The IAA CARBOXYLMETHYLTRANSFERASE1 (IAMT1) gene encodes a methyltransferase that methylates IAA in vitro (Qin et al., 2005), but only in C. vulgaris does an ortholog seem to be present (Fig. 2; Supplemental Fig. S21).
In conclusion, it appears that auxin metabolism is not prominently present in Chlorophyta. While it was suggested that P. patens might not conjugate IAA to Ala, Leu, Asp, or Glu, consistent with empirical data and the absence of the respective gene families (Rensing et al., 2008), our analyses suggest at least the presence of putative orthologs of IAR4, IAR3, ILR1, and ILL1 to ILL6 in the investigated Chlorophyta (Fig. 2; Supplemental Fig. S18 and S20).
Auxin Transport in Green Algae
An important feature of auxin signaling is active auxin transport toward specific tissues and even single cells, resulting in auxin gradients (Petrásek and Friml, 2009). In Arabidopsis and other land plants, three major auxin transport protein families have been described: PINs, mediating auxin efflux (Petrásek et al., 2006; Křeček et al., 2009); AUX1-LAXs, mediating auxin influx (Yang et al., 2006; Swarup et al., 2008); and ABCB/PGP/MDRs, mediating auxin transport (Geisler and Murphy, 2006; Verrier et al., 2008; Yang and Murphy, 2009).
Here, in addition to the Chlorophyta genomes, we also analyzed EST libraries of two green algae within the Streptophyta, C. orbicularis and S. pratensis (Fig. 1; Timme and Delwiche, 2010), allowing us to close in on the origin of auxin transport.
PINs
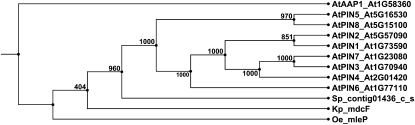
Notwithstanding that intercellular PIN-dependent auxin efflux might seem to be of limited importance in single-celled green algae, the endoplasmic reticulum (ER)-localized PIN5-like subclade was shown to be evolutionarily older than the plasma membrane-localized (PIN1-like) proteins that function in intercellular auxin transport (Mravec et al., 2009). Such ER-localized PIN5-like proteins might exist in unicellular algae, where they could play a similar role as suggested in land plants, namely mediating auxin flow from the cytosol to the lumen of the ER to regulate auxin homeostasis (Mravec et al., 2009). Here, in agreement with earlier studies, we did not identify PIN orthologs in Chlorophyta (Křeček et al., 2009; Mravec et al., 2009; Fig. 2). However, in contrast to earlier studies, which proposed that the most archaic PIN proteins were found in P. patens (Křeček et al., 2009; Mravec et al., 2009), we identified a putative PIN ortholog in S. pratensis (Figs. 2 and 5; Supplemental Fig. S22). However, while this observation starts filling the gap on when PIN proteins emerged in evolution, it is not clear whether they represent ER- or plasma membrane-localized proteins. The short hydrophilic loop, reminiscent of the PIN5-like subclass of PIN proteins (Křeček et al., 2009; Mravec et al., 2009), hints that the PIN orthologs in S. pratensis might localize to the ER (Supplemental Fig. S22). Similarly, the transporters MleP and MdcF of Oenococcus oeni and Klebsiella pneumoniae, respectively, also lack a lengthy central hydrophilic domain (Kerr and Bennett, 2007) and might represent the ancestral state for PIN proteins. In the future, investigation of the localization of the PIN-like proteins identified in Streptophyta and basal land plants will presumably shed light on their function.
Figure 5.
UPGMA tree of PIN proteins in Arabidopsis and a PIN-like protein in S. pratensis. At, Arabidopsis; Kp, K. pneumoniae; Oe, O. oeni; Sp, S. pratensis.
AUX1/LAXs
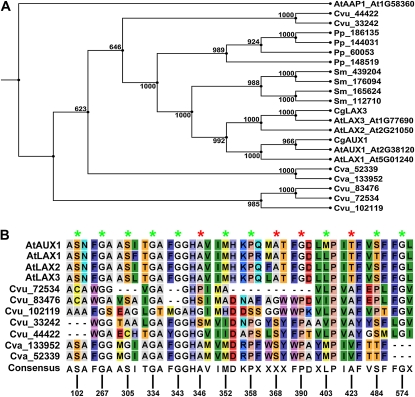
While in most investigated Chlorophyta and Streptophyta no orthologs of AUX1 and the LAX proteins could be identified, Chlorella species appear to contain some putative candidates (Figs. 2 and 6A). A comparison of protein sequences shows that the N- and C-terminal sequences are the most divergent, while the central sequence is highly similar (Supplemental Fig. S23). Out of 14 amino acids that have been shown to be important for the activity of auxin influx carriers (Swarup et al., 2004; Péret et al., 2007), 10 are conserved in at least one of the Chlorella species orthologs (Fig. 6B). At present, it is unclear why, from the investigated green algae, only Chlorella species possess putative AUX1/LAXs orthologs.
Figure 6.
A, Phylogenetic analyses of AUX1-LAX orthologs in land plants and Chlorella species. B, Alignment of AUX1/LAX proteins, highlighting the conserved residues in C. vulgaris (Cvu) and C. variabilis (Cva). Asterisks indicate residues conserved in at least one putative ortholog (green) or not conserved (red). Residue colors are according to the rasmol color scheme, which is consistent with traditional amino acid properties. At, Arabidopsis; Cg, Casuarina glauca; Pp, P. patens; Sm, S. moellendorffii. [See online article for color version of this figure.]
ABCB/PGPs
The ABCB/PGP/MDR family of ABC transporters was only recently shown to be involved in auxin transport (Geisler and Murphy, 2006; Verrier et al., 2008; Yang and Murphy, 2009), and we could identify orthologs for ABC transporters in the investigated Chlorophyta and Streptophyta (Fig. 2; Supplemental Fig. S24). However, notwithstanding that these Chlorophyta and Streptophyta proteins group within the correct subfamily of ABC transporters, the functional diversity within ABC proteins (Rea, 2007) makes sequence-based assignment of functional orthologs nearly impossible.
Auxin Response in Green Algae
The key factors in canonical auxin signaling are AUX/IAAs and ARFs (Lau et al., 2008). In Arabidopsis, there are 29 AUX/IAA family members, which usually contain four (I–IV) conserved domains and display strong genetic redundancy, and 23 ARF genes encoding proteins that contain a B3 DNA-binding domain, a repression or activation domain, and two domains similar to domains III and IV of AUX/IAAs (Lau et al., 2008; Fig. 7A). Critically, AUX/IAAs are bound by TIR1 via their domain II and form heterodimers with ARF transcription factors through domains III and IV (Lau et al., 2008).
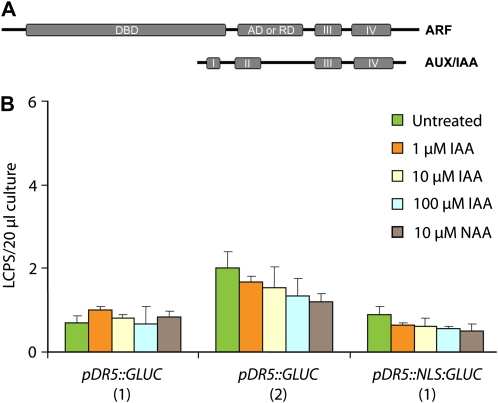
Figure 7.
Auxin response in Chlorophyta. A, Schematic representation of important domains of ARFs and AUX/IAAs. ARFs contain domains for the interaction with AUX-IAAs and ARFs (III and IV), an activation or repression domain (AD or RD), and a B3 DNA-binding domain (DBD). AUX-IAAs contain domains for the interaction with ARFs (III and IV), a repression domain (I), and a domain that contains a degron sequence (II). B, Effect of auxin on the expression of GLUC from the DR5 promoter in C. reinhardtii. As an example, representative data for two transgenic clones expressing the pDR5::GLUC construct (1 and 2) and one clone expressing the pDR5::NLS:GLUC construct (1) are shown. Luciferase activity was measured 7 h after auxin addition (for details, see “Materials and Methods”). No auxin-induced expression of pDR5::GLUC or pDR5::NLS:GLUC is detectable in C. reinhardtii. The luminescence units are presented as luminescence counts per second (LCPS). Control strains lacking the Luc gene had very low luminescence (0.1 LCPS per 20 μL of culture). Bars represent means of three independent experiments, and sd is indicated. [See online article for color version of this figure.]
AUX/IAA-ARF-TIR1/AFB-Dependent Auxin Signaling in Chlorophyta
The auxin signaling and response pathway has undergone considerable functional diversification within vascular plants since they diverged from bryophytes (Paponov et al., 2009; Romanel et al., 2009). We have previously shown that the AUX/IAA-ARF-TIR1/AFB-dependent auxin signaling machinery is not present in various Chlorophyta (Lau et al., 2009). In addition, we show here that there are no orthologs for the transcriptional corepressor TOPLESS (TPL) and related proteins in the investigated Chlorophyta, whereas those seem to be present in P. patens and S. moellendorffii (Fig. 2; Supplemental Fig. S25).
DR5 Promoter Activity in C. reinhardtii
In land plants, the AUX/IAA-ARF-dependent auxin-responsive promoter DR5, consisting of tandem repeats comprising the auxin-responsive element TGTCTC fused to a reporter gene, is widely used for monitoring auxin responses in planta (Ulmasov et al., 1997; Benková et al., 2003; Bierfreund et al., 2003; Friml et al., 2003; Yamamoto et al., 2007; Fujita et al., 2008; Gallavotti et al., 2008). Here, to corroborate the genomic evidence for the absence of an AUX/IAA-ARF-dependent auxin response mechanism in Chlorophyta, we analyzed the auxin inducibility of the DR5 promoter fused to the codon-optimized Gaussia Luciferase (GLUC) in C. reinhardtii (Shao and Bock, 2008). Since transgene silencing represents a common problem in C. reinhardtii transformation (Manuell and Mayfield, 2006), multiple transformants were analyzed. These experiments revealed no detectable auxin-inducible expression of the very lowly expressed pDR5::GLUC and pDR5::NLS:GLUC constructs in independently generated clones of 3-d-old C. reinhardtii cultures incubated with 1, 10, or 100 μm IAA or 10 μm naphthylacetic acid (NAA) for 7 h compared with the 0 μm IAA control (Fig. 7B). Obviously, due to the lack of auxin-inducible gene expression in C. reinhardtii, no strict positive control could be included in these experiments (in addition to the constitutively expressed and heat shock-inducible GLUC constructs; Shao and Bock, 2008). Nonetheless, these data further corroborate the absence of the typical land plant components of the auxin signaling cascade at least in C. reinhardtii.
Alternative Mechanisms in Chlorophyta
We have previously suggested that Chlorophyta might convey an auxin response through alternative mechanisms involving ABP1 and/or IBR5 (Lau et al., 2009). To further support this notion, we used reciprocal BLAST, and we could show that IBR5 is present in Chlorophyta (Fig. 2; Supplemental Fig. S26). Similarly, using this approach, we could corroborate the presence of ABP1 in some Chlorophyta, like C. variabilis NC64A, and C. reinhardtii, while in C. vulgaris and the other investigated species, no or only ABP1-like proteins appear to be present, respectively (Fig. 2; Supplemental Fig. S27), which is in agreement with a recent report (Tromas et al., 2010). While crucial amino acids are conserved in ABP1 orthologs (Lau et al., 2009; Tromas et al., 2010), the elucidation of their cellular localization will require further investigation. For example, the presence of a typical ER retention signal is not clear (Tromas et al., 2010). Interestingly, these mechanisms, together with the canonical AUX/IAA-ARF pathway, appear to be absent from E. siliculosus (Cock et al., 2010; Le Bail et al., 2010; data not shown), raising the question of how the brown algae control auxin response.
Auxin Response in Algae of the Streptophyta Clade
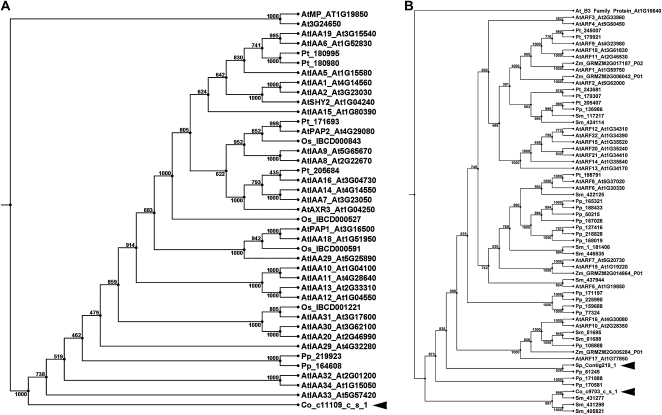
Previously, separate domains that make up AUX/IAAs and ARFs, such as the degron sequence of the AUX–IAA domain II or the ARF B3 domain, were identified in some Chlorophyta (Lau et al., 2009; Romanel et al., 2009). Here, we analyzed EST libraries of the Streptophyta algae C. orbicularis and S. pratensis (Fig. 1; Timme and Delwiche, 2010) and identified regions with striking similarities to domains III and IV of AUX/IAAs and ARFs (Fig. 8; Supplemental Figs. S28 and S29). One AUX/IAA-like protein, which lacks domain I and likely domain II, was identified in C. orbicularis (Fig. 8A). Since this is also true for AtIAA33 (Supplemental Fig. S30), this protein could be a primitive version of an AUX/IAA. On top, C. orbicularis and S. pratensis contain ARF-like proteins that show similarities to the putative ARFs with a large N-terminal truncation in S. moellendorffii (Fig. 8B; Supplemental Fig. S29; Paponov et al., 2009).
Figure 8.
Phylogenetic analysis of the putative AUX/IAAs and ARFs in Streptophyta. UPGMA trees show subsets of the AUX/IAA (A) and ARF (B) families. The sequences from C. orbicularis and S. pratensis are indicated with arrowheads. At, Arabidopsis; Co, C. orbicularis; Os, Oryza sativa; Pp, P. patens; Pt, Populus trichocarpa; Sp, S. pratensis; Sm, S. moellendorffii; Zm, Zea mays.
Finally, TPL-related proteins were represented in Streptophyta (Fig. 2; Supplemental Fig. S25).
CONCLUSION
Chlorophyta appear to encode at least some of the proteins necessary for auxin biosynthesis and metabolism, which correlates with auxin measurements in members of this division of green algae (Cooke et al., 2002; Lau et al., 2009; Ross and Reid, 2010). On top, putative auxin transport proteins such as AUX1-like and ABCB/PGP-like proteins appear to be present in Chlorophyta, while PIN-like proteins are only encoded in some Streptophyta. While single-celled Chlorophyta seem to possess the potential to transport auxin, at present it is not clear if they import and export auxin from and to the environment, respectively. In the absence of PIN-like auxin carriers, the AUX1-like and ABCB/PGP-like proteins could facilitate putative auxin fluxes in Chlorophyta. While our analyses of S. pratensis and C. orbicularis EST libraries suggest that PIN proteins emerged in the Streptophyta clade, functional data will be required to determine if directional PIN-mediated auxin transport emerged to coordinate filamentous and/or thalloid growth of green algae and not necessarily with the transition to land. The availability of more genomic data from Streptophyta and multicellular Chlorophyta would help to address this question.
Whereas the complete known “auxin machinery” is present from “basal” land plants onward (Rensing et al., 2008), Chlorophyta do not have the necessary components that we find in land plants to control the canonical auxin response. Chlorophyta appear to possess alternative ways to respond to auxin, such as ABP1 and/or IBR5 (Lau et al., 2009; Tromas et al., 2010). This also indicates that more complex auxin signaling elements emerged somewhere between Chlorophyta and early land plants (such as P. patens). Since, at present, no fully sequenced and annotated genomes of green algae within the Streptophyta clade are available, we made use of EST libraries for some Streptophyta members to investigate where the origin of auxin signaling might lie. Here, we did not retrieve full-length proteins involved in auxin response, but it appears that partial and/or ancestral forms are present. When these full-length proteins might have emerged is not clear, but it seems that the canonical auxin response is not required for building multicellular filamentous or mat-forming life forms. Hence, this suggests that the formation of more complex, thalloid forms or, indeed, as proposed earlier, that the “move” to land (Rensing et al., 2008) required an expansion of auxin signaling mechanisms. In this respect, it is also interesting that the investigation of the E. siliculosus genome, a multicellular, thalloid brown alga, revealed that canonical auxin machinery genes are absent (Cock et al., 2010; Le Bail et al., 2010).
As it appears that there are species that contain auxin but no or only limited auxin response machinery, the presence of auxin might predate the emergence of such response mechanisms. This raises some questions. If many of the key players or domains of those key proteins are already present in the Chlorophyta and Streptophyta but are obviously not yet integrated into a complex response system, what, if any, are their respective functions in these green algae? Is the absence of an auxin response system responsible for them being small and/or unicellular? Was auxin initially just a metabolite (as possibly indicated by the fact that it can be synthesized and metabolized in Chlorophyta), a side product without any function, or does it have an unknown physiological function? On the other hand, indole, and even IAA itself, can act as a microbial signaling molecule (Spaepen et al., 2007; Lee and Lee, 2010), and this might be the basis from which auxin as a signaling molecule in land plants could have evolved.
Future genome analyses will also reveal if the transitions from unicellular to multicellular form (filamentous or thalloid) in Chlorophyta coincided with the emergence of auxin signaling, or if this only developed in the Streptophyta lineage, and whether these components can also be found in single-celled Streptophyta. In this respect, it will be intriguing to investigate if it was the freshwater lifestyle of the Streptophyta, and the possibly associated physiological preadaptation to terrestrial habitats (Becker and Marin, 2009), including the development of auxin signaling, that allowed the origin of land plants. In contrast, macroalgae (such as Ulvophyceae, red and brown seaweed), which are major species in marine environments, never conquered land, and at least E. siliculosus appears to lack auxin signaling (Cock et al., 2010; Le Bail et al., 2010).
Finally, while we are aware that defining functional orthologs for the diverse proteins, and especially enzymes, involved in auxin signaling, based on only genomic evidence, is difficult, our data provide a starting point for further in-depth, functional analyses by molecular and biochemical studies and should not be taken as proof for the existence of these proteins in algae, at least not in the function as we know it from land plants. At this stage, our data merely serve as a starting point for exploring the evolution of auxin biology; also, our study helps, together with recent advances on other phytohormones (Pils and Heyl, 2009; Hartung, 2010; Ross and Reid, 2010), to clarify the evolution of complex signaling pathways in land plants. In the future, it will be interesting to functionally test the putative orthologs identified in this study: for example, the AUX1 orthologs for their potential to rescue the Arabidopsis aux1 mutant, which is impaired in lateral root formation and gravitropic response (Bennett et al., 1996; Marchant et al., 2002), and the PIN ortholog for its localization and auxin efflux abilities.
MATERIALS AND METHODS
Data Sources and Candidate Retrieval
The protein sequences for Arabidopsis (Arabidopsis thaliana) were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/). For Physcomitrella patens, Selaginella moellendorffii, and the Chlorophyta genomes, protein sequences were identified from the Joint Genome Institute (http://genome.jgi-psf.org/): Ostreococcus tauri (http://genome.jgi-psf.org/Ostta4/Ostta4.home.html; Derelle et al., 2006), Ostreococcus lucimarinus (http://genome.jgi-psf.org/Ost9901_3/Ost9901_3.home.html; Palenik et al., 2007), Micromonas pusilla strain CCMP1545 (http://genome.jgi-psf.org/MicpuC2/MicpuC2.home.html) and strain RCC299 (http://genome.jgi-psf.org/MicpuN3/MicpuN3.home.html; Worden et al., 2009), Chlamydomonas reinhardtii (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html; Merchant et al., 2007), Volvox carteri (http://genome.jgi-psf.org/Volca1/Volca1.home.html), Chlorella vulgaris (http://genome.jgi-psf.org/Chlvu1/Chlvu1.home.html), and Chlorella variabilis NC64A (http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.home.html; Blanc et al., 2010). Using the Arabidopsis protein sequences as bait and the assumed/annotated proteins of the respective species as target, we performed BLASTP analyses on the respective Web sites of the Joint Genome Institute. For the Streptophyta species Coleochaete orbicularis and Spirogyra pratensis, we used recent data from EST libraries (Timme and Delwiche, 2010), and to retrieve putative orthologs, Clustal alignment analyses were performed using assumed/annotated protein sequences from these species together with the protein sequences from Arabidopsis and the retrieved Chlorophyta sequences. The sequences of putative PIN orthologs in various land plants were obtained from the Phytozome repository of genomic sequences (http://www.phytozome.org/). The AUX/IAA and ARF protein sequences in various land plants were retrieved from the Plant Transcription Factor Database (http://plntfdb.bio.uni-potsdam.de/v3.0/).
Alignment and Phylogenetic Analyses
Retrieved sequences were aligned using a progressive alignment algorithm (Feng and Doolittle, 1987) in order to create multiple alignments with the CLC DNA Workbench 5.1 using the following settings: Gap open cost (10), Extension cost (1), End gap cost (as any other), Alignment (very accurate; see http://www.clcbio.com/index.php?id=502). The residue colors in these alignments are according to the rasmol color scheme (http://www.openrasmol.org/doc/rasmol.html), which is consistent with traditional amino acid properties. The phylogenetic trees were built using the UPGMA algorithm and a bootstrap analysis with 1,000 replicates within the CLC DNA Workbench 5.1. Subsequently, putative orthologs were considered when they grouped in the clade of the protein of interest and when the protein of interest was retrieved in the top five or with an E-value < 2e-09 for the reciprocal blast in Arabidopsis. The sequences finally taken into consideration can be found in the Supplemental Sequences S1.
Luciferase Assay
The codon-optimized GLUC (Shao and Bock, 2008) was fused with the DR5 auxin-responsive promoter with (pDR5::NLS:GLUC) or without a nuclear localization signal derived from SV40 (pDR5::GLUC). pDR5 and pDR5::NLS were amplified with 5′-GCGGCCGCGAGACAAAAGGGAGAC-3′ and 5′-ACCATATGGGATCCCCTGTAATTGTAATTGTAAATAG-3′ or 5′-TTCATATGCACCTTGCGCTTCTTCTTGGGGGCCAT-3′ from GIIK DR5rev::SV40:3xGFP (Weijers et al., 2006) and cloned into PpsaD::GLUC by replacing PpsaD using NotI and NdeI restriction sites. The constructs were introduced into the C. reinhardtii nuclear genome by glass bead-mediated transformation using standard protocols. A total of 101 transformants were initially screened for luciferase activity after incubation with 10 μm NAA for 6 h. For each construct, luciferase activity was then assayed in three independent transgenic clones after incubation with 0, 1, or 10 μm and 100 μm IAA or NAA for 0, 3, 8, and 48 h. To this end, algal cultures grown in liquid Tris-acetate-phosphate medium (Harris, 1989) under normal light conditions (55 μE m−2 s−1) for 3 d to a final cell density of 3 to 4 × 106 cells mL−1 (determined with a Z2 Coulter Counter cell counter; Beckman Coulter) were incubated with IAA or NAA followed by measurement of their luciferase activities with a luminometer (MicroBeta TriLux; Perkin-Elmer) after adding the substrate (0.01 mm coelenterazine). Growth tests with the wild-type strain confirmed that concentrations of 1 to 100 μm IAA or NAA did not exhibit any toxic effects on C. reinhardtii cells. The data shown in Figure 7 were based on measurements 7 h after auxin addition with three transgenic clones and three replicates each.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. UPGMA tree for ASB1 orthologs.
Supplemental Figure S2. UPGMA tree for ASA1 and ASA2 orthologs.
Supplemental Figure S3. UPGMA tree for PAT1 orthologs.
Supplemental Figure S4. UPGMA tree for PAI1, PAI2, and PAI3 orthologs.
Supplemental Figure S5. UPGMA tree for IGPS orthologs.
Supplemental Figure S6. UPGMA tree for TSA1 orthologs.
Supplemental Figure S7. UPGMA tree for TSB1 and TSB2 orthologs.
Supplemental Figure S8. UPGMA tree for TAA1, TAR1, TAR2, TAR3, and TAR4 orthologs.
Supplemental Figure S9. UPGMA tree for SUR1 orthologs.
Supplemental Figure S10. UPGMA tree for UGT74B1 and UGT84B1.
Supplemental Figure S11. UPGMA tree for ST5A orthologs.
Supplemental Figure S12. UPGMA tree for TGG1 and TGG2 orthologs.
Supplemental Figure S13. UPGMA tree for ESP orthologs.
Supplemental Figure S14. UPGMA tree for ESM1 orthologs.
Supplemental Figure S15. UPGMA tree for YUC orthologs.
Supplemental Figure S16. UPGMA tree for AMI1 orthologs.
Supplemental Figure S17. UPGMA tree for NIT orthologs.
Supplemental Figure S18. UPGMA tree for ILL1 to ILL6, ILR1, and IAR3 orthologs.
Supplemental Figure S19. UPGMA tree for GH3s.
Supplemental Figure S20. UPGMA tree for IAR4 orthologs.
Supplemental Figure S21. UPGMA tree for IAMT1 orthologs.
Supplemental Figure S22. Alignment of AtPIN proteins and putative orthologs.
Supplemental Figure S23. Alignment of AUX1-LAXs and putative orthologs.
Supplemental Figure S24. UPGMA tree for PGP orthologs.
Supplemental Figure S25. UPGMA tree for TPL, TPR1, TPR2, TPR3, and TPR4 orthologs.
Supplemental Figure S26. UPGMA tree for IBR5 orthologs.
Supplemental Figure S27. UPGMA tree for ABP1 orthologs.
Supplemental Figure S28. Alignment of AUX/IAAs and putative orthologs.
Supplemental Figure S29. Alignment of ARFs and putative orthologs.
Supplemental Figure S30. Alignment of IAA33 and putative orthologs.
Supplemental Sequences S1. Sequences of putative orthologs and those used for generating the tree.
Supplementary Material
Acknowledgments
We thank Pierre Rouzé and Olivier De Clerck for critical reading and useful suggestions as well as two anonymous referees for useful suggestions.
References
- Basu S, Sun H, Brian L, Quatrano RL, Muday GK. (2002) Early embryo development in Fucus distichus is auxin sensitive. Plant Physiol 130: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B. (2009) Streptophyte algae and the origin of embryophytes. Ann Bot (Lond) 103: 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer LL, Boyd ES, Peters JW, Posewitz MC. (2009) Engineering algae for biohydrogen and biofuel production. Curr Opin Biotechnol 20: 264–271 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L. (1998) Algal phylogeny and the origin of land plants. Plant Physiol 116: 9–15 [Google Scholar]
- Bierfreund NM, Reski R, Decker EL. (2003) Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep 21: 1143–1152 [DOI] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. (2010) The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Cooke TJ, Poli D, Sztein AE, Cohen JD. (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49: 319–338 [PubMed] [Google Scholar]
- Costacurta A, Vanderleyden J. (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21: 1–18 [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, et al. (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng DF, Doolittle RF. (1987) Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol 25: 351–360 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fujita T, Sakaguchi H, Hiwatashi Y, Wagstaff SJ, Ito M, Deguchi H, Sato T, Hasebe M. (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10: 176–186 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D. (2008) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. (2006) The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett 580: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Hao C, Chen SL, Mu J, Xiao PG. (2009) Molecular phylogeny, long-term evolution, and functional divergence of flavin-containing monooxygenases. Genetica 137: 173–187 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Hartung W. (2010) The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct Plant Biol 37: 806–812 [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ. (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Kerr ID, Bennett MJ. (2007) New insight into the biochemical mechanisms regulating auxin transport in plants. Biochem J 401: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk DL. (1999) Evolution of multicellularity in the volvocine algae. Curr Opin Plant Biol 2: 496–501 [DOI] [PubMed] [Google Scholar]
- Kirk DL, Nishii I. (2001) Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Dev Growth Differ 43: 621–631 [DOI] [PubMed] [Google Scholar]
- Křeček P, Skupa P, Libus J, Naramoto S, Tejos R, Friml J, Zazímalová E. (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Zimmer AD, Rensing SA, Reski R. (2008) Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci 13: 542–549 [DOI] [PubMed] [Google Scholar]
- Lau S, Jürgens G, De Smet I. (2008) The evolving complexity of the auxin pathway. Plant Cell 20: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Shao N, Bock R, Jürgens G, De Smet I. (2009) Auxin signaling in algal lineages: fact or myth? Trends Plant Sci 14: 182–188 [DOI] [PubMed] [Google Scholar]
- Le Bail A, Billoud B, Kowalczyk N, Kowalczyk M, Gicquel M, Le Panse S, Stewart S, Scornet D, Cock JM, Ljung K, et al. (2010) Auxin metabolism and function in the multicellular brown alga Ectocarpus siliculosus. Plant Physiol 153: 128–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee J. (2010) Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34: 426–444 [DOI] [PubMed] [Google Scholar]
- Lehmann T, Hoffmann M, Hentrich M, Pollmann S. (2010) Indole-3-acetamide-dependent auxin biosynthesis: a widely distributed way of indole-3-acetic acid production? Eur J Cell Biol 89: 895–905 [DOI] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM. (2004) Green algae and the origin of land plants. Am J Bot 91: 1535–1556 [DOI] [PubMed] [Google Scholar]
- Manuell AL, Mayfield SP. (2006) A bright future for Chlamydomonas. Genome Biol 7: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG. (2004) Charophyte algae and land plant origins. Trends Ecol Evol 19: 661–666 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA. (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37: 770–777 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA 104: 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K. (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup R, Jansen L, Devos G, Auguy F, Collin M, Santi C, Hocher V, Franche C, Bogusz D, et al. (2007) Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pils B, Heyl A. (2009) Unraveling the evolution of cytokinin signaling. Plant Physiol 151: 782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Barkawi LS, Fan KT, Cohen JD, Gray WM. (2009) Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol 150: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ. (2001) Higher plant glycosyltransferases. Genome Biol 2: S3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Reid JB. (2010) Evolution of growth-promoting plant hormones. Funct Plant Biol 37: 795–805 [Google Scholar]
- Schmitt R. (2003) Differentiation of germinal and somatic cells in Volvox carteri. Curr Opin Microbiol 6: 608–613 [DOI] [PubMed] [Google Scholar]
- Shao N, Bock R. (2008) A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet 53: 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztein AE, Cohen JD, Cooke TJ. (2000) Evolutionary patterns in the auxin metabolism of green plants. Int J Plant Sci 161: 849–859 [Google Scholar]
- Timme RE, Delwiche CF. (2010) Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paponov I, Perrot-Rechenmann C. (2010) AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci 15: 436–446 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, et al. (2008) Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J. (2007) Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G. (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ. (2010) An outlook on microalgal biofuels. Science 329: 796–799 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS. (2009) Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J 59: 179–191 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2008) The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11: 16–22 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.