Abstract
This paper describes a method by which molecules that are impermeable to cells are encapsulated in dye-sensitized lipid nanocapsules for delivery into cells via endocytosis. Once inside the cells, the molecules are released from the lipid nanocapsules into the cytoplasm with a single nanosecond pulse from a laser in the far red (645nm). We demonstrate this method with the intracellular release of the second messenger IP3 in CHO-M1 cells, and report that calcium responses from the cells changed from a sustained increase to a transient spike when the average number of IP3 released is decreased below 50 molecules per nanocapsule. We also demonstrate the delivery of a 23 kDa AGT fusion protein into Ba/F3 cells to inhibit a key player BCR-ABL in the apoptotic pathway. We show that an average of ~ 8 molecules of the inhibitor is sufficient to induce apoptosis in the majority of Ba/F3 cells.
Keywords: Lipids, Dye-Sensitized, Nanocapsule, Intracellular, Photolysis
To uncover the spatiotemporal dynamics of cellular function, we need techniques that allow us to perturb cells over space and time in a controlled fashion. The most common approach for the delivery of bioactive molecules to cells in a highly spatiotemporally-resolved fashion is the two-photon release of a chemically-caged compound.1–5 Caged molecules, however, suffer from a number of drawbacks. The design and synthesis of a new caged compound can be tedious and the caging of large protein molecules can be difficult, if not impossible. For intracellular delivery, the caged molecules must be permeable to the cell; this requirement, in turn, makes it difficult to control the precise concentration of the caged molecules inside the cell because of their preferential partitioning into different cellular organelles and membranes. These drawbacks of caged compounds have prompted us to develop light-addressable lipid nanocapsules as a general platform for caging a wide range of bioactive molecules.6–12 Nanocapsules based on lipid vesicles represent an emerging class of physical cages,, some of which also have been used recently for intracellular release.13–18
Our first nanocapsules were lipid vesicles which we manipulated with optical tweezers and photolyzed using a single nanosecond laser pulse in the UV.6,8,10 This capability was sufficient for delivering molecules extracellularly in a cell-culture setting. Unfortunately, the lipid vesicles were fragile and could not be stored in solution for more than a few days. This practical constraint prompted us to explore a range of more robust non-lipid based nanocapsules, including ones formed by colloidal templating in silica and polymer-based nanocapsules.7,9 However, we recently overcame the storage issue for lipid nanocapsules by using an optimized lyophilization procedure that allows us to store lipid nanocapsules over long periods of time at −20 °C as a lyophilized powder.19 As a result, we returned to developing light-addressable lipid nanocapsules as a general caging platform for bioactive molecules.
By sensitizing the shell of lipid nanocapsules with dyes, we were able to tune the one-photon photolysis wavelength to the far red,11 which to date was not possible for chemically-caged molecules because of the photophysical constraints of the caging groups. By doping the shell of nanocapsules with dyes that have high two-photon absorption cross sections, we were also able to achieve two-photon uncaging from lipid nanocapsules with 100% efficiency.11 This capability opens up new applications for lipid nanocapsules in tissue slices or in vivo because of the optical advantages offered by far-red and near-IR light.
Unlike chemically caged compounds which become uncaged via a pathway involving radicals, we determined the mechanism for uncaging lipid nanocapsules was photothermal.12 The advantages of uncaging bioactive molecules in the far red or near-IR via a photothermal pathway inside cells are significant. The cell is largely transparent to these long wavelengths and is not damaged by radicals, which can be an issue with chemically caged compounds.12 Because lipid nanocapsules physically isolate the bioactive molecules from the cell until they are released, the concentration of bioactive molecules can be controlled and preserved within the nanocapsule even after the nanocapsules have been delivered into the cell. This feature is important because, as we demonstrate in this paper, the cellular response can be quite different, depending on the copy number of molecules released into the cytoplasm.
In addition to these advantages, lipid nanocapsules also act as a carrier for cell-impermeable molecules into the cellular interior. As we show in this paper, a single nanosecond laser pulse in the far red was sufficient to release the encapsulated molecules into the cytoplasm of the cell. Finally, because single lipid nanocapsules could be photolyzed with great precision over space and time,6–8,11 we can accurately control the spatiotemporal profile of release from individual nanocapsules. We demonstrated the release of small molecules (Alexa 488 and IP3) and a conjugate of a protein and small molecule inhibitor (AGT-PP1-1) with our new approach. IP3 binds to the IP3 receptors located on the endoplasmic reticulum, which then causes release of internal calcium stores into the cytosol. AGT-PP1-1 acts as an inhibitor of BCR-ABL, a kinase, which when inhibited can lead to apoptosis. We observed intriguing cellular responses following the intracellular release of these molecules.
Results and Discussion
Delivery of lipid nanocapsules inside cells
There are several common approaches for the delivery of membrane-impermeable molecules or nanoparticles to interiors of cells. For molecules, one popular approach is microinjection. For nanoparticles, endocytosis is the most common pathway into the cell. However, in our studies with lipid nanocapsules, the initial starting point was microinjection, not endocytosis, because it is well known that endocytosed nanoparticles often cannot escape the endosome. We thought we would have difficulty in releasing the molecules from the lipid nanocapsule into the cytoplasm if we followed the endocytosis route. Indeed, we found microinjection was effective in delivering the lipid nanocapsules into single cells. Unfortunately, we also found microinjection to be rather tedious.
This constraint prompted us to re-evaluate endocytosis and other alternative pathways for introducing lipid nanocapsules into cells. We found endocytosis was efficient in delivering lipid nanocapsules in parallel into all the cells in the culture. Most importantly, we found the application of a single nanosecond light pulse was sufficient to release the contents of the lipid nanocapsules into the cytoplasm of the cells.
The mechanism of endocytosis of nanoparticles is well studied. Different methods are available to promote the uptake of nanoparticles and lipid vesicles by cells, including molecular recognition and electrostatic interactions.20–24 For charge-induced endocytosis, different cell types prefer different charges on the nanoparticle. For example, CHO-M1, Ba/F3, HeLa, and HEK 293 preferentially take up positively-charged vesicles; others cell types, such as CV1 and MCF-7, prefer negatively-charged vesicles.21, 24 In our experiments, we chose 100-nm-diameter lipid vesicles composed of 9:1 DPPC:DOPC + 4 mol% DiD. The positive charges on the vesicles come primarily from the membrane dye, DiD, and also from DiO when present.
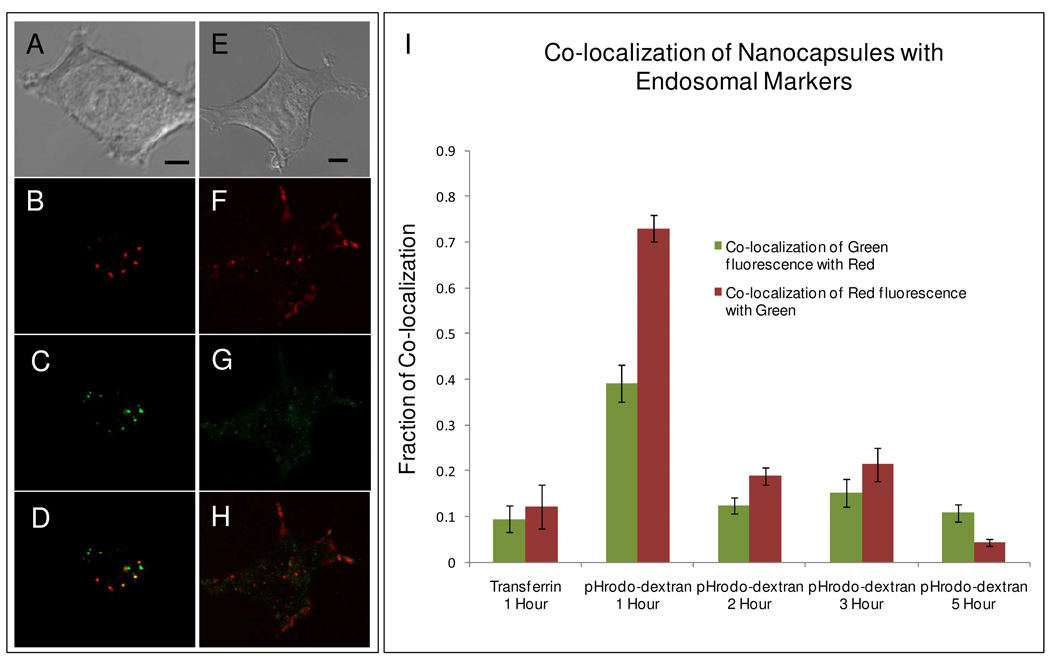
We first studied the pathway by which cells take up our vesicles. Figure 1A–D shows images of HEK 293 cells (taken from a slice of a confocal z-scan) simultaneously loaded with lipid nanocapsules (which fluoresce red because of the presence of DiD) and the common endosomal marker, pHrodo-dextran (10,000 MW) shown in green. Figure 1E–H shows HEK 293 cells simultaneously loaded with lipid nanocapsules (red) and another common endosomal marker, transferrin-Alexa 488 (green). In both cases, the cells were simultaneously loaded with lipid nanocapsules and their respective endosome markers in serum-free media for a total of one hour. The cells were then washed twice and immediately imaged, with subsequent images collected every hour for five hours. Image analysis was performed using the co-localization program Jacop, and the Manders co-efficient was calculated and reported (Figure 1I).25 In Figure 1I, the green bars represent the amount of co-localization of the endosomal markers with DiD and the red bars indicate the amount of co-localization of DiD with the endosomal markers.
Figure 1. Co-localization study of lipid nanocapsules with endosomal markers.
Images of HEK 293 cells that were loaded for 1 hour with 100-nm-diameter 9:1 DPPC:DOPC+4mol% DiD nanocapsules and endosomal markers (A–D) pHrodo-dextran and (E–H) transferrin-Alexa 488. The cells were then washed and imaged immediately. Red fluorescence is from DiD and green fluorescence is from the endosomal marker. Scale bar represents 4 µm. (I) Histogram plot of Mander’s coefficient displaying co-localization of lipid nanocapsules with the respective endosomal markers. Again, cells were loaded with lipid nanocapsules and endosomal markers for one hour, washed, then imaged immediately and then 2–5 hours after the start of incubation. For each case 30–35 cells were imaged for data collection.
Transferrin has been implicated in the endocytosis of nanoparticles.26–27 In our experiments, we saw little or no co-localization of transferrin-Alexa with lipid nanocapsules, indicating that the nanocapsules were not endocytosed via the transferrin pathway. In contrast, the lipid nanocapsules showed excellent co-localization with pHrodo-dextran, which is commonly used as a marker for following endosomes formed by fluid-phase endocytosis. As expected, the red bar was much higher than the green bar because not all endosomes contained a lipid nanocapsule but nearly all lipid nanocapsules were located within an endosome.
The high degree of co-localization was observed after one hour, indicating the lipid nanocapsules were likely taken up through fluid-phase endocytosis. We also followed the degree of co-localization over five hours. Co-localization was markedly decreased starting at hour 2. There are two possibilities that may account for this observation: (1) a loss of fluorescent signal from pHrodo due to pH changes in the endosome; indeed, we observed that the number of pHrodo-labeled endosomes decreased by half 2 hours post incubation, or (2) the lipid nanocapsules somehow escaped from the endosome. We will discuss this point in more detail in a later section. Additionally, we found that the size of the cell seems to play an important role in how many lipid nanocapsules are endocytosed. In the case of the smallest cell type (Ba/F3) typical cells contained roughly 20–40 lipid nanocapsules while the largest (HEK 293) sometimes contained >100 lipid nanocapsules. Additional images are available in the supporting information (Figure S1).
Far-red photolysis of individual lipid nanocapsules inside cells and intracellular release of IP3
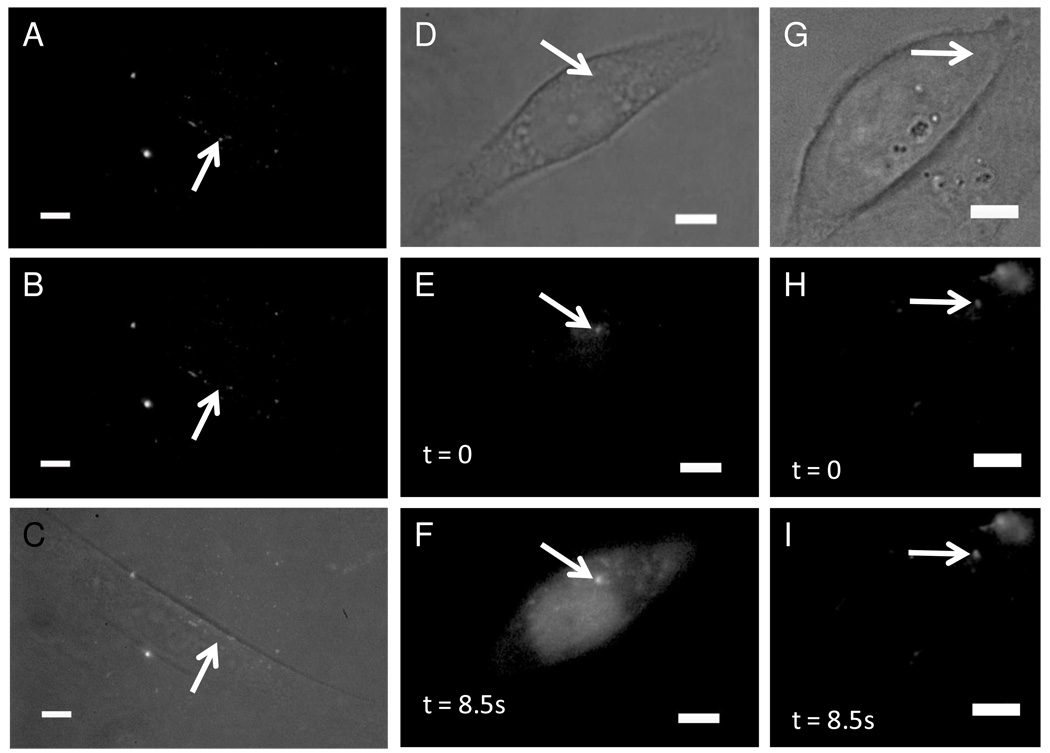
We next demonstrated the release of an encapsulated dye, Alexa-488 (Figure 2A–C). Chinese hamster ovary (CHO-M1) cells were loaded with the same 9:1 DPPC:DOPC + 4 mol% DiD 100-nm nanocapsules used on the HEK 293 cells. These lipid nanocapsules were loaded with 50µM Alexa-488. Based on the encapsulation procedure and past calculations of encapsulated contents,28 each lipid nanocapsule should have contained approximately 12 molecules of Alexa dye. We saw a complete loss of fluorescent signal for successful photolysis events. It is possible that the lipid nanocapsule was photolyzed inside the endosome and the loss of signal occurred because the dye molecules became too dilute, but we have evidence in the form of IP3 activation that the encapsulated dye was released into the cytosol of the cell. We note we were still able to visualize the Alexa loaded lipid nanocapsules 24 hours later, indicating that our lipid nanocapsules appear to be stable over this time period.
Figure 2. Release of encapsulated contents from endocytosed lipid nanocapsules into the cytoplasm.
The lipid nanocapsules were photolyzed by a single 3-ns pulse of 645-nm light after 2 hours of incubation followed by 2 washing steps. (A–C) CHO-M1 cells loaded with 100-nm-diameter nanocapsules consisting of 9:1 DPPC:DOPC+4mol% DiD and 50µM Alexa-488. (D–F) CHO-M1 cells loaded with the calcium indicator dye fluo-3 and 100-nm-diameter nanocapsules made of 9:1 DPPC:DOPC+4mol% DiD+1mol% DiO and 730µM IP3. (G–I) CHO-M1 cells loaded with fluo-3 and empty 100-nm-diameter nanocapsules made of 9:1 DPPC:DOPC+4mol% DiD+1mol% DiO. Fluorescence is from 488nm excitation in the epifluorescence configuration; the scale bar is 5µm.
To demonstrate delivery of the contents of the lipid nanocapsules into the cytosol, we released D-myo-inositol 1,4,5-trisphosphate (IP3) inside CHO-M1 cells. IP3 is a well-studied second messenger that binds to the IP3 receptors (IP3R) on the endoplasmic reticulum. The binding event releases calcium from the cell’s internal stores.29 Visualization of successful intracellular release of IP3 can be monitored by loading the cells with fluo-3, which has a 40-fold increase in fluorescent signal upon binding calcium. Cells were loaded for two hours with 100-nm-diameter nanocapsules that consisted of 9:1 DPPC:DOPC + 4 mol% DiD + 0.5 mol% DiOC18 and included 730 µM IP3. When we photolyzed single lipid nanocapsules containing IP3, we nearly always observed an increase in fluorescence from fluo-3 (Figure 2D–F). In our control experiments, where all the conditions were identical except there was not any IP3 in the lipid nanocapsules (Figure 2G–I), we never saw an increase in fluorescence from fluo-3, which was expected because IP3 was not present to bind to the IP3R to trigger a calcium increase. Because calcium signaling is also triggered when cells are stressed, this experiment showed that our technique does not cause any measureable cell stress, unlike uncaging initiated with UV light pulses.11
From the IP3 experiment above, we knew IP3 was successfully released from the lipid nanocapsules into the cytoplasm. It was still unclear, however, whether the lipid nanocapsule photolysis took place inside the endosome (in which case the endosome must have ruptured during photolysis) or the nanocapsules escaped the endosomes by the time we photolyzed them. To distinguish between the two possibilities, we compared the percentage of cell activation (calcium increase) by IP3 at 1 and 2.5 hours after we initiated the loading of lipid nanocapsules into the cells. At 1 hour, over 80% of the lipid nanocapsules were still located inside the endosome (Figure 1I), but at 2.5 hours, the majority of lipid nanocapsules no longer exhibited co-localization with the endosomal marker. The rationale follows that if the percentage of cell activation between the 1-hour and 2.5-hour time points was comparable then the photolysis of the lipid nanocapsule must have been sufficient to rupture the endosome as well. If the percentage of cell activation was markedly less at the 1-hour time point, then the lipid nanocapsules must have escaped the endosomes by the 2.5-hour time point.
We observed a dramatic increase in the number of cell activations for the later time point (83%) than the earlier one (15%). This result indicates the lipid nanocapsules had likely escaped the endosomes within 2–3 hours after being endocytosed. The mechanism of endosomal escape by our lipid nanocapsules is unclear, but once escaped, the contents of the nanocapsule could be easily released into the cytosol with a single nanosecond laser pulse.
Dose-dependent calcium responses
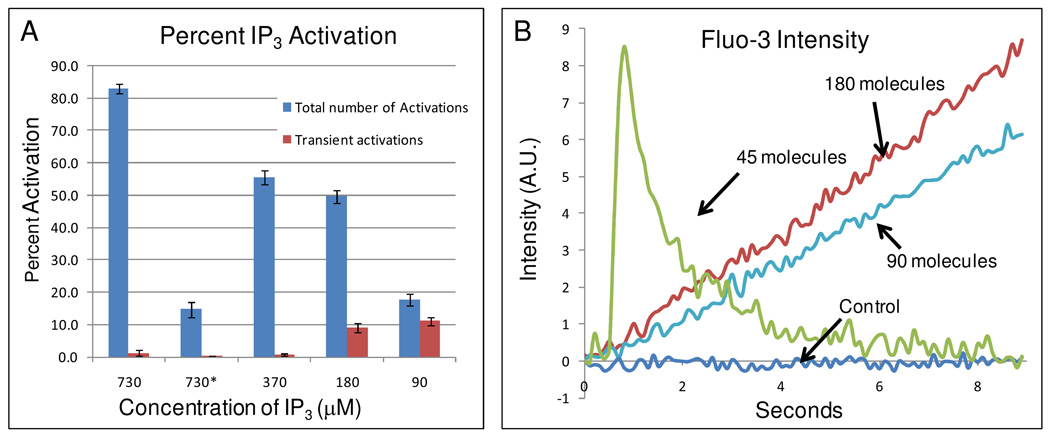
Next, we varied the concentration of IP3 inside the lipid nanocapsules and photolyzed them individually inside the cell. To quantify the cellular calcium response, we plotted over time the average fluorescent intensity of a 1 µm2 area of the CHO-M1 cell that was located 1 µm from the region where the lipid nanocapsule was photolyzed. As the average concentration of IP3 inside each lipid nanocapsule was dropped from 730 to 370 to 180 to 90 µM, we observed a corresponding decrease in the frequency of successful calcium activations (Figure 3A). Surprisingly, we also observed a different type of calcium response. Based on our previous measurements,28 these concentrations of IP3 corresponded to an average of 180, 90, 45, and 22 molecules, respectively. For high concentrations of IP3, we observed a long, sustained increase in calcium (Figure 3B). However, as the number of IP3 molecules in the lipid nanocapsules decreased to below ~50 molecules, we began to observe more frequently a transient calcium increase (Figure 3B inset). This difference in calcium response could lead to different downstream signaling pathways.30–33 When only an hour of incubation time (i.e. loading of cells with nanocapsules) was allowed (bar labeled 730*), we saw a decrease in the number of activations as compared to longer incubation time. We believe this is further evidence that our lipid nanocapsules are escaping the endosomes.
Figure 3. Dose dependent response from single cells after cytoplasmic delivery of IP3.
(A) Histogram plot showing the percent calcium activation as measured using fluo-3 versus the average concentrations of IP3 contained within the 100nm lipid nanocapsules. The blue bars show the fraction of cells exhibiting a calcium response, including both sustained and transient calcium increases. The red bars show only those cells that exhibited a transient calcium increase. At the 90 µM of IP3, the majority of calcium responses was transient, in sharp contrast to the 370 µM result where nearly all responses were sustained. All measurements were performed at 2.5 hours after the start of lipid nanocapsule loading, with the exception of the 730* results, which were obtained at the 1-hour time point after loading was initiated. Total N = 150 cells for each bar (50 cell activation attempts over 3 days for each bar). (B) Plots showing the increase in calcium as a function of time, illustrating examples of sustained and transient responses. The control indicates photolysis of an empty vesicle. The cells used for these experiments were CHO-M1.
Intracellular release of a protein-small molecule inhibitor conjugate (AGT-PP1-1) against the tyrosine kinase BCR-ABL
In order to determine the diversity of molecules compatible with our intracellular delivery method, we aimed to release a protein-small molecule conjugate that inhibited kinases into Ba/F3 cells. Ba/F3 is an interleukin-3 (IL-3) dependent, murine cell line that has been used to determine the intracellular potency of a number of protein kinase inhibitors.34 Ba/F3 cells expressing the constitutively active tyrosine kinase, BCR-ABL, become IL-3 independent. Because of the independence from IL-3, growth arrest and apoptosis can be readily detected with intracellular kinase inhibition. In these cells, we utilized AGT-PP1-1, a potent and selective inhibitor of BCR-ABL tyrosine kinase.35
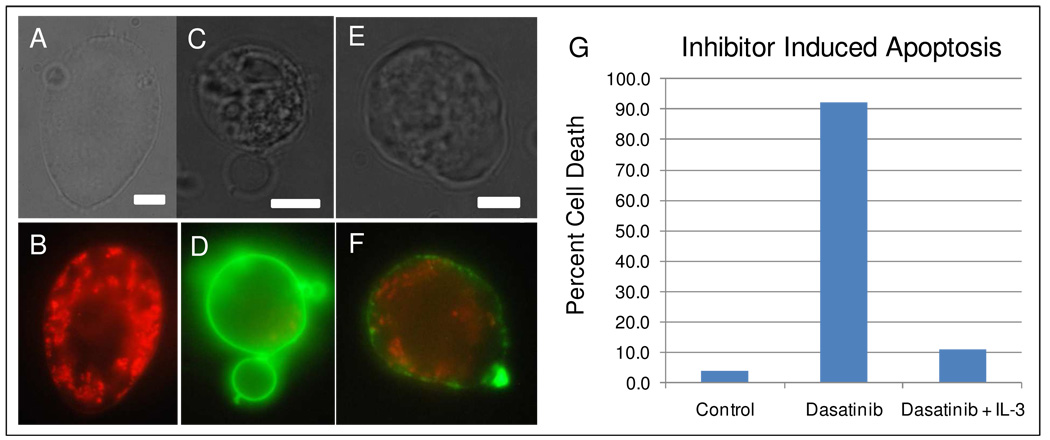
The effects of kinase inhibition were determined with Ba/F3 cells expressing p210 BCR-ABL adhering to fibronectin-coated cover slips. Adherent Ba/F3 cells (Figure 4A,B) in the absence of IL-3 appeared smooth and elongated. Treatment with a 5µM solution of the clinically-approved kinase inhibitor, dasatinib36–37, caused >95% of the adherent cells to undergo apoptosis within six hours (Figure 4G; bar labeled “Dasatinib”). Drug-treated apoptotic cells underwent a fairly drastic morphological change, blebbing (Figure 4C,D) and/or a rough morphology (Figure 4E,F), which was observed using oblique illumination. An assay utilizing MitoTracker Red and Annexin V Alexa488 confirmed apoptosis of the cells (Figure D,F). In this assay, healthy cells will be stained red, and apoptotic cells will show green fluorescence. Cells treated with 5µM dasatinib in the presence of IL-3 were rescued from cell death, consistent with BCR-ABL inhibition causing apoptosis (bar labeled “Dasatinib + IL3”).
Figure 4. Apoptosis of Ba/F3 cells induced by inhibition of BCR-ABL.
(A–B)Healthy Ba/F3 cell stained with MitoTracker Red and Annexin-Alexa 488. (C–F)Apoptotic cells stained with MitoTracker Red and Annexin-Alexa 488. Scale bar is 5 µm. (G) Histogram of cell death in Ba/F3 cells. From left to right, percent cell death of unperturbed (control), cells exposed to 5µM Dasatinib, and cells exposed to 5µM Dasatinib and 1 ng/ml IL-3. For each bar in the histogram, N=100 cells.
Next, we encapsulated AGT-PP1-1 (supplementary information scheme 1) in our lipid nanocapsules and delivered this protein-small molecule conjugate into adherent Ba/F3 cells. After 2 hours of loading and an additional 6 hours of incubation, 95% of the Ba/F3 cells remained viable (Figure 5C, bar labeled “Unphotolyzed”). Different numbers of lipid nanocapsules were then photolyzed inside the cells. The cells were tracked for over 6 hours to determine their viability (supplementary info video). Because apoptotic cells lose their adhesion to the fibronectin surface, cell death was confirmed with the observation of morphological changes using oblique illumination, which did not require washing steps that would disrupt the location of the cells on the coverslip. As in Figure 4, healthy cells were smooth and round (Figure 5A) while cells that died appeared rough and/or displayed signs of significant blebbing (Figure 5B).
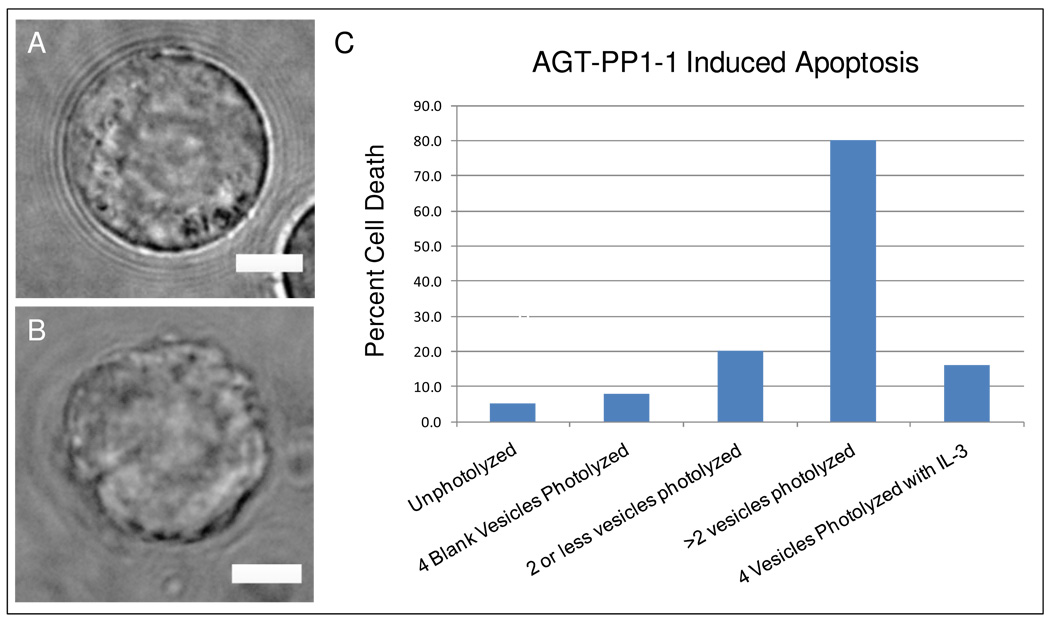
Figure 5. Apoptosis is induced in the majority of Ba/F3 cells with only eight molecules of the 23 kDa protein inhibitor of BCR-ABL.
Images of a (A)healthy and (B)dying cell observed using oblique illumination. Scale bar is 5 µm. (C) Histogram of cell death in Ba/F3 cells. From left to right, cells loaded with 100nm lipid nanocapsules containing AGT-PP1-1 for six hours and left unphotolyzed; cells in which 4 empty lipid nanocapsules were photolyzed; cells in which 1–2 lipid nanocapsules containing AGT-PP1-1 were photolyzed; cells in which 3–4 lipid nanocapsules containing AGT-PP1-1 were photolyzed; and cells in which 4 lipid nanocapsules containing AGT-PP1-1 were photolyzed in the presence of 1 ng/ml IL-3. For each bar in the histogram, N=25 cells.
Interestingly, a strong correlation was observed between intracellular inhibitor release and cell death. Twenty percent of cells that had 1 or 2 photolyzed lipid nanocapsules underwent cell death but 80% of cells with 3–4 lipid nanocapsules photolyzed underwent cell death. When empty lipid nanocapsules (bar labeled “4 Blank Vesicles Photolyzed”) were photolyzed, only 8% of cells died; the cell death rate was comparable to that in our control experiments where cells were cultured on fibronectin-coated coverslips in RPMI media without any further additions or perturbations (Figure 4G; bar labeled “Control”). Finally, to confirm that cell death was due to intracellular inhibition of BCR-ABL, four lipid nanocapsules containing AGT-PP1-1 were photolyzed in adherent Ba/F3 cells in the presence of IL-3. Under these conditions, the cell death rate decreased to 16%, indicating a significant increase in cell viability.
We were interested in how many AGT-PP1-1 molecules were loaded in each lipid nanocapsule after observing a dose-dependent onset of cell death based on the number of nanocapsules photolyzed. It was not clear whether large protein molecules had the same encapsulation efficiency as we previously determined for small molecules.28 Therefore, to determine the average number of AGT-PP1-1 contained in lipid nanocapsules, we labeled AGT-PP1 with Alexa488 and quantified the number of AGT-PP1-Alexa488 molecules per lipid nanocapsule using a single-molecule counting technique we previously developed.38 From this experiment, we found there were, on average, 2.4 +/− 0.2 molecules of AGT-PP1-Alexa488 per lipid nanocapsule. This means approximately 8 molecules of AGT-PP1-1 are sufficient to induce apoptosis in the majority of Ba/F3 cells.
Conclusions
Light-addressable lipid nanocapsules for intracellular delivery offer several important advantages: (1) The lipid nanocapsules act as carriers to deliver cell-impermeable molecules into the cell where a single light pulse releases their contents into the cytoplasm; (2) There is zero or near-zero background biological activity over the timescale of the experiments (~ 6 hours) from the encapsulated molecules because the lipid nanocapsules prevent physical contact between the encased molecules and the cell; (3) Lipid nanocapsules allow facile tuning of the photolysis wavelength to the far red or near IR; (4) Photolysis occurs via a photothermal pathway (which does not cause cell stress as monitored using a calcium indicator), eliminating concerns with radical-induced cell stress or damage; (5) It is straightforward to control and vary the concentration of molecules encapsulated within lipid nanocapsules which, in turn, determines the copy number of molecules released into the cell; (6) Photolysis of individual lipid nanocapsules using single-focused laser pulses allows stimuli to be released in the cell with high spatial and temporal precision. We believe these advantages, coupled with the ability to target cell-impermeant molecules (e.g. drugs, metabolites, siRNA, DNA, proteins) into cells using lipid nanocapsules, will open new possibilities in studying cellular signaling and screening drugs against intracellular targets.
Materials and Methods
Materials
The mini-extruder used to generate vesicles was purchased from Avestin (Ottawa, Ontario). All lipids were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). All dyes were purchased from Invitrogen, Molecular Probes (Carlsbad, CA). AGT-PP1 and SNAP-Surface™ Alexa Fluor® 488 were all purchased from New England BioLabs (Ipswich, MA). The culture medium, RPMI-1460, was obtained from atcc.org. All other materials and reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culturing
Chinese Hamster Ovary (CHO) and HEK 293 cells were purchased from atcc.org, and Ba/F3 cells were graciously provided by the Shah lab at UCSF. CHO-M1 and Ba/F3 cells were cultured in RMPI-1460 medium supplemented with glutamine, 10% fetal bovine serum (FBS), and 1% Penn/Strep. HEK 293 cells were cultured in DMEM supplemented with 10% FBS and 1% PennStrep. Ba/F3 cells were selected for their BCR-ABL dependence. Inhibition of BCR-ABL reverts the cells to interleukin 3 (IL-3) dependence and results in apoptosis when IL-3 is not present. This cell line serves as a model for chronic myelogenous leukemia (CML).
Optical Setup
The setup for visualization and photolysis of vesicles has been described in detail elsewhere.12 The optical setup used for TIRF analysis also has been described in detail.38 Confocal images were taken using a Zeiss 510META Confocal microscope.
Design and expression of His-(AGT)-SGSG-APTYSPPPPP (AGT-PP1)
Design and synthesis were completed as previously described.35 Briefly, a mutant form of the human gene O6-alkylguanine-DNA-alkyltransferase (AGT) was amplified and extended from the pSS26b plasmid (Covalys) by a two-step process using the following primers:
-
Step 1,
FWD: 5’- GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT AAT GGA CAA AGA TTG ACG A-3’
RVS: 5’- CGG GCT ATA GGT CGG CGC GCT ACC GCT GCC TCC CAG ACC CGG TTT ACC CAG-3’
-
Step 2
FWD: 5’- GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT AAT GGA CAA AGA TTG ACG A-3’
RVS: 5’- GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC CTA CGG CGG CGG CGG CGG GCT ATA GGT CGG CGC-3’
AGT-PP1 was directly incorporated into a pDONR™ 221 vector (Invitrogen) using Gateway® technology. AGT-PP1 was then placed into the vector PDEST-527, which contains an N-terminal His6 tag, using Gateway® technology. The sequence of the final construct was confirmed by sequencing of the entire gene. AGT-PP1 was expressed in BL21 E. coli cells and purified using Ni·NTA resin. Purity was confirmed by SDS-PAGE.
Synthesis of Compound 1
Compound 1 (supplementary info, scheme1) was synthesized as previously described.35 Briefly, the quinazoline portion, Boc protected at the pipirizine, was deprotected in 30% Trifluoroacetic acid/methylene chloride for 3 hours. All volatiles were removed in vacu and the product was used without further purification. The deprotected quinazoline portion was reacted with the glutaric anhydride linked benzylguanine portion in DMF with HOBT·H2O, DIPEA, and EDCI overnight while stirring at room temperature. The reaction was then purified using reverse phase preparatory HPLC to yield the product. The product was characterized using analytical HPLC, mass spectrometry, and 1H NMR.
Labeling of AGT-PP1 with Compound 1
AGT-PP1 was labeled with 1 using the following conditions. A purified AGT fusion protein (15 µM) was incubated with 1 (22.5 µM) in labeling buffer (50 mM Tris buffer, pH 7.5, 100 mM NaCl, 0.1% Tween 20 and 1 mM DTT) for 1.5 h at 25 °C. The protein-small molecule conjugate was then purified (using the conditions described below), quantified, and used directly.
Labeling of AGT-PP1 with SNAP-Surface™ Alexa Fluor® 488
AGT-PP1 was labeled with SNAP-Surface™ Alexa Fluor® 488. A purified AGT fusion protein (15 µM) was incubated with SNAP-Surface™ Alexa Fluor® 488 (22.5 µM) in labeling buffer (50 mM Tris buffer, pH 7.5, 100 mM NaCl, 0.1% Tween 20 and 1 mM DTT) for 1.5 hours at 25 °C. The protein-small molecule conjugate was then purified (using the conditions described below), quantified, and used directly.
Purification of AGT-PP1-Small Molecule Conjugates
Bio Rad Micro Bio-Spin® columns equilibrated with Tris buffer were used to purify the protein-small molecule conjugates. All AGT-small molecule conjugate labeling reactions were purified twice using two Bio Rad Micro Bio-Spin® columns according to the manufacturer’s procedure
Nanocapsule Synthesis
Small unilamellar vesicles (SUVs) were formed by extrusion. Chloroform solutions of lipids and membrane dyes were combined in a vial and dried under a stream of N2 for ~30 min. Samples were then further dried under vacuum for a minimum of two hours to remove any residual chloroform. Once the chloroform was removed, the dried lipid material was then rehydrated with a biological buffer (pH = 7.4) containing (if any) the contents to be encapsulated to a final lipid concentration of 2.0 mM. This solution was then vortexed for about 1 minute to remove the lipid cake and to create multi-lamellar vesicles. SUVs were formed by passing the solution of multi-lamellar vesicles through a mini extruder that contained 100-nm pores at a temperature above the melting temperature (Tm) of the lipid membrane at least 25 times. Because the loaded contents are not membrane permeable and non-encapsulated materials are diluted to concentrations in the nM range when introduced to the cells, no further purification step was utilized.
Loading Lipid Nanocapsules into Cells
Cells were grown to confluence on No. 1 (for photolysis) and No. 1 ½ coverslips (for confocal imaging). For photolysis experiments, a solution of concentrated vesicles was added to the media for endocytosis for 2 hours, washed gently with fresh media, and returned to the incubator for an additional 30 minutes. For the confocal images, vesicles were loaded for 1 hour simultaneously with an endosome marker, pHrodo-dextran or transferrin. Cells were then gently washed and returned to fresh media.
Intracellular Photolysis
After various loading times, cells were washed twice with media and returned to the incubator for an additional 30 minutes. Vesicles were photolyzed with a single 645-nm, 3-ns pulse of 600nJ/pulse measured at the back of the objective. For IP3 experiments, CHO-M1 cells were washed twice with a biological buffer that didn’t contain Ca2+.
Tracking Cells with Photolyzed Vesicles
For the inhibition experiments, cells were photolyzed in very small wells (diameter ~1mm) and tracked via brightfield illumination with a 10× objective for a period of six hours with a snapshot taken every 90 seconds in order to monitor movement. After six hours, positions were noted, and morphology was determined using a 100× superfluor objective.
Supplementary Material
Acknowledgements
The authors would like to thank the Shah lab at UCSF for graciously providing the Ba/F3 cell line. K. Gregersen acknowledges support from the Center of Nanotechnology at the University of Washington for an IGERT fellowship, and NIH Training Program No.1T32CA138312-01. This work is also supported by the National Institutes of Health (NIH; Grant AG 029574) and the NSF (CHE-0924320).
Footnotes
Supporting Information Available: Structure of Compound 1. Additional images of cells loaded with lipid nanocapsules and labeled with pHrodo-dextran and trasnferrin-Alexa488. Video recording of Ba/F3 cell death. This material is available free of charge via the Intrnet at http://pubs.acs.org
References
- 1.Gug S, Charon S, Specht A, Alarcon K, Ogden D, Zietz B, Leonard J, Haacke S, Bolze F, Nicoud JF, et al. M. Photolabile Glutamate Protecting Group with High One- and Two-Photon Uncaging Efficiencies. ChemBioChem. 2008;9:1303–1307. doi: 10.1002/cbic.200700651. [DOI] [PubMed] [Google Scholar]
- 2.Matsuzaki M, Hayama T, Kasai H, Ellis-Davies GCR. Two-Photon Uncaging of Gamma-Aminobutyric Acid in Intact Brain Tissue. Nat. Chem. Biol. 2010;6:255–257. doi: 10.1038/nchembio.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kantevari S, Matsuzaki M, Kanemoto Y, Kasai H, Ellis-Davies GCR. Two-Color, Two-Photon Uncaging of Glutamate and GABA. Nat. Methods. 2010;7:123–125. doi: 10.1038/nmeth.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warther D, Bolze F, Leonard J, Gug S, Specht A, Puliti D, Sun XH, Kessler P, Lutz Y, Vonesch JL, et al. Live-cell One- and Two-Photon Uncaging of a Far-Red Emitting Acridinone Fluorophore. J. Am. Chem. Soc. 2010;132:2585–2590. doi: 10.1021/ja9074562. [DOI] [PubMed] [Google Scholar]
- 5.Nikolenko V, Poskanzer KE, Yuste R. Two-Photon Photostimulation and Imaging of Neural Circuits. Nat. Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- 6.Sun B, Chiu DT. Spatially and Temporally Resolved Delivery of Stimuli to Single Cells. J. Am. Chem. Soc. 2003;125:3702–3703. doi: 10.1021/ja029942p. [DOI] [PubMed] [Google Scholar]
- 7.Sun BY, Chiu DT. Synthesis, Loading, and Application of Individual Nanocapsules for Probing Single-Cell Signaling. Langmuir. 2004;20:4614–4620. doi: 10.1021/la0364340. [DOI] [PubMed] [Google Scholar]
- 8.Sun BY, Lim DSW, Kuo JS, Kuyper CL, Chiu DT. Fast Initiation of Chemical Reactions with Llaser-Induced Breakdown of a Nanoscale Partition. Langmuir. 2004;20:9437–9440. doi: 10.1021/la048444m. [DOI] [PubMed] [Google Scholar]
- 9.Sun B, Mutch SA, Lorenz RM, Chiu DT. Layered Polyelectrolyte-Silica Coating for Nanocapsules. Langmuir. 2005;21:10763–10769. doi: 10.1021/la0513297. [DOI] [PubMed] [Google Scholar]
- 10.Sun BY, Chiu DT. Determination of the Encapsulation Efficiency of Individual Vesicles Using Single-Vesicle Photolysis and Confocal Single-Molecule Detection. Anal. Chem. 2005;77:2770–2776. doi: 10.1021/ac048439n. [DOI] [PubMed] [Google Scholar]
- 11.Dendramis KA, Allen PB, Reid PJ, Chiu DT. Spectrally Tunable Uncaging of Biological Stimuli from Nanocapsules. Chemical Communications. 2008;39:4795–4797. doi: 10.1039/b806685j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dendramis KA, Chiu DT. Laser Photolysis of Dye-Sensitized Nanocapsules Occurs via a Photothermal Pathway. J. Am. Chem. Soc. 2009;131:16771–16778. doi: 10.1021/ja904976r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G, Mikhailovsky A, Khant HA, Fu C, Chiu W, Zasadzinski JA. Remotely Triggered Liposome Release by Near-Infrared Light Absorption via Hollow Gold Nanoshells. J. Am. Chem. Soc. 2008;130:8175–8177. doi: 10.1021/ja802656d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volodkin DV, Skirtach AG, Mohwald H. Near-IR Remote Release from Assemblies of Liposomes and Nanoparticles. Angew. Chem. Int. Ed. 2009;48:1807–1809. doi: 10.1002/anie.200805572. [DOI] [PubMed] [Google Scholar]
- 15.Troutman TS, Leung SJ, Tomanowski M. Light-Induced Content Release from Plasmon-Resonant Liposomes. Adv. Mat. 2009;21:2334–2338. doi: 10.1002/adma.200900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skirtach AG, Javier AM, Kreft O, Kohler K, Alberola AP, Mohwalkd H, Parak WJ, Sukhorkov GB. Laser-Induced Release of Encapsulated Materials inside Living Cells. Angew. Chem. Int. Ed. 2006;45:4612–4617. doi: 10.1002/anie.200504599. [DOI] [PubMed] [Google Scholar]
- 17.Palankar R, Skirtach AG, Kreft O, Bedard M, Garstka M, Gould K, Mohwald H, Sukhorukov GB, Winterhalter M, Springer S. Intracellular Transport. Small. 2009;5:2168–2176. doi: 10.1002/smll.200900809. [DOI] [PubMed] [Google Scholar]
- 18.Munoz Javier A, del Pino P, Bedard MF, Ho D, Skirtach AG, Sukhorukov GB, Plank C, Parak WJ. Photoactivated Release of Cargofrom the Cavity of Polyelectrolyte Capsules to the Cytosol of Cells. Langmuir. 2008;24:12517–12520. doi: 10.1021/la802448z. [DOI] [PubMed] [Google Scholar]
- 19.Smith BP, Dendramis KA, Chiu DT. Investigating Lyophilization of Lipid Nanocapsules with Fluorescence Correlation Spectroscopy. Langmuir. 2010;26:10218–10222. doi: 10.1021/la1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai JH, Song SX, Rui MJ, Liu D, Ding Q, Peng JL, Xu YH. A Synthetic Peptide Mediated Active Targeting of Cisplatin Liposomes to Tie2 Expressing Cells. J. Control. Release. 2009;139:174–181. doi: 10.1016/j.jconrel.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 21.Miller CR, Bondurant B, McLean SD, McGovern KA, O'Brien DF. Liposome-Cell Interactions in vitro: Effect of Liposome Surface Charge on the Binding and Endocytosis of Conventional and Sterically Stabilized Liposomes. Biochemistry. 1998;37:12875–12883. doi: 10.1021/bi980096y. [DOI] [PubMed] [Google Scholar]
- 22.Peetla C, Labhasetwar V. Effect of Molecular Structure of Cationic Surfactants on Biophysical Interactions of Surfactant-Modified Nanoparticles with a Model Membrane and Cellular Uptake. Langmuir. 2009;25:2369–2377. doi: 10.1021/la803361y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cressman S, Dobson I, Lee JB, Tam YYC, Cullis PR. Synthesis of a Labeled RGD-Lipid, its Incorporation into Lliposomal Nanoparticles, and Their Trafficking In Cultured Endothelial Cells. Bioconjugate Chem. 2009;20:1404–1411. doi: 10.1021/bc900041f. [DOI] [PubMed] [Google Scholar]
- 24.Corsi F, De Palma C, Colombo M, Allevi R, Nebuloni M, Ronchi S, Rizzi G, Tosoni A, Trabucchi E, Clementi E, et al. Towards Ideal Magnetofluorescent Nanoparticles for Bimodal Detection of Breast-Cancer Cells. Small. 2009;5:2555–2564. doi: 10.1002/smll.200900881. [DOI] [PubMed] [Google Scholar]
- 25.Bolte S, Cordelieres FPA. Guided Tour into Ssubcellular Colocalization Analysis in Light Microscopy. J. Microsc.-Oxf. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang PH, Sun XS, Chiu JF, Sun HZ, He QY. Transferrin-Mediated Gold Nanoparticle Cellular Uptake. Bioconjugate Chem. 2005;16:494–496. doi: 10.1021/bc049775d. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Weise S, Hafner M, Rocker C, Zhang F, Parak WJ, Nienhaus GU. Quantitative Analysis of the Protein Corona on FePt Nanoparticles Formed by Transferrin Binding. Journal of the Royal Society Interface. 7:S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuyper CL, Kuo JS, Mutch SA, Chiu DT. Proton Permeation into Single Vesicles Occurs via a Sequential Two-Step Mechanism and is Heterogeneous. J. Am. Chem. Soc. 2006;128:3233–3240. doi: 10.1021/ja057349c. [DOI] [PubMed] [Google Scholar]
- 29.Berridge MJ, Bootman MD, Lipp P. Calcium - A Life and Death Signal. Nature. 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 30.Negulescu PA, Shastri N, Cahalan MD. Intracellular Calcium-Dependence of Gene-Expression in Single T-Lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2873–2877. doi: 10.1073/pnas.91.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmerman LA, Clipstone NA, Ho SN, Northrop JP, Crabtree GR. Rapid Shuttling of NF-AT in Discrimination of Ca2+ Signals and Immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 32.Zitt C, Obukhov AG, Strubing C, Zobel A, Kalkbrenner F, Luckhoff A, Schultz G. Expression of TRPC3 in Chinese Hamster Ovary Cells Results in Calcium-Activated Cation Currents not Related to Store Depletion. Journal of Cell Biology. 1997;138:1333–1341. doi: 10.1083/jcb.138.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandadi S, Numazaki M, Tominaga M, Bhat MB, Armati PJ, Roufogalis BD. Activation of Protein Kinase C Reverses Capsaicin-Induced Calcium-Dependent Desensitization of TRPV1 Ion Channels. Cell Calcium. 2004;35:471–478. doi: 10.1016/j.ceca.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 Cells and Their Use in Kinase Drug Discovery. Curr. Opin. Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 35.Hill ZB, Perera BGK, Maly DJA. Chemical Genetic Method for Generating Bivalent Inhibitors of Protein Kinasese. J. Am. Chem. Soc. 2009;131:6686–6688. doi: 10.1021/ja900871y. [DOI] [PubMed] [Google Scholar]
- 36.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding Imatinib Resistance with a Novel ABL Kinase Inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 37.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, et al. Dasatinib in Imatinib-Resistant Philadelphia Chromosome-Positive Leukemias. New England Journal of Medicine. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 38.Mutch SA, Fujimoto BS, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT. Deconvolving Single-Molecule Intensity Distributions for Quantitative Microscopy Measurements. Biophys. J. 2007;92:2926–2943. doi: 10.1529/biophysj.106.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.