Abstract
Plants are constantly confronted by multiple types of stress. Despite their distinct origin and mode of perception, nutrient deprivation and most stresses have an impact on the overall energy status of the plant, leading to convergent downstream responses that include largely overlapping transcriptional patterns. The emerging view is that this transcriptome reprogramming in energy and stress signaling is partly regulated by the evolutionarily conserved energy sensor protein kinases, SNF1 (sucrose non-fermenting 1) in yeast, AMPK (AMP-activated protein kinase) in mammals and SnRK1 (SNF1-related kinase 1) in plants. Upon sensing the energy deficit associated with stress, nutrient deprivation and darkness, SnRK1 triggers extensive transcriptional changes that contribute to restoring homeostasis, promoting cell survival and elaborating longer-term responses for adaptation, growth and development.
Linking energy and stress
Controlling energy and metabolic homeostasis is a challenge for all organisms, and an intimate relationship exists between energy availability and stress tolerance, survival, cell growth and longevity [1,2]. Energy deprivation is likely to be a consequence of most types of stress regardless of their site and mode of perception. Often associated with stress is a reduction in photosynthesis and/or respiration, which in turn results in energy deprivation and ultimately in growth arrest and cell death [2,3]. This suggests that stress is partly decoded as an energy-deficiency signal that triggers convergent responses independently of the origin of its cause [2]. Comparative studies of public microarray data have uncovered that this is in part accomplished through changes in gene expression, with many genes being induced or repressed by multiple stress stimuli [2,4–7]. Although several plant stress-signaling cascades have been dissected in detail, the intersection points between different signaling pathways, as well as the identity of the signaling intermediates and key regulators, remain largely unknown.
In this review we will first provide an overview of the physiological and molecular responses associated with plant energy deficit. Although traditionally associated with sugar deprivation and darkness, energy deficit is, to varying degrees, probably triggered by all adverse conditions that impinge on cellular energy and metabolite levels. Based on recent findings, the role of energy signaling mediated by the SnRK1 (SNF1 [sucrose non-fermenting 1]-related kinase 1) protein kinases (PKs) in the orchestration of the transcriptional responses will be discussed. Finally, we will introduce the emerging view that other nutrient and metabolic signaling pathways might interact either directly or through cross-talk with SnRK1 and contribute to energy and stress responses.
Convergent responses to energy deficit
Fluctuation of energy status or energy deprivation is an inherent part of plants’ lifestyle and can be caused by alterations of the normal day–night cycle due to shading or to an extension of the night hours [3,8]. It can occur in secondary sink tissues when competing for photoassimilate with primary sink organs like seeds and fruits [3] or in etiolated seedlings and tissues before the acquirement of full photosynthetic competency [9]. An energy deficit can also be triggered by carbon hijack by pathogens or by many adverse conditions such as drought, extreme temperatures, pollutants or flooding that interfere with carbon assimilation and/or respiration [2].
The consequences of energy deprivation have been studied for decades, and a common set of physiological, metabolic and molecular events have been uncovered, including immediate cessation of growth, activation of catabolic pathways to provide alternative nutrient, metabolite and energy sources, and a decline in the activity of biosynthetic enzymes to preserve energy [2,8,10,11] (for earlier references, see [12,13]). A reduction in the cellular energy pool is often coupled to the activation of vacuolar autophagy [10,11,14], a process by which nonselective bulk degradation of the cytosolic content takes place to recycle nutrients. This is accomplished through the generation of a double membranous structure, the autophagosome, which sequesters and transports portions of the cytoplasm to the vacuole for its degradation [15]. Autophagy also plays an essential role in plant immunity to viral infection [16].
Metabolic and structural modifications are often accompanied by changes in gene expression. Initial studies focused on individual genes that are induced upon removal of sugar from the culture medium or upon dark-induced starvation [12,13,17]. Recently, large-scale transcriptome profiling has revealed that the effect of sugar deprivation or prolonged darkness is not restricted to a few genes and that it impacts more than a thousand gene targets [2,8,10,11,18–25]. In general, carbon depletion induces genes involved in remobilization of alternative nutrient, metabolite and energy sources and represses those related to biosynthetic processes, growth and proliferation [2,8,10,11,18–25]. Feeding metabolizable sugars, on the other hand, has an opposite effect on gene expression [18–22]. The physiological relevance of this response is supported by the fact that a largely similar transcriptional pattern is associated with differential endogenous sugar levels induced by differential photosynthetic rates [21].
Many studies employed high sugar concentrations (over 100 mM) and/or long treatments (in the range of hours to days), raising questions about the sensitivity and kinetics of the responses and possible secondary effects. To examine early sugar responses, a recent study investigated global gene expression changes within 30 min of the addition of 15 mM sucrose to Arabidopsis (Arabidopsis thaliana) seedlings starved for two days [22]. This experiment led to the identification of a set of 165 rapidly responsive genes with marked transcript changes that are enriched in transcription factors (TFs), redox regulators, components of the proteasome and trehalose metabolism [22]. Most interestingly, many of these genes, including the genes for the trehalose phosphate synthase-like protein (TPS8, TPS9, TPS10 and TPS11) and the autophagy gene ATG8e (AUTOPHAGY 8E), which are repressed by sucrose within 30 min, are also repressed early on in the light period and are rapidly induced during the extended night, suggesting that a small reduction in the carbon status is sufficient to trigger changes in their expression [23]. In fact, even a two to four hour extension of the night can result in acute energy deprivation, launching a response similar to that of prolonged starvation affecting not only transcription but also polysome loading, translation rates and cell proliferation [2,3,23,26]. A clear coupling between sugar availability and cell growth is indeed observed for the regulation of cell cycle, where the G1/S transition is blocked in sugar-starved cells. One reason for this inhibition is that the G1/S transition relies on cyclin 3.1, a protein whose levels are strongly dependent on sugar availability [27,28].
The finding that the response to energy fluctuation is triggered before a complete energy deficit occurs is particularly important considering that even short periods of severe carbon starvation lead to an inhibition of growth, as exemplified by the seed abortion associated with episodes of heat and drought or other stress [3,29]. To avoid such deleterious consequences and in the context of the diurnal cycle, it has been proposed that plants respond to decreasing carbon in an acclimatory manner, adjusting their growth, storage and carbon mobilization to maximize biomass production, but at the same time preventing extreme situations of starvation [3]. Such a scenario provides a compelling physiological explanation for the sensitivity of the response to small energy changes resulting from the progressive depletion of starch during the night [3,30]. Despite our increased understanding of the consequences of energy deficit at the physiological and gene expression levels, it is not known how energy availability is linked to the regulation of growth and how plants cope with unpredicted environmental perturbations that cause sudden depletion of their energy sources at the cellular and molecular levels.
SnRK1 as master regulator of transcription in energy signaling
Recently, two Arabidopsis PKs, KIN10 and KIN11 (also known as Arabidopsis kinase 10 [AKIN10] and AKIN11), have been identified as central regulators of the transcriptome in response to darkness and multiple types of stress signals, providing new insight into the molecular mechanisms underlying energy signaling and new tools and directions of research [2]. KIN10 and KIN11 are the Arabidopsis orthologs of yeast (Saccharomyces cerevisiae) SNF1 and mammalian AMPK (AMP-activated protein kinase), all members of a highly conserved eukaryotic PK family and collectively named SnRKs in plants [31–35]. The large Arabidopsis SnRK superfamily consists of three distinct subfamilies. The SnRK1 subfamily members KIN10 and KIN11 are the closest relatives of SNF1 and AMPK, and the SnRK2 and SnRK3 subfamilies comprise 35 more divergent and plant-specific PKs involved in stress and abscisic acid signaling in Arabidopsis and other plant species [31,32,34,36,37].
In response to glucose limitation, yeast SNF1 is known to control genes involved in the metabolism of alternative carbon sources, respiration, gluconeogenesis, nutrient transport and meiosis [38]. Both transcription repressors and activators, as well as histone H3 and RNA polymerase II, are modulated by SNF1 [38]. In mammals, AMPK is activated by hypoxia, ischemia, heat shock and exercise, which increase the AMP/ATP ratio, thereby switching off energy-consuming processes and activating catabolism [35,39]. Recent research has uncovered that this is implemented not only through direct enzyme regulation but also through transcriptional control of metabolism, cell signaling, growth, proliferation, immunity, transcription and apoptosis. Many AMPK targets involved in transcriptional regulation have been identified in diverse organs and cell types [35,39]. However, full characterization of the gene networks regulated by AMPK activation remains to be established.
As in mammals, research on plant SnRK1s has traditionally centered around enzyme regulation [31,33,34,40–42]. Excellent reviews cover information on the SnRK1 control of metabolic enzymes, as well as details on the subunit composition, regulation by upstream kinases and the evolution of the SnRK1, SNF1 and AMPK systems [31–35,38,39]. However, few SnRK1 target genes have been identified; two examples of such genes are a potato (Solanum tuberosum) sucrose synthase gene activated by sucrose and a wheat α-amylase gene repressed by glucose [43,44]. Recently, rice (Oryza sativa) SnRK1 has been shown to regulate the activation of the α-amylase gene promoter by MYBS1 (v-myb avian myeloblastosis viral oncogene homolog involved in sugar signaling) under glucose starvation [45]. Remarkably, regulation of gene expression by SnRK1 is not restricted to a few genes, as transient activation of the Arabidopsis ortholog KIN10 triggers extensive reprogramming of transcription, affecting over a thousand genes in mesophyll cells [2]. Most importantly, the transcriptional profile induced by KIN10 activity largely overlaps with the profiles obtained under a variety of dark and starvation conditions in suspension culture cells and whole plants [2,8,10,24,25], and it is the opposite of profiles obtained from sucrose- or glucose-fed seedlings [2,18,20] and seedlings with maximal versus limited carbon assimilation owing to differential CO2 concentrations [2,21]. Such extensive transcriptome overlaps provide compelling evidence for the physiological relevance of the SnRK1-mediated transcriptional responses and highlight the universal nature of energy signaling [2]. Significantly, double kin10 kin11 deficiency abrogates the transcriptional switch in darkness and stress signaling and impairs growth and starch degradation. Thus, KIN10 and KIN11 are likely to have pivotal roles in linking stress, sugar and developmental signals for the global regulation of plant metabolism, energy balance, growth and survival [2].
SnRK1 target genes in metabolism
Although many enzymes and genes for central metabolic pathways have been extensively studied in plants, little is known about their regulatory mechanisms. The surprisingly large and broad spectrum of Arabidopsis genes as KIN10 targets is discussed below. These Arabidopsis genes [2,8,10,24,25] (Figures 1 and 2) are likely to be conserved in most plant species, therefore validating investigations with a model system for understanding the regulation of plant metabolic pathways.
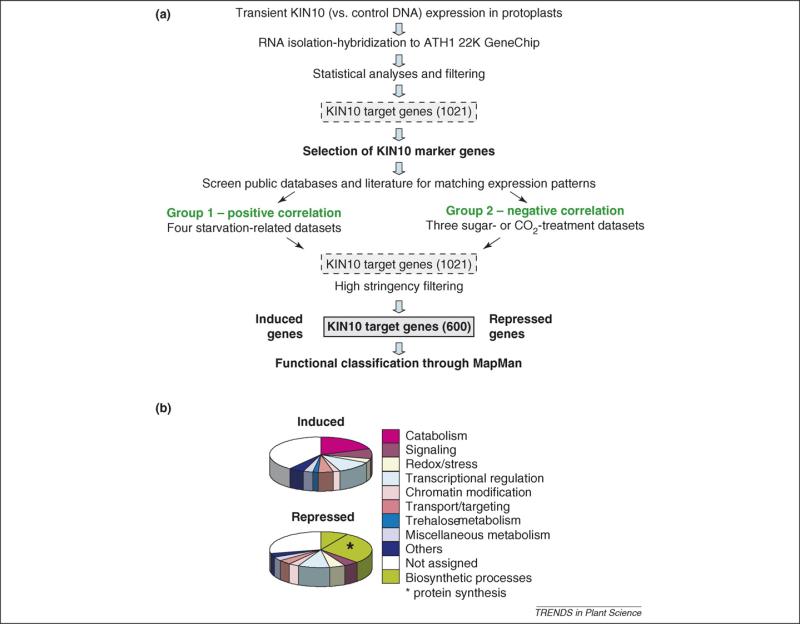
Figure 1.
Transcriptome response to energy deprivation mediated by SnRK1 activation. Extensive transcriptional reprogramming is an important part of the convergent responses to stress, nutrient starvation and darkness [2,8,10,11,24,25]. Energy deficiency is sensed by the SnRK1 PKs that trigger the induction (280 genes) and repression (320 genes) of genes involved in a wide variety of cellular processes [2]. (a) The SnRK1 target gene list was generated by filtering overlapped genes controlled by transient KIN10 activation in Arabidopsis mesophyll protoplasts and by various starvation conditions in cultured cells, seedlings and leaves. The microarray datasets were independently generated and are publicly accessible [2]. These 600 SnRK1 target genes are regulated in an opposite manner by sugar availability because glucose and sucrose inactivate SnRK1. (b) The functional categories for the SnRK1 target genes in the pie chart were assigned based on the classification in the MapMan program [8] and sorted in Excel.
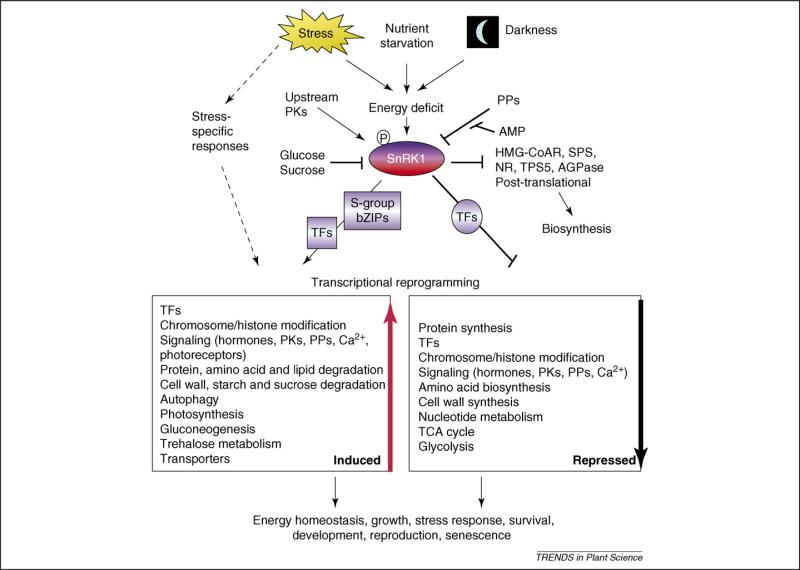
Figure 2.
Decoding diverse stress as convergent energy signaling. Besides triggering different stress-specific responses, multiple types of stress ultimately converge and generate energy-deficiency signals that result in the activation of the SnRK1 energy sensors [2,8,10,24,25]. Conversely, sugars have a repressive effect [2,18,20,21]. Phosphorylation at a conserved threonine residue (e.g. T175 in Arabidopsis KIN10, T210 in yeast SNF1 and T172 in human AMPKα) is required for SnRK1 activity, but the ultimate metabolic signal responsible for SnRK1 activation remains enigmatic. Upstream protein kinases (PKs), protein phosphatases (PPs), and additional regulatory subunits might contribute to the fine-tuning of the system and possibly confer tissue and cell-type specificity [33]. Activated SnRK1 initiates an energy-saving program at several levels, including massive transcriptional reprogramming that targets a wide range of cellular processes. The S-group bZIP (basic leucine zipper) transcription factors (TFs) mediate some SnRK1 activated genes [2]. In addition to contributing to the maintenance of cellular energy homeostasis and tolerance to stress, SnRK1 has profound effects at the whole-organism level, influencing growth, viability, reproduction and senescence, and is thus proposed to be central in the integration of metabolic, stress and developmental signals [2]. SnRK1 also phosphorylates and regulates enzymes mostly involved in carbon (C) and nitrogen (N) metabolism [31,33,34,40–42]. Abbreviations: AGPase, ADP-glucose pyrophosphorylase; HMGCoAR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; NR, nitrate reductase; SPS, sucrose phosphate synthase; TCA, tricarboxylic acid cycle; TPS5, trehalose-6-phosphate synthase 5.
Biosynthetic processes
Over 300 genes involved in various biosynthetic processes, including amino acid, cell wall, lipid, nucleotide, protein, sucrose and starch synthesis, are repressed through SnRK1 in energy-deprivation conditions [2,8,10,24,25] (Figures 1 and 2). The most prominent targets of repression are genes involved in protein synthesis, including a large set of genes encoding ribosomal proteins, and the nucleolin AtNUC-L1, a major regulator of ribosome biogenesis [46].
Catabolic processes
In response to energy deficit, SnRK1 mediates the induction of nearly 300 genes related to multiple nutrient remobilization processes, consistent with a global metabolic switch induced by SnRK1 to provide alternative sources of metabolites and energy supplies through amino acid catabolism, sucrose, starch, cell wall and polysaccharide hydrolysis, lipid mobilization and β-oxidation [2,8,10,24,25] (Figures 1 and 2). Under conditions of carbon deprivation, protein degradation is a key process for recycling cellular components, and it is likely to proceed as part of a larger autophagy program [14,15]. Indeed, starvation modulates specific amino acid accumulation and the activation of many genes related to amino acid catabolism [3,8,10,11,23–25], suggesting enhanced rates of protein degradation. Launching of autophagy, however, might be reflected in the induction of several ATG8 genes. The upregulation of these genes as molecular markers for autophagy [14] coincides with vacuolar autophagy triggered by sucrose starvation in Arabidopsis suspension culture cells [10]. The ATG8/ATG12 conjugation pathways are conserved in plants for starvation responses and might have important roles during development and stress [15].
Upon starvation or dark treatment, enhanced hydrolytic activities can be measured that are accompanied by a decrease in some of the cell wall components [47]. This is consistent also with extensive transcriptional changes in genes related to cell wall modification, synthesis and degradation, suggesting a role for the cell walls as an alternative energy source and perhaps as a means of restricting cell growth and elongation. Changes in metabolism genes are also accompanied by increased expression of genes encoding carbohydrate-, amino acid-, peptide- and ion-transporters and aquaporins, presumably to facilitate mobilization and recycling of these molecules.
Alternative metabolic pathways
SnRK1 activity triggers the coordinated induction of genes encoding phosphoenol pyruvate carboxykinase (PEPCK), pyruvate phosphate dikinase (PPDK), pyruvate kinase (PK), glutamate dehydrogenase (GDH) and asparagine synthetase (ASN1, the corresponding gene also known as DARK-INDUCED 6 [DIN6]) [2]. These genes have been suggested to be part of a novel cycle that generates asparagine for more energy-economical nitrogen remobilization under darkness, stress and starvation conditions [24,48]. The importance of the catabolic activity of GDH during starvation is demonstrated by the enhanced susceptibility of the double gdh1-2/gdh2-1 mutant to starvation in darkness [49], a phenotype shared by mutants impaired in autophagy [14].
Another consequence of energy deprivation is the upregulation of the mitochondrial pathway mediated by the ETF (electron transfer flavoprotein) and ETFQO (ETF:ubiquinone oxidoreductase) enzymes [50,51]. The ETFQO transcript is dramatically induced by sugar starvation, whereas the genes encoding the α- and β-subunits of ETF are constitutively expressed [50,51]. The corresponding knockout mutants undergo accelerated senescence during prolonged darkness and accumulate significant amounts of isovaleryl-CoA (an intermediate of leucine catabolism), phytanoyl-CoA (an intermediate of chlorophyll degradation), leucine, valine and isoleucine. These studies indicate that ETF and ETFQO function together during amino acid and chlorophyll catabolism, probably as a way to fuel respiration with alternative substrates and to avoid accumulation of branched chain amino acids that can be toxic to the cell [50,51].
SnRK1 target genes in regulatory processes
SnRK1 appears to play a central and previously unrecognized regulatory role because a large number of genes encoding putative TFs as well as histones and histone deacetylases are highly activated or repressed by KIN10 [2,8,10,24,25] (Figures 1 and 2). In addition, several hormone metabolism and hormone responsive genes, as well as many genes encoding other signal transduction components, including PKs, protein phosphatases, and calcium modulators, are affected by KIN10 [2].
SnRK1 target genes in general stress signaling
Noticeably, comparative studies using a large compendium of microarray profiles associated with very diverse types of stress have identified several of the SnRK1-induced genes as ‘multi-stress’ responsive genes (Table 1) [2,4–6]. Intriguingly, some of them are conserved and are implicated in general stress responses in other organisms. RSH2, for instance, is one of the three Arabidopsis homologs of RelA/SpoT, a central regulator of the ‘stringent response’ that represses bacterial transcription and ultimately growth in response to stress [52]. The zinc finger TF AZF2 (Arabidopsis zinc finger [C2H2 type] protein 2) was originally identified as a suppressor of the yeast snf4 mutation [53] and is related to the zinc finger TFs Msn2 (multicopy suppressor of SNF1 mutation 2) and Msn4, two global regulators of the transcriptional stress response in yeast [54]. It is possible that the overall function of these regulators could be conserved from yeast to plants because AZF2 and other closely related TFs of its family seem to be involved in defense [55] and in the response to cold, salt and drought [56].
Table 1.
SnRK1 target genes involved in general stress signalinga
| Gene | Annotation |
|---|---|
| At3g14050 | RSH2 (RelA/SpoT homolog), ppGpp metabolism |
| At3g19580 | AZF2 (Arabidopsis zinc finger [C2H2 type] protein 2), TF |
| At4g11330 | MPK5 (MAP kinase 5) |
| At3g55840 | HSPRO1 (heat-shock-like protein 1), nematode-resistance gene |
| At2g40000 | HSPRO2 (heat-shock-like protein 2), nematode-resistance protein |
| At5g22920 | PGPD14 (pollen-germination-related protein), zinc finger (C3HC4-type RING) family, TF |
| At4g35770 | SEN1 (senescence-associated protein 1) |
| At4g27260 | GH3.5, auxin inducible |
| At2g33830 | Auxin-regulated protein |
| At3g61060 | Lectin-related protein |
| At2g18700 | TPS11 (a putative trehalose-6-phosphate synthase) |
| At3g48360 | AtBT2, BTB/POZ domain, TF |
| At5g06690 | Thioredoxin-like protein |
| At3g26740 | CCL (CCR [cold- and circadian-regulated]-like) |
| At3g47160 | RNA-binding-protein-like protein |
| At3g02140 | TMAC2 (two or more ABREs-containing gene 2) |
| At1g25400 | Unknown protein |
| At3g15630 | Unknown protein |
| At1g15010 | Unknown protein |
| At1g01240 | Unknown protein |
| At2g36220 | Unknown protein |
| At1g23710 | Unknown protein |
| At2g15890 | Unknown protein |
| At1g27100 | Unknown protein |
A large impact of SnRK1 on transcription, including stress-related gene expression, has been reported in pea (Pisum sativum) embryos, where SnRK1 silencing was induced by conditional antisense expression [57]. Consistent with these findings, Arabidopsis plants overexpressing KIN10 are more tolerant to nutrient deprivation [2], and antisense lines of potato StubGAL83 (Solanum tuberosum homolog of the glucose repression protein GAL83, a regulatory β-subunit of the SnRK1 complex) are hypersensitive to salt [58]. A link of SnRK1 to biotic stress signaling is also supported by the enhanced resistance of tobacco (Nicotiana tabacum) plants overexpressing SnRK1 to geminivirus infection [59], the interaction of the chimeric βγ-regulatory subunit with proteins involved in resistance to nematodes [60], and the role of the GAL83 β-subunit in resource allocation to roots in response to herbivory [61].
Metabolic regulation of SnRK1s
Although it is well established that SNF1 and AMPK are activated under glucose depletion (high AMP/ATP) conditions, direct activation of plant SnRK1 and yeast SNF1 by AMP has not been observed [31,38]. However, AMP blocks SnRK1 inactivation by preventing dephosphorylation of a critical and conserved residue in the T loop [62]. The fact that SnRK1 target genes are activated under a wide variety of starvation and stress conditions suggests a common signal reflective of energy deficiency that awaits to be elucidated (Figure 2).
It has been shown that glucose-6-phosphate (G6P) can block SnRK1 activity [63], consistent with effective suppression of SnRK1-mediated target gene activation by glucose and sucrose in seedlings and leaf cells [2,18–20,22,47]. However, several studies have reported SnRK1 activation by sucrose [43,64,65]. The various observations might be due to different growth conditions, long-term sugar depletion, stress signaling and difficulties in measuring overt changes in SnRK1 activities, suggesting technical limitations and differences in regulatory mechanisms compared to other organisms [2,9,31,66,67]. Another important consideration is that the plant SnRK1 family has greatly expanded in some species (especially cereals but also potato) [31,34]. Some of its members are not able to complement the yeast Δsnf1 mutants, indicating a clear functional divergence [43,65,68]. Although specific sucrose-signaling pathways do exist in plants [69,70], regulation of SnRK1 seems to rely on the overall energy status of the cell, independently of specific sugar molecules [2,47].
Possible links to SnRK1-mediated energy signaling
Connection to hexokinase
Both sucrose and glucose have a repressive effect on SnRK1 target genes [2], and the product of glucose metabolism through hexokinase (HXK), G6P, can block SnRK1 activity [63]. However, in addition to its central metabolic function, Arabidopsis HXK1 has a distinct signaling function [71,72] (Figure 3). This is reflected in its split distribution between the mitochondria for glycolysis [73,74] and the nucleus for signaling [72]. Despite having significant glucose phosphorylation activity resulting from other HXKs, HXK1 null mutants (gin2 [glucose-insensitive2]) are unable to promote growth under light conditions that favor photosynthesis and thus energy production [71]. It is thus tempting to suggest a scenario where upon sensing glucose, the ‘growth-promoting’ HXK1 pathway would antagonize the ‘stress and growth-restricting’ SnRK1 pathway and vice versa (Figure 3). Even though sugar regulation of some KIN10 target genes is unaffected in the gin2 mutant [2], it is still plausible that there is some level of cross-talk between these two nutrient- and energy-sensing pathways during the stress response, and their possible interaction needs to be further addressed.
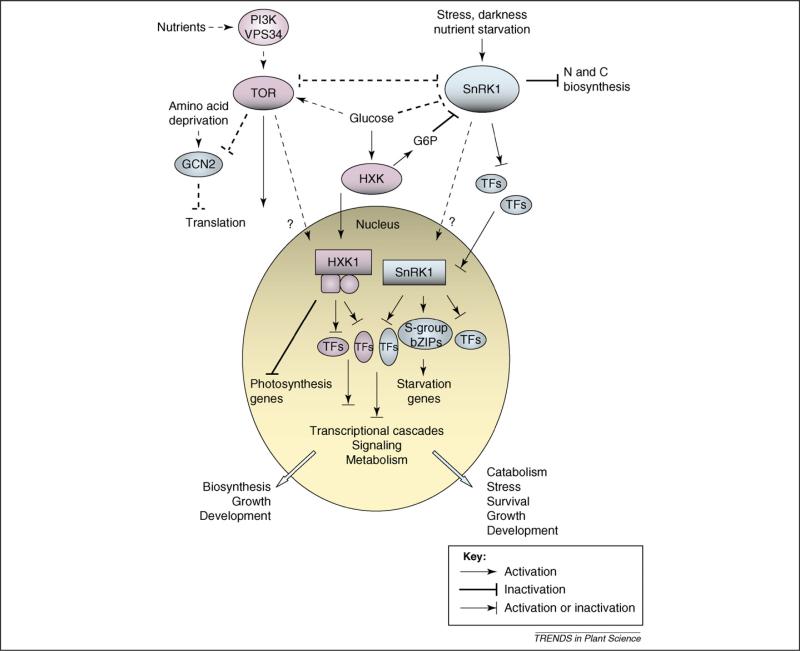
Figure 3.
A model depicting interactions of putative nutrient and energy signaling components. The blue components are part of a network that, upon sensing nutrient and/or energy deficiency, restricts growth and promotes nutrient remobilization, survival and tolerance to stress. The pink components form a hypothetical antagonistic network that couples nutrient and/or energy availability with growth. In response to energy deprivation, SnRK1 orchestrates an energy-saving program through direct enzyme regulation and through extensive transcriptional reprogramming that involves at least the S-group bZIP (basic leucine zipper) transcription factors (TFs) [2]. SnRK1 response is blocked by sugars, partly through the product of hexokinase (HXK) activity, glucose-6-phosphate (G6P) [63]. In addition to its catalytic role, HXK1 has a distinct signaling function in the nucleus, where it regulates the expression of photosynthetic genes, among others [71,72]. HXK might act in a cooperative manner with the TOR (target of rapamycin) PK (protein kinase) in the growth-promoting network. As in mammals [75], nutritional information might be conveyed to plant TOR through the PI3K/VPS34 protein [78]. In nutrient-rich conditions, mTOR (mammalian TOR) promotes growth partly through regulation of the translational machinery [79] and blocks the translation-inhibitory pathway mediated by the amino-acid-deficiency-sensing GCN2 PK. Similar functions seem to apply to the plant TOR [76,77] and to some extent to GCN2 [81,82]. Plants could have evolved unique modes of interplay between the SnRK1 and TOR pathways. A scenario is proposed where various growth-promoting and growth-limiting pathways interact to regulate metabolism, stress tolerance and development in response to the environment and nutrient availability. Solid lines denote proven connections in plants, whereas broken lines represent connections described for other organisms that might or might not exist in plants.
Interaction with TOR
Sugars have been shown to play a central role in the regulation of gene expression, outweighing that of other major nutrients such as nitrogen [18,20]. Nevertheless, an extensive interaction exists between sugar and nitrogen nutrients because the expression of many of the sugar-regulated genes is strongly affected by nitrogen [18,20]. It is likely that energy- and glucose-dependent metabolic sensors, such as SNF1/AMPK/SnRK1 and HXK1, interact with systems sensing the nitrogen status (Figure 3).
In mammals, AMPK is known to negatively regulate TOR (target of rapamycin), a central PK that promotes cell growth and proliferation in response to amino acids and insulin [75]. The function of TOR in coupling cell growth and proliferation seems to be conserved also in Arabidopsis [76], and its expression levels correlate with root and shoot growth, cell size and seed yield [77]. However, little is known thus far about the nutrient regulation of plant TOR or its putative interplay with other metabolic signaling pathways. Nutritional information is conveyed to mammalian TOR (mTOR) through two pathways, the TSC1/2 (tumor suppressor complex 1/2)–Rheb (small GTPase) axis and the hVPS34 (human vacuolar protein sorting 34)/PI3K (phosphoinositide 3-kinase) protein [75]. Arabidopsis lacks any obvious TSC1/2–Rheb functional orthologs, but the C-terminal third of the yeast VSP34 can be replaced with the sequence from AtVPS34/PI3K to complement the yeast Δvps34 mutant [78], raising it as a candidate input pathway for amino acid signals (Figure 3). In the presence of nutrients, TOR blocks the action of GCN2 (general control nonrepressed 2) [79], a PK that inhibits translation initiation upon sensing the uncharged transfer RNAs that accumulate during amino acid limitation [80]. Interestingly, AtGCN2 complements the corresponding yeast mutant [81], and yeast GCN2 phosphorylates wheat eIF2α [82], suggesting that the GCN2 function might be conserved in plants (Figure 3). Future studies might unravel the molecular interactions among TOR, HXK1 and SnRK1 in plant nutrient, energy and stress signaling.
Concluding remarks and perspectives
Energy deprivation in plants is a consequence not only of direct nutrient shortage but also of prolonged darkness and probably of most biotic and abiotic stresses that interfere with photosynthesis and/or respiration. Even short periods of starvation result in growth arrest and a delayed resumption of growth after normal conditions are restored [3]. Therefore, mechanisms have evolved to prevent such extreme situations by responding to changes in the energy status, providing nutrients, metabolites and energy from alternative sources and allowing the adjustment of growth and development based on the available resources and environmental conditions. The finding that different stresses are partly decoded as an energy-deficiency signal sensed by SnRK1s provides new insight into the molecular mechanisms underlying stress responses and possibly stress cross-tolerance. Ultimately, shifting the focus from TFs to more global upstream regulators might open novel directions in crop engineering and improvement. Our knowledge of the nutrient and energy sensing and signaling systems in plants is still very limited. Future work needs to address the actual signals that activate or repress each system, the molecular mechanism of orchestrated transcription regulation and the possible interplay in the convergent transcriptional stress responses. Energy and nutrient signals interact with light and circadian clock inputs in a complex signaling network that is only now beginning to be understood [2,3,21,23,71,72]. These interplays, as well as a possible cross-talk with hormone signaling pathways, should be assessed and elucidated to understand how the cellular energy signaling is fully integrated into whole-plant adaptation and regulation of growth and development.
Acknowledgements
We apologize to all authors whose work could not be cited here because of space constraints. This work was supported by grants from the US National Science Foundation and the National Institutes of Health.
References
- 1.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Baena-Gonzalez E, et al. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 3.Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 4.Walley JW, et al. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007;3:e172. doi: 10.1371/journal.pgen.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilian J, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 6.Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 2007;8:R49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita M, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 9.Rolland F, et al. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 10.Contento AL, et al. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 2004;135:2330–2347. doi: 10.1104/pp.104.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HJ, et al. Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol. Biol. 2007;63:441–463. doi: 10.1007/s11103-006-9100-4. [DOI] [PubMed] [Google Scholar]
- 12.Koch KE. Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 13.Yu SM. Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 1999;121:687–693. doi: 10.1104/pp.121.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson AR, Vierstra RD. Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Bassham DC, et al. Autophagy in development and stress responses of plants. Autophagy. 2006;2:2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Fujiki Y, et al. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 2001;111:345–352. doi: 10.1034/j.1399-3054.2001.1110312.x. [DOI] [PubMed] [Google Scholar]
- 18.Palenchar PM, et al. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol. 2004;5:R91. doi: 10.1186/gb-2004-5-11-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price J, et al. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell. 2004;16:2128–2150. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osuna D, et al. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007;49:463–491. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- 23.Usadel B, et al. Global transcript levels respond to small changes of the carbon status during a progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 2008;46:1834–1861. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JF, Wu SH. Molecular events in senescing Arabidopsis leaves. Plant J. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan-Wollaston V, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 26.Nicolai M, et al. Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol. 2006;141:663–673. doi: 10.1104/pp.106.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riou-Khamlichi C, et al. Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 2000;20:4513–4521. doi: 10.1128/mcb.20.13.4513-4521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menges M, et al. The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell. 2006;18:893–906. doi: 10.1105/tpc.105.039636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilioni L, et al. High temperature and water deficit may reduce seed number in field pea purely by decreasing plant growth rate. Funct. Plant Biol. 2003;30:1151–1164. doi: 10.1071/FP03105. [DOI] [PubMed] [Google Scholar]
- 30.Gibon Y, et al. Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004;39:847–862. doi: 10.1111/j.1365-313X.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- 31.Halford NG, et al. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- 32.Hrabak EM, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Halford NG, et al. Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J. Exp. Bot. 2004;55:35–42. doi: 10.1093/jxb/erh019. [DOI] [PubMed] [Google Scholar]
- 35.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 36.Boudsocq M, et al. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 37.Gong D, et al. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGee SL, Hargreaves M. AMPK and transcriptional regulation. Front. Biosci. 2008;13:3022–3033. doi: 10.2741/2907. [DOI] [PubMed] [Google Scholar]
- 40.Sugden C, et al. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiessen A, et al. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell. 2002;14:2191–2213. doi: 10.1105/tpc.003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harthill JE, et al. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 2006;47:211–223. doi: 10.1111/j.1365-313X.2006.02780.x. [DOI] [PubMed] [Google Scholar]
- 43.Purcell P, et al. Antisense expression of sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J. 1998;14:195–203. [Google Scholar]
- 44.Laurie S, et al. Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an α-amylase (α-Amy2) gene promoter in cultured wheat embryos. J. Exp. Bot. 2003;54:739–747. doi: 10.1093/jxb/erg085. [DOI] [PubMed] [Google Scholar]
- 45.Lu CA, et al. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell. 2007;19:2484–2499. doi: 10.1105/tpc.105.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kojima H, et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 47.Lee EJ, et al. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 2007;48:405–413. doi: 10.1093/pcp/pcm009. [DOI] [PubMed] [Google Scholar]
- 48.Lam HM, et al. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 49.Miyashita Y, Good AG. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 2008;59:667–680. doi: 10.1093/jxb/erm340. [DOI] [PubMed] [Google Scholar]
- 50.Ishizaki K, et al. The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell. 2005;17:2587–2600. doi: 10.1105/tpc.105.035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishizaki K, et al. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 2006;47:751–760. doi: 10.1111/j.1365-313X.2006.02826.x. [DOI] [PubMed] [Google Scholar]
- 52.van der Biezen EA, et al. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3747–3752. doi: 10.1073/pnas.060392397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinow T, et al. Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 2000;23:115–122. doi: 10.1046/j.1365-313x.2000.00809.x. [DOI] [PubMed] [Google Scholar]
- 54.Causton HC, et al. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pauwels L, et al. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1380–1385. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto H, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radchuk R, et al. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006;140:263–278. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovas A, et al. Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J. 2003;33:139–147. doi: 10.1046/j.1365-313x.2003.016015.x. [DOI] [PubMed] [Google Scholar]
- 59.Hao L, et al. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell. 2003;15:1034–1048. doi: 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gissot L, et al. AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol. 2006;142:931–944. doi: 10.1104/pp.106.087718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwachtje J, et al. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12935–12940. doi: 10.1073/pnas.0602316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugden C, et al. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 1999;19:433–439. doi: 10.1046/j.1365-313x.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- 63.Toroser D, et al. Regulation of a plant SNF1-related protein kinase by glucose-6-phosphate. Plant Physiol. 2000;123:403–412. doi: 10.1104/pp.123.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhalerao RP, et al. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiessen A, et al. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003;35:490–500. doi: 10.1046/j.1365-313x.2003.01823.x. [DOI] [PubMed] [Google Scholar]
- 66.Xiao B, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 67.Thelander M, et al. Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovas A, et al. Functional diversity of potato SNF1-related kinases tested in Saccharomyces cerevisiae. Gene. 2003;321:123–129. doi: 10.1016/j.gene.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Wiese A, et al. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 2004;16:1717–1729. doi: 10.1105/tpc.019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiou TJ, Bush DR. Sucrose is a signal molecule in assimilate partitioning. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moore B, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 72.Cho YH, et al. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 73.Granot D. Role of tomato hexose kinases. Funct. Plant Biol. 2007;34:564–570. doi: 10.1071/FP06207. [DOI] [PubMed] [Google Scholar]
- 74.Karve A, et al. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta. 2008;228:411–425. doi: 10.1007/s00425-008-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Avruch J, et al. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 76.Menand B, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welters P, et al. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11398–11402. doi: 10.1073/pnas.91.24.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wek RC, et al. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, et al. Molecular cloning of an Arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta. 2003;217:668–675. doi: 10.1007/s00425-003-1025-4. [DOI] [PubMed] [Google Scholar]
- 82.Chang LY, et al. Specific in vitro phosphorylation of plant eIF2α by eukaryotic eIF2α kinases. Plant Mol. Biol. 1999;41:363–370. doi: 10.1023/a:1006379623534. [DOI] [PubMed] [Google Scholar]