Abstract
Increasing awareness of the possible role of internal dynamics in protein function has led to the development of new methods for experimentally characterizing protein dynamics across multiple time scales, especially using NMR spectroscopy. A few analyses of the conformational dynamics of proteins ranging from nonallosteric single domains to multi-domain allosteric enzymes are now available; however, demonstrating a connection between dynamics and function remains difficult on account of the comparative lack of studies examining both changes in dynamics and changes in function in response to the same perturbations. In previous work, we characterized changes in structure and dynamics on the ps-ns time scale resulting from hydrophobic core mutations in chymotrypsin inhibitor 2 and found that there are moderate, persistent global changes in dynamics in the absence of gross structural changes [Whitley MJ, et al. Biochemistry 2008; 47:8566-8576]. Here, we assay those and additional mutants for inhibitory ability toward the serine proteases elastase and chymotrypsin to determine the effects of mutation on function. Results indicate that core mutation has only a subtle effect on CI2 function. Using chemical shifts, we also studied the effect of complex formation on CI2 structure and found that perturbations are greatest at the complex interface but also propagate toward CI2’s hydrophobic core. The structure-dynamics-function data set completed here suggests that dynamics plays a limited role in the function of this small model system, although we do observe a correlation between nanosecond-scale reactive loop motions and inhibitory ability for mutations at one key position in the hydrophobic core.
Keywords: NMR, serine protease, chemical shift perturbation, protein complex, enzyme kinetics, inhibition constant, mutations
INTRODUCTION
The steady development of both experimental and theoretical methods for the analysis of internal dynamics in biomolecules has resulted in a paradigm shift in our understanding of proteins. Once considered to fold into discrete, static conformations, proteins are now known to undergo conformational fluctuations (structural dynamics) over time scales ranging from faster than picoseconds to seconds and even slower. With the dynamical nature of proteins across multiple time scales well established, the obvious question then becomes whether intrinsic dynamics can be related directly to protein function in some way.
In principle, the relationship between dynamics and function is in many cases difficult to address because establishing a relationship requires paired measurements of both dynamics and function for a series of perturbations, and, ideally, sufficient information to exclude structural effects as a dominant influence. Such structure-dynamics-function data sets exist1-4, though they are rare. More generally, the set of proteins for which a systematic experimental analysis of changes in dynamics in response to various perturbations includes: analyses of the influence of temperature on the dynamics of calmodulin5-6 and ribonuclease H7; the effects of mutation on the dynamics of calmodulin8, eglin c9-11, chymotrypsin inhibitor 2 (CI2)12, cyclophilin A3, and PDZ domains13; and the effects of peptide binding on the dynamics of calmodulin2, Pin114 and PDZ domains15. Even for these relatively few systems in which the effects of various perturbations on internal dynamics have been characterized, it is commonly the case that data on the corresponding effects of a given perturbation on function are not available.
In previous work, we have experimentally characterized the effects of multiple core mutations on dynamics and internal communication in the small serine protease inhibitors eglin c and chymotrypsin inhibitor 2 (CI2).9-10,12 These are small, ~70 residue globular proteins of the potato I inhibitor family that inhibit serine proteases through the presentation of a “reactive site” loop that protrudes from the core globule and binds in the protease active site (Figure 1).16 The loop, which is moderately flexible, is structurally distinct from the core, and hence core mutations would not be expected to affect the reactive loop. However, in CI2, and to a lesser extent in eglin c, mutation of core residues induces global changes in ps-ns dynamics in the protein (detected by NMR relaxation), including the reactive loop; these changes occur in the absence of detectable structural perturbations.9,12 Modulation of dynamics on slower timescales does not appear to be at work, since there is little evidence of significant μs-ms motions in WT or L68A (see below). In addition to the NMR findings, ensemble-based theoretical studies of CI2 have showed that connectivity exists between the core and loop.17-18 Thus, the question arises as to whether coupling between the core and loop regions of CI2 plays a role in CI2 function.
Figure 1.

The structure of chymotrypsin inhibitor 2 (PDB 2CI2) in the free state. The positions of mutated side chains studied in the kinetics experiments are labeled and colored in black.
To resolve the relevance of core-loop coupling in CI2 function and to complete a novel structure-dynamics-function data set, we have assayed the ability of a panel of CI2 (and eglin c) mutants to inhibit elastase and chymotrypsin proteases. Because core mutations induce changes in dynamics that propagate throughout CI2 without significant changes in structure12, this represents an opportunity to link dynamics directly with function. Although few studies probing long-range dynamic effects on function have been reported, there is some precedent for this idea, as distal mutations in a PDZ domain were demonstrated to exert long-range effects on the kinetics of peptide binding19. Our results from the mutants studied here indicate that, overall, core mutations have only a subtle or no effect on inhibitory function. However, one of the more perturbing mutations, L68A, appears to have a weak anti-correlation between nanosecond-scale loop motions and inhibitory power. To further probe the nature of the inhibitory complex, we have measured and assigned NMR chemical shifts of CI2 in complex with active chymotrypsin and used this information to assess how the association of CI2 with the much larger protease in solution affects CI2 structure. To our knowledge, this is the first NMR study of a natural macromolecular inhibitor bound to a serine protease. We conclude with a discussion of how these findings might be related to the question of the role of internal dynamics in the function of more complicated but also more biologically or biomedically interesting proteins.
MATERIALS AND METHODS
Proteins
All variants of the protease inhibitors CI2 and eglin c were created, expressed, and purified as described in published work10,12. For proteins used in NMR experiments, E. coli cultures were grown in media supplemented with U-13C-glucose, 15NH4Cl, and D2O as necessary to yield suitably labeled proteins. Unlabeled proteins were used in all enzyme assays. Both bovine α-chymotrypsin and porcine pancreatic elastase were purchased from Worthington Biochemical Corporation (Lakewood, NJ, USA) in the purest grade available.
Determination of the apparent inhibition constant, Ki,app, for inhibition of two serine proteases
Enzyme assays were generally carried out according to a published protocol20. Briefly, the steady-state rate of enzymatic hydrolysis of a chromogenic substrate was measured both before and after the addition of inhibitor by monitoring the increase in absorbance of the reaction mixture at 410 nm as a function of time. Using the slopes of the absorbance curves before and after inhibition addition as proxies for the uninhibited and inhibited reaction rates, respectively, the apparent inhibition constant can be calculated from the equation , where Vo and Vi are respective rates of substrate hydrolysis in the absence and presence of inhibitor, and [I] is the concentration of inhibitor in the assay mixture. For each inhibitor variant, multiple independent measurements of Ki,app were made at different concentrations, and we report the results as the average value ± standard deviation (with 5 ≤ n ≤ 15). Linear regressions are shown in Supplementary Information.
Assays for porcine pancreatic elastase inhibition were carried out in a total volume of 1 mL of buffer comprised of 100 mM Tris (pH 8.0), 5 mM CaCl2, and 0.05% v/v Tween-80. The chromogenic substrate AAA (N-succinyl-Ala-Ala-Ala-p-nitroanilide) (Sigma-Aldrich, Inc.) was used at an initial concentration of 1 mM. In each reaction, the concentration of elastase was fixed at 1 nM, whereas the inhibitor concentration was varied through a range surrounding the approximate value of Ki,app, from 30 to 75 nM. Assays of α-chymotrypsin inhibition were carried out similarly, except that AAPF (N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide) (Sigma-Aldrich, Inc.) was used as the chromogenic substrate. The enzyme concentration in each reaction was 100 pM, and inhibitor concentrations ranged from ~150 to ~500 nM. All measurements were made using a Shimadzu UV-1601 spectrophotometer equipped with a six cell changer and a temperature controller used to maintain the reaction temperature at 25 °C. The change in absorbance was monitored for at least 45 min both before and after addition of inhibitor, and the data were analyzed using a script written for MATLAB R2008b.
Nuclear magnetic resonance spectroscopy
We have used NMR spectroscopy to probe the structural response of WT CI2 upon binding chymotrypsin in solution. Backbone resonance assignments of free CI2 are known from our previous work; assignments of HN, N, Cα and Cβ atoms of CI2 in complex with chymotrypsin were obtained during the present work using a combination of 3D HNCA and HNCACB spectra in conjunction with a 2D 1H-15N HSQC spectrum and the assignments of free CI2. NMR spectra were recorded on a 700 MHz Varian INOVA spectrometer equipped with a cryogenic probe and z-axis pulsed field gradients. NMR samples consisted of approximately 750 μM CI2 with at least a two-fold excess of protease in a buffer consisting of 75 mM HEPES, pH 8.0, 50 mM NaCl, 5 mM CaCl2, and 10% D2O v/v. The data were processed using NMRPipe/NMRDraw21 and analyzed using NMRView22. The NMR spectra indicate that the inhibitor-protease complex was stable for approximately 15 hours before signs of degradation began to appear, consistent with known rates of CI2 cleavage by serine proteases23. The assignments for protease-bound CI2 were used along with those for free CI2 to assess and localize structural perturbations upon protease binding via a backbone chemical shift perturbation analysis. The chemical shift perturbation for each pair of backbone HN-N backbone atoms was calculated according to the equation , where Δδtotal represents the total perturbation in ppm at the backbone amide position and δx,y is the measured chemical shift for nucleus x in state y.
RESULTS
Non-reactive loop mutants show little variability in inhibitory ability as reflected in Ki,app
To explore whether amino acid positions that can alter internal dynamics can also affect function in CI2 and eglin c, we have determined the apparent inhibition constant, Ki,app, for a total of 13 CI2 variants and 5 eglin c variants with porcine pancreatic elastase and also for 5 variants of CI2 with bovine α-chymotrypsin. The location of these mutants in the 3D structure of CI2 is shown in Figure 1, with most mutations located away from the inhibitory binding loop (“reactive site” loop), which competitively inhibits by binding in the protease active site. An example of the primary cleavage velocity data used to calculate the apparent inhibition constant for each mutant is shown in Figure 2. In addition to making multiple independent measurements for each variant studied, positive and negative controls were interspersed regularly with the experimental measurements. The positive control consisted of a reaction mixture containing protease and substrate but no inhibitor; the slope of the absorbance trace remains the same throughout the experiment, assuming that there is always a large excess of substrate, and thus demonstrate that the protease does not become degraded or otherwise functionally compromised over the length of the whole measurement (Figure 2, reaction 6). The negative control consisted of a reaction containing substrate and inhibitor but no protease. The absorbance of such a sample should not change over time, thus ensuring that protease contamination is insignificant and that the substrate itself does not undergo significant autohydrolysis over the course of the experiment.
Figure 2.

Representative data used in the determination of Ki,app for L68A CI2 in complex with porcine pancreatic elastase. The graph shows the absorbance at 410 nm as a function of both time and inhibitor concentration. The black arrow indicates time = 45 min, the point at which inhibitor was added to each reaction cell. Reactions 1 and 6 are negative and positive controls, respectively. Reaction 1 contains no protease, and the absorbance therefore does not increase significantly throughout the experiment. Reaction 6 contains no inhibitor, and therefore the slope remains large and constant over the course of the measurement; in fact, the absorbance of this reaction increases to a very large value that is hidden by the scaling of the z-axis in this figure. L68A CI2 was added to reactions 2-5 to final concentrations of 62.5, 53, 49, and 40 nM, respectively; the different inhibitor concentrations lead to the different levels of substrate cleavage and thus to different maximum absorbances achieved during the measurement time. Individual 2D plots of the data from reactions 2-5, as well as linear regressions, can be found in Supplementary Figures 1 and 2.
The specific variants of each inhibitor studied were selected for particular reasons. CI2 mutants V66A, V28A, I76V, L68V, and L68A were chosen so that a possible connection between inhibitory function and changes in side-chain flexibility on the ps-ns time scale observed in our earlier work12 could be probed. The other CI2 mutants were selected for their positions within the CI2 structure: E34D is located at the N-terminus of CI2’s α-helix, and shortening this side chain may strain electrostatic interactions at this location, A35G removes the side chain of the most important residue of CI2’s folding nucleus24-25, I39V shortens the side chain of a residue pointing into the protein’s hydrophobic core, and the N-terminal truncations should weaken the formation of secondary structures at this terminus. Finally, the R65A mutation was chosen to serve as an internal control. This arginine side chain makes a critical hydrogen bond to CI2’s reactive loop that is known to be critical for maintaining the loop’s architecture; abolishing the hydrogen bonding capability at this position is known to severely disrupt inhibitor function in both CI226 and C. maxima trypsin inhibitor V27, a fellow member of the potato I inhibitor family. The mutants of eglin c under study were also chosen because of the availability of data on the dynamical consequences of these mutations9-10. Furthermore, the eglin c mutants V54A and V62A correspond to positions 68 and 76 in CI2, thus allowing the functional consequences of the mutations in the two homologous proteins to be compared.
The results of the elastase inhibition experiments with CI2 and eglin c are presented in Table I. The measured apparent inhibition constant of 12 nM for WT CI2 is similar to a previously published value of 35 nM obtained in slightly different buffer conditions20. Viewed as a whole, the most striking result of the CI2 experiments is that most mutants show little variability in inhibitory ability. Most mutant Ki,app values fall within 35% of the WT value, but the greatest measured change was a factor of 8 for the Δ3NT mutant, not considering the R65A CI2 control mutant, which is an approximately 78-fold worse inhibitor than WT CI2 on account of the structural deformations resulting from the loss of hydrogen bonding capability at this position.
Table I.
Measured apparent inhibition constants, Ki,app, of variants of CI2 and eglin c in complex with porcine pancreatic elastase
| Variant | Ki,app (nM) | St. Dev. (nM) | Rel. Error (%) | Variant/WT | |
|---|---|---|---|---|---|
| WT CI2 | 12 | ± | 0.4 | 3.6 | 1.00 |
| I76V CI2 | 11 | ± | 2 | 16.0 | 0.88 |
| V66A CI2 | 13 | ± | 1 | 6.7 | 1.11 |
| E34D CI2 | 14 | ± | 1 | 9.5 | 1.12 |
| V28A CI2 | 14 | ± | 2 | 10.9 | 1.14 |
| I39V CI2 | 14 | ± | 1 | 8.6 | 1.16 |
| A35G CI2 | 15 | ± | 1 | 6.6 | 1.26 |
| D2NT CI2 | 16 | ± | 1 | 7.4 | 1.30 |
| L68V CI2 | 16 | ± | 0.4 | 2.4 | 1.35 |
| L68A CI2 | 24 | ± | 1 | 5.6 | 2.02 |
| D3NT CI2 | 92 | ± | 4 | 4.6 | 7.68 |
| R65A CI2 | 936 | ± | 81 | 8.7 | 77.90 |
| WT eglin c | 10 | ± | 1 | 7.2 | 1.00 |
| V54A eglin c | 7 | ± | 1 | 9.5 | 0.76 |
| V18A eglin c | 15 | ± | 1 | 6.6 | 1.26 |
| V34A eglin c | 13 | ± | 1 | 10.3 | 1.07 |
| V62A eglin c | 14 | ± | 2 | 12.6 | 1.17 |
Notes: Δ2NT and Δ3NT represent CI2 N-terminal truncations of two and three residues, respectively. For all experiments, the listed value of Ki,app is the average of no fewer than five independents measurements.
A closer inspection of the CI2 data shows that the results can be subdivided into two groups based on the ratio of mutant to WT Ki,app. Most CI2 mutants are part of a group in which the mutant/WT ratio is at most 1.35, but the mutants L68A, Δ3NT, and R65A have ratios of 2.0, 7.7, and 78.0, respectively. The increased Ki,app for R65A is easily rationalized because of the known structural consequences of mutating this position, but the other two mutants are known to be stable, well-folded proteins (data not shown). In essence, the L68A and Δ3NT mutations, both distal to the reactive loop, result in a small but measurable decrease in inhibitory ability.
In contrast to the majority of mutants tested (Table 1), some additional activity patterns at core residue L68 are suggestive of correlation between loop dynamics and inhibitory function. Although the effects are small, the L68V and L68A mutations resulted in larger increases in Ki,app (1.35 and 2.0 fold, respectively) than for any other single point mutation tested, excluding the control R65A. Residue L68 is thought to be a key energetic linchpin connecting CI2’s hydrophobic core to its reactive loop17. Interestingly, L68V and L68A also display dynamical properties distinct from the other mutants tested previously; namely, an expanded suite of NMR-based 2H relaxation measurements revealed the increasing presence of side-chain motions on the nanosecond time scale concentrated in the reactive loop (residues 53-63) as the side chain at position 68 is shortened from L to V to A12. Specifically, ns-scale side-chain dynamics were detected at M59 in the L68V mutant and at V53, T55, I56, T58, M59, and I63 in the L68A mutant. Dynamical analysis of the other mutants tested (V66A, I76V, V28A) did not reveal the presence of ns-scale side-chain dynamics in the loop, which correlates with no change in Ki,app for those three mutants (Table 1). On the slower μs-ms time scale, CI2 shows almost no motion from model-free analysis of 15N relaxation data (see ref. 12). Furthermore, measurement of Rex from relaxation dispersion experiments indicates no detectable Rex in wild-type and an Rex value of 6 s−1 for I63 in L68A; all other Rex values in CI2 are essentially zero (Supplementary Tables I and II). While it is possible that the μs-ms motion at I63 in L68A contributes to a change in inhibitory function since this residue is at the base of the loop, the lack of significant Rex elsewhere in the loop (or even outside it) is problematic for implicating concerted hinge-like motions. It is also possible that the appearance of Rex at I63 in L68A corresponds to a slowing of motion at this position upon mutation that would be somehow correlated with slowing of ns-scale motion of the methyl residues mentioned above. Taken together, the structural, dynamical, and functional data available concerning position 68 confirm that it is of special importance in this protein, as suggested by computational analysis17. The results indicate a correlation between slowing of the reactive loop dynamics (a shift from ps-scale to ns-scale processes) and a loss of inhibitory ability reflected by the increased Ki,app for the L68 mutants.
Several mutants of CI2 were also assayed against α-chymotrypsin, the results of which can be seen in Table II. The numerical values of Ki,app for WT CI2 and mutants I76V, V66A, L68V, and L68A with chymotrypsin fall in the same order as they do when assayed against elastase, with I76V yielding a very similar value to WT CI2, V66A being a slightly worse inhibitor, and the L68 mutants again having the greatest negative impact on inhibitory power. We note that our measured value of Ki,app for WT CI2 in conjunction with chymotrypsin is an order of magnitude larger than the value reported in an earlier study20, although the previously reported values were determined in slightly different buffer conditions and at elevated temperature (37 °C compared to 25 °C in the present study). In light of the fact that we are mostly interested in the differences among the assayed mutants and not the absolute values themselves, it is clear that the subtle mutations tested have only a subtle impact on CI2 inhibition of bovine α-chymotrypsin, as observed for the same experiments with pancreatic elastase.
Table II.
Measured apparent inhibition constants, Ki,app, of variants of CI2 in complex with bovine α-chymotrypsin
| Variant | Ki,app (nM) | St. Dev. (nM) | Rel. Error (%) | Variant/WT | |
|---|---|---|---|---|---|
| WT CI2 | 202 | ± | 17 | 8.4 | 1.00 |
| I76V CI2 | 217 | ± | 15 | 6.9 | 1.08 |
| V66A CI2 | 248 | ± | 42 | 17.0 | 1.23 |
| L68V CI2 | 259 | ± | 23 | 8.8 | 1.28 |
| L68A CI2 | 282 | ± | 9 | 3.1 | 1.40 |
Note: For all experiments, the listed value of Ki,app is the average of no fewer than five independents measurements.
Finally, we also measured Ki,app for several mutants of eglin c in order to discover whether mutation at analogous positions results in similar functional consequences. All eglin c mutants were low nM inhibitors of elastase, and little difference was detected among the five variants tested (Table 1), just as for CI2. In eglin c, V54 occupies a position analogous to L68 in CI2. In CI2, L68 mutations resulted in less effective inhibitors, but mutating the analogous V54 in eglin c appears to slightly increase inhibitory function. The opposite appears to be true for the pair V62A and I76V in eglin c and CI2, respectively. I76V appears to be a neutral mutation regarding CI2 function, but V62A has the greatest impact on eglin c inhibition of elastase observed in this small set of mutants. Thus, homologous mutations in the two inhibitors does not translate into the same functional effect. The two inhibitors are known to experience distinct dynamical perturbations upon mutation at these analogous positions9-12, which suggests that dynamics (or dynamics in conjunction with subtle, differential structural perturbations) does play a role in determining functional properties in these model systems. Overall, however, it is clear that these conservative larger-to-smaller aliphatic point mutations in CI2 and eglin c have limited impact on their inhibitory ability toward porcine pancreatic elastase.
CI2 chemical shift perturbation upon binding chymotrypsin radiates from the reactive loop toward CI2’s hydrophobic core
To probe CI2-based inhibition from a more structural perspective, we initiated NMR studies of a CI2-protease complex. Crystal structures are available for CI2 and mutants in complex with subtilisin BPN’23,26,28 and subtilisin novo29, and similarly, structures of eglin c have been determined in complex with subtilisin novo30 and chymotrypsin31. However, ironically, there is no structure of CI2 in complex with chymotrypsin. We therefore decided to apply NMR spectroscopy to perform an initial characterization of the 32 kDa CI2-chymotrypsin complex in order to gain some insight into the structural consequences of complex formation in solution. To our knowledge, this is the first solution NMR analysis of a natural macromolecular protease inhibitor in complex with a serine protease target.
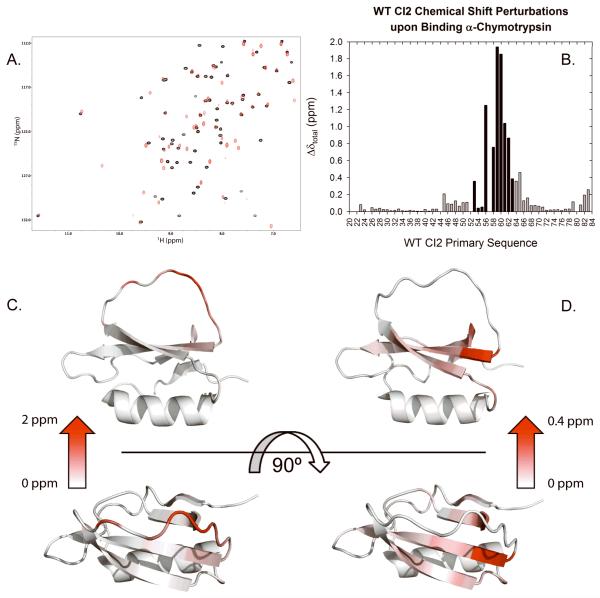
Using triple resonance NMR methods, assignments were made for the HN, N, Cα, and Cβ resonances of CI2 labeled uniformly with 13C and 15N and deuterated to approximately 80% while in complex with chymotrypsin. An overlay of the free and bound CI2 spectra can be seen in Figure 3A. NMR chemical shifts are exquisitely sensitive to the local environment surrounding a particular NMR-active nucleus, and thus changes in the local structural environment are reflected by changed chemical shifts. The chemical shift perturbations experienced by WT CI2 upon binding chymotrypsin are shown in Figure 3B. The largest perturbations are found in the reactive loop, reaching a maximum at the scissile bond (residues 59-60) and generally decreasing moving along the primary sequence in either direction. Upon complex formation, these residues become buried, though the conformation of the reactive loop remains similar to the free-state conformation; thus, the large perturbations in the reactive loop are due primarily to new contacts with chymotrypsin. Figure 3C shows the entire range of chemical shift perturbations plotted onto the structure, making clear the impact of complex formation on the reactive loop of CI2. Figure 3D shows a rescaled view of the chemical shift perturbations with the reactive loop residues excluded. By excluding these residues, significant perturbations that propagate beyond the reactive loop into CI2’s β-sheet are more easily seen. Chymotrypsin binding clearly has long distance effects, as the chemical shift perturbation at A45 is 0.2 ppm even though A45 is approximately 15 Å from the nearest atoms the protease (using 1LW6). Of the residue positions mutated and assayed against chymotrypsin (Table II), V66 and L68, both located on the central β-strand, show the greatest change in Ki,app. In summary, the perturbations are highest within the reactive loop, but also significant along the β-strands.
Figure 3.
Analysis of HN-N chemical shift perturbations in WT CI2 upon binding bovine α-chymotrypsin. Panel A shows the overlaid 1H-15N HSQC spectra for CI2 in the free (black) and chymotrypsin-bound (red) states. Panel B shows the calculated chemical shift perturbation upon binding as a function of CI2 residue. The black bars represent CI2’s reactive loop, the site of binding to its protease targets. Panel C shows the data from panel B plotted onto the CI2 structure (PDB 2CI2). The shading from white to red makes it visually clear that the strongest perturbations are located in the reactive loop. Panel D is the same as panel C, except that the data for the reactive loop (residues 53-63) perturbations have been removed so as to reveal smaller but still significant changes that are visually masked by the dominant reactive loop perturbations in panel C.
Because the L68A CI2 mutant shows the largest difference in Ki,app of any of the single point mutations compared to the WT value for both chymotrypsin and elastase, and because its side chain points from the β-strand into the hydrophobic core, we wondered whether differential structural effects incurred upon protease binding might be responsible. To answer this question, CI2 chemical shift assignments were used for both free and bound WT and for free L68A in conjunction with a 1H-15N HSQC spectrum of the L68A CI2-chymotrypsin complex to assign 45 of 64 total residues in the bound L68A mutant. The results of chemical shift perturbation calculations for the transition from free to complexed L68A CI2 are nearly identical to the chemical shift perturbations experienced by WT CI2 upon binding chymotrypsin with regard to both the overall pattern of chemical shift perturbations and the magnitude of the perturbation for each individual residue (data not shown). This suggests that differential conformational perturbations upon binding the protease are not responsible for the variations in the measured values of Ki,app for the variants tested and thus that the dominant conformation of the free state ensemble determines inhibitory ability. The chemical shift results taken together lead us to conclude that (i) the primary structural consequences upon complex formation in solution are confined to CI2’s reactive loop, (ii) binding the protease at the reactive loop is sensed by the core of the protein, as reflected by the significant detected chemical shift perturbations throughout CI2’s β-sheet, and (iii) the chemical shift perturbation analysis of L68A CI2 binding to chymotrypsin results in nearly identical chemical shift perturbations compared to chymotrypsin complex formation with WT CI2. This result suggests that complex formation with the protease affects each of the tested CI2 variants in a similar manner.
DISCUSSION
In this paper, basic functional and structural features of serine protease inhibitors CI2 and eglin c were added to the existing knowledge base of structure and dynamics on these inhibitors and their corresponding functional complexes. Specifically, we assayed the inhibitory ability of a set of variants of CI2 and eglin c toward the serine proteases elastase and α-chymotrypsin with the aim of creating a unique mutational structure-dynamics-function data set to gain insight into the possible roles of dynamics and structure in the function of these small model protein systems. We also characterized the conformational perturbations to WT and L68A CI2 upon binding chymotrypsin to determine whether differential structural perturbations among the various mutants upon binding a protease might also play a role in inhibitory ability.
Overall, the functional assays revealed little significant change in the apparent inhibition constant for the tested single-site variants of CI2 and eglin c. Still, one noticeable result concerns mutations at position L68 in CI2. Our previous work showed that shortening this side chain from L➔V➔A leads to increasing levels of ns-scale side-chain dynamics, a phenomenon not detected to the same degree in the other mutants studied.12 Interestingly, the same mutations L68V and L68A have larger effects on inhibitory ability than any of the other single point mutations tested in the hydrophobic core of CI2, even though the effects on function are a mere factor of two. We note that some known allosteric systems do no better; upon exposure to light, a photoallosteric PAS domain fused to DHFR results in an enhancement of DHFR catalysis by a factor of two32. The L68 mutant results lend credence to the previously suggested notion that L68 mediates a connection between the hydrophobic core “scaffold” of CI2 and the functional reactive loop that binds serine protease targets17-18. Furthermore, it suggests that propagated dynamics to the loop might serve as the mechanism for this connection. It is also interesting to point out that when four variants of CI2 were assayed against chymotrypsin, including L68A and L68V, the rank order of reduction of inhibition was identical to that against elastase (Table 2). This suggests that these CI2 core variants are indeed distinct from one another with regard to functional ability, notwithstanding the only slight changes in Ki,app. Based on principle, it is intriguing that such subtle “allosteric-like” phenomena can be observed through functional measurements even in simple model systems like CI2 that are classically non-allosteric. Taken as a whole, the fact that global changes in dynamics detected in previous work results in only fine changes in inhibitory ability in the variants tested leads us to conclude that dynamics, at least in these single-site substitutions, plays at most a subtle role in the inhibitory function of these model protein systems. Nevertheless, we did identify two CI2 mutants, L68A and Δ3NT, the effects of which were sensed at the reactive loop and resulted in at least a twofold change in Ki,app. Long range effects are the foundation of classical allostery, and it is therefore intriguing to find such effects present in CI2 in response to relatively subtle perturbations and despite it being a classically nonallosteric protein.
Because of the exquisitely sensitive dependence of the NMR chemical shift on the local environment surrounding an NMR-active nucleus, NMR chemical shift perturbations are often used as indications of structural perturbations when a protein binds another molecule. We selected WT and L68A CI2 for an analysis of chemical shift perturbations upon binding chymotrypsin, as these two variants showed the largest numerical difference in Ki,app. As expected, chemical shift perturbations were extensive at the reactive loop, but there were also significant perturbations along the β-strands, even though they are not in direct contact with the protease, thus indicating that the force of binding propagates along the reactive loop and is ultimately “sensed” in the core of the protein and outward to the edges of the β-sheet. Importantly, however, the chemical shift perturbation analysis for the L68A CI2 mutant in complex with chymotrypsin yielded results very similar to the WT analysis. This suggests that, whatever perturbations are experienced upon binding between WT CI2 and chymotrypsin, similar perturbations are experienced by the L68A mutant. Thus, differential structural responses upon protease binding can be ruled out as a major determinant of functional ability. Because dynamics appears not to play a dominant role in determining CI2 function and because structural perturbations upon binding appear to be similar even among the most functionally-distinct CI2 variants, we conclude that the architecture of the CI2 reactive loop in the unbound state plays the major role in CI2 function.
We also make the intriguing observation that the first third of the protein (from the N-terminus until approximately residue 44) appears to be a “blind spot” to backbone structural and dynamical perturbations. Our previous work demonstrated that mutations in the hydrophobic core had little to no effect on backbone structure and dynamics, and the present work reveals that binding to target proteases also results in no chemical shift perturbations in this region. This represents nice example of an obvious pattern of correspondence between lack of chemical shift changes and lack of dynamical changes upon perturbation. It thus appears that perturbations in CI2, whether structural or dynamical, are directed toward or localized in the functional region, namely the reactive loop and its β-sheet support structure, a phenomenon also observed via statistical analysis of cooperative interactions among residues in CI218. CI2 and the other potato I inhibitor family members are classically non-allosteric, but the fact that distal perturbations result in effects in the functional region is reminiscent of allostery, albeit without major functional changes as the defining characteristic.
Finally, we return to the general question of what role dynamics might play in protein function. In the present work, we detected no sizeable role for dynamics in CI2 function, but that does not preclude that such a connection exists. It could simply be the case that the conservative mutations we made in CI2’s hydrophobic core were not severe enough to result in dynamical perturbations significant enough to clearly impact function, or it could also be the case that perhaps multiple mutations are necessary instead of simple single point mutations. The accumulation of incremental mutations to achieve a particular trait, such as the emergence of a dynamics-based regulation of function, is the primary mechanism of protein evolution. In this light, the subtle functional consequences resulting from conservative mutations in CI2 could be considered to be a model of the early stages in the evolution of allostery. The foundation of functional allostery is long range communication and action at a distance – a perturbation at one site results in a functional change elsewhere in the protein. Mutations in the hydrophobic core of CI2 result in dynamical changes propagated from the reactive loop into the inhibitor’s core. In summary, CI2 exhibits the capacity for long range communication, but it appears that CI2 has not evolved to harness this ability to enhance function.
Supplementary Material
Acknowledgments
Funding Acknowledgement: This work was supported by NIH grant GM066009 to A.L.L. and an NSF Graduate Research Fellowship to M.J.W.
REFERENCES
- 1.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 2.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 4.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee AL, Sharp KA, Kranz JK, Song XJ, Wand AJ. Temperature dependence of the internal dynamics of a calmodulin-peptide complex. Biochemistry. 2002;41:13814. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 6.Lee AL, Wand AJ. Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature. 2001;411:501. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 7.Mandel AM, Akke M, Palmer AG., 3rd. Dynamics of ribonuclease H: temperature dependence of motions on multiple time scales. Biochemistry. 1996;35:16009. doi: 10.1021/bi962089k. [DOI] [PubMed] [Google Scholar]
- 8.Igumenova TI, Lee AL, Wand AJ. Backbone and side chain dynamics of mutant calmodulin-peptide complexes. Biochemistry. 2005;44:12627. doi: 10.1021/bi050832f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarkson MW, Lee AL. Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry. 2004;43:12448. doi: 10.1021/bi0494424. [DOI] [PubMed] [Google Scholar]
- 11.Boyer JA, Lee AL. Monitoring aromatic picosecond to nanosecond dynamics in proteins via 13C relaxation: expanding perturbation mapping of the rigidifying core mutation, V54A, in eglin c. Biochemistry. 2008;47:4876. doi: 10.1021/bi702330t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitley MJ, Zhang J, Lee AL. Hydrophobic core mutations in CI2 globally perturb fast side-chain dynamics similarly without regard to position. Biochemistry. 2008;47:8566. doi: 10.1021/bi8007966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of Energetic and Dynamic Coupling Networks in a PDZ Domain Protein. J. Mol. Biol. 2006;364:337. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 14.Namanja AT, et al. Substrate recognition reduces side-chain flexibility for conserved hydrophobic residues in human Pin1. Structure. 2007;15:313. doi: 10.1016/j.str.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 2004;335:1105. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 16.McPhalen CA, James MN. Crystal and molecular structure of the serine proteinase inhibitor CI-2 from barley seeds. Biochemistry. 1987;26:261. doi: 10.1021/bi00375a036. [DOI] [PubMed] [Google Scholar]
- 17.Hilser VJ, Dowdy D, Oas TG, Freire E. The structural distribution of cooperative interactions in proteins: analysis of the native state ensemble. Proc. Natl. Acad. Sci. USA. 1998;95:9903. doi: 10.1073/pnas.95.17.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T, Whitten ST, Hilser VJ. Functional residues serve a dominant role in mediating the cooperativity of the protein ensemble. Proc. Natl. Acad. Sci. USA. 2007;104:4347. doi: 10.1073/pnas.0607132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gianni S, et al. Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering. Structure. 2006;14:1801. doi: 10.1016/j.str.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Roesler KR, Rao AG. Conformation and stability of barley chymotrypsin inhibitor-2 (CI-2) mutants containing multiple lysine substitutions. Protein Eng. 1999;12:967. doi: 10.1093/protein/12.11.967. [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 23.Radisky ES, Kwan G, Karen Lu CJ, Koshland DE., Jr. Binding, proteolytic, and crystallographic analyses of mutations at the protease-inhibitor interface of the subtilisin BPN’/chymotrypsin inhibitor 2 complex. Biochemistry. 2004;43:13648. doi: 10.1021/bi048797k. [DOI] [PubMed] [Google Scholar]
- 24.Itzhaki LS, Otzen DE, Fersht AR. The structure of the transition state for folding of chymotrypsin inhibitor 2 analysed by protein engineering methods: evidence for a nucleation-condensation mechanism for protein folding. J. Mol. Biol. 1995;254:260. doi: 10.1006/jmbi.1995.0616. [DOI] [PubMed] [Google Scholar]
- 25.Shakhnovich E, Abkevich V, Ptitsyn O. Conserved residues and the mechanism of protein folding. Nature. 1996;379:96. doi: 10.1038/379096a0. [DOI] [PubMed] [Google Scholar]
- 26.Radisky ES, Lu CJ, Kwan G, Koshland DE., Jr. Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2. Biochemistry. 2005;44:6823. doi: 10.1021/bi047301w. [DOI] [PubMed] [Google Scholar]
- 27.Cai M, Gong YX, Wen L, Krishnamoorthi R. Correlation of binding-loop internal dynamics with stability and function in potato I inhibitor family: relative contributions of Arg(50) and Arg(52) in Cucurbita maxima trypsin inhibitor-V as studied by site-directed mutagenesis and NMR spectroscopy. Biochemistry. 2002;41:9572. doi: 10.1021/bi0258952. [DOI] [PubMed] [Google Scholar]
- 28.Radisky ES, Koshland DE., Jr. A clogged gutter mechanism for protease inhibitors. Proc. Natl. Acad. Sci. USA. 2002;99:10316. doi: 10.1073/pnas.112332899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McPhalen CA, James MN. Structural comparison of two serine proteinase-protein inhibitor complexes: eglin-c-subtilisin Carlsberg and CI-2-subtilisin Novo. Biochemistry. 1988;27:6582. [PubMed] [Google Scholar]
- 30.Heinz DW, Priestle JP, Rahuel J, Wilson KS, Grutter MG. Refined crystal structures of subtilisin novo in complex with wild-type and two mutant eglins. Comparison with other serine proteinase inhibitor complexes. J. Mol. Biol. 1991;217:353. doi: 10.1016/0022-2836(91)90549-l. [DOI] [PubMed] [Google Scholar]
- 31.Frigerio F, et al. Crystal and molecular structure of the bovine alpha-chymotrypsin-eglin c complex at 2.0 A resolution. J. Mol. Biol. 1992;225:107. doi: 10.1016/0022-2836(92)91029-o. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, et al. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.