Abstract
Cu, Zn superoxide dismutase (SOD1) is a dimeric metal binding enzyme responsible for the dismutation of toxic superoxide to hydrogen peroxide and oxygen in cells. Mutations at dozens of sites in SOD1 induce amyotrophic lateral sclerosis (ALS), a fatal gain-of-function neurodegenerative disease whose molecular basis is unknown. To obtain insights into effects of the mutations on the folded and unfolded populations of immature monomeric forms whose aggregation or self-association may be responsible for ALS, the thermodynamic and kinetic folding properties of a set of disulfide-reduced and disulfide-oxidized Zn-free and Zn-bound stable monomeric SOD1 variants were compared to the wild-type (WT) protein. The most striking effect of the mutations on the monomer stability was observed for the disulfide-reduced metal-free variants. Whereas the WT and S134N monomers are >95% folded at neutral pH and 37 °C, A4V, L38V, G93A, and L106V ranged from 50% to ∼90% unfolded. The reduction of the disulfide-bond was also found to reduce the apparent Zn affinity of the WT monomer by 750-fold, into the nanomolar range where it may be unable to compete for free Zn in the cell. With the exception of the S134N metal-binding variant, the Zn affinity of disulfide-oxidized SOD1 monomers showed little sensitivity to amino acid replacements. These results suggest a model for SOD1 aggregation where the constant synthesis of ALS-variants of SOD1 on ribosomes provides a pool of species in which the increased population of the unfolded state may favor aggregation over productive folding to the stable native dimeric state.
Keywords: Disulfide bond, Zn binding, protein folding, aggregation
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that results in the death of motor neurons.1 An inherited form of ALS has been linked to mutations in the gene encoding for the Cu, Zn superoxide dismutase (SOD1).2,3 To date, over 150 amino acid replacements, insertions, deletions and truncations throughout the sequence have been discovered that give rise to this deleterious gain-of-function disease (http://alsod.iop.kcl.ac.uk/Als/Index.aspx).4 Although a number of mechanisms have been proposed for toxicity,5 ALS variants of SOD1 often lead to the formation of macroscopic aggregates in the motor neurons of afflicted individuals. Recognizing that these types of sequence alterations typically destabilize proteins, it is reasonable to hypothesize that marginally soluble forms of SOD1 self-associate to produce toxic small oligomers or larger aggregates.6 Nevertheless, the conformations of SOD1 responsible for aggregation are unclear, and a common molecular explanation for the formation of these aggregates has not been discovered.
SOD1 is a cytosolic protein responsible for the dismutation of superoxide, a toxic byproduct of metabolism, into hydrogen peroxide and oxygen through the cyclical oxidation and reduction of its catalytic Cu ion.7 In its mature form, SOD1 is a homodimer with one Zn and one Cu ion bound to each subunit. The fold of each monomer can be described as a β-sandwich composed of eight β-strands supporting two large loops. The Zn-binding loop binds the structurally-important Zn ion, and the electrostatic loop plays a role in guiding the superoxide anion to the active site. In addition, each monomer contains an intramolecular disulfide bond between C57 in the Zn-binding loop and C146 in β8 (Figure 1).
Figure 1.
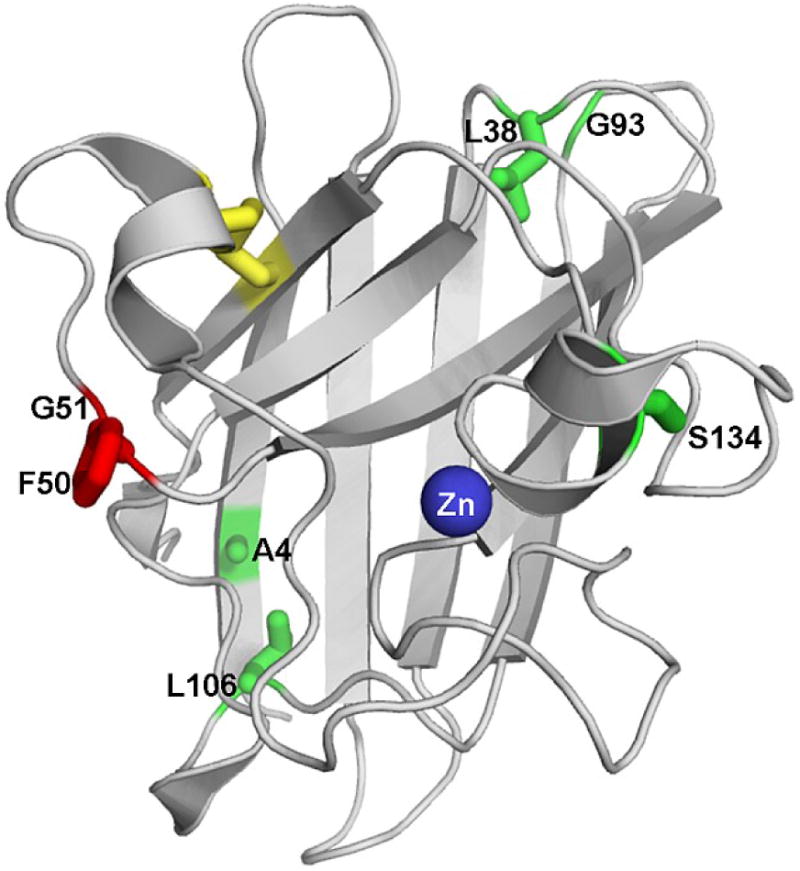
Crystal structure of holo-SOD1 (PDB: 2C9V). The Cu ion and the other monomer have been omitted. The disulfide bond is highlighted in yellow, and the Zn ion in blue. The sites of ALS mutations studied are highlighted by the green side chains for the native amino acids. The two residues replaced by glutamic acid to create the monomer model, F50 and G51, are shown in red. The ALS-inducing variants studied are A4V, L38V, G93A, L106V and S134N.
There has been growing interest in the role of immature monomeric species, i.e. metal-free and/or disulfide-reduced, in aggregation.8 Monomeric species of SOD1 may be more prone to aggregation9,10,11 because they are likely to be partially folded and marginally soluble.12,13,14,15 This hypothesis is consistent with the three-state folding mechanism of SOD1, 2U⇆2M⇆N2, in which the rate-limiting monomer folding reaction from the unfolded state, U, to the folded state, M, is followed by the rapid association of the monomers to form the dimeric native state, N2.16,17 As a consequence, the folded monomer population is minimized during the folding reaction of the WT protein. Also supporting this view is the observation that several ALS variants destabilize the native state, leading to increased populations of folded and unfolded monomeric species at equilibrium.18,19,20 These studies focused on forms of SOD1 and its variants that contain the intramolecular disulfide bond, in essence, the mature species closely connected with the native conformation.
It has previously been shown that the reduction of the disulfide bond leads to the dissociation of the dimer in the absence of metal ions; zinc binding reverses this process.21 The reduction of the disulfide bond in several ALS-inducing variants decreases the apparent melting point below physiological temperature.22 However the irreversibility of the thermal unfolding reaction and the requirement for a stabilizing osmolyte render this conclusion ambiguous. To obtain unambiguous quantitative insight into the thermodynamic coupling between disulfide bond, Zn binding and amino acid replacements on immature SOD1, a combined thermodynamic and kinetic analysis of the reversible folding reactions of a monomeric version of SOD1 (mAS-SOD1), C6A/F50E/ G51E/C111A, and five ALS variants introduced into this background (Figure 1) was performed. The A4V mutation increases the side chain volume in a tightly packed hydrophobic cluster inside the β-sandwich. Both L38 and L106 serve as hydrophobic capping residues for the β-sandwich, and replacement with valine at these positions decreases the side chain volume and replaces a γ-branched side chain with a β-branched side chain. The main chain of G93 is in a tight loop, and the dihedral angles are disfavored for all but glycine at this position. Finally, S134 is involved in the hydrogen bonding network that links the electrostatic loop to the Zn-binding loop, and its replacement with asparagine leads to decreased metalation (For a general review see Valentine et al.5).
With the exception of S134N, both oxidized and reduced monomeric forms of the remaining variants exhibited a significant loss of thermodynamic stability for their metal-free forms. By contrast, S134N was the only variant studied which exhibited a significantly weakened apparent Zn affinity. Surprisingly, the reduction of the disulfide bond decreased the affinity of mAS-SOD1 for Zn by ∼750-fold. The substantially increased populations of the unfolded forms in four of the five disulfide-reduced ALS-variants of SOD1 at physiological temperature may provide the common link between mutations at many positions and protein aggregation. The reduction in the apparent Zn affinity of the S134N variant would allow this variant to more readily sample the metal-free folding free energy surface whose folded and/or unfolded monomeric populations might be sufficient to induce aggregation even for WT SOD1.
Results
The thermodynamic stability of the Zn-bound and/or disulfide-reduced states of monomeric WT, A4V, L38V, G93A, L106V and S134N SOD1 variants was determined by chemical denaturation experiments on the C6A/F50E/G51E/C111S model of SOD1 (Figure 1).23 In this pseudo-WT model, the free cysteines, C6 and C111, have been replaced with non-oxidizable side chains to eliminate intramolecular disulfide-interchange in the unfolded state and intermolecular disulfide crosslinking in the native state. The F50 and G51 residues at the dimer interface have been replaced with glutamic acid to prevent dimerization.24,25 The structure of this stable monomer is very similar to that for the monomer in the context of the dimer,25,26 as is the thermodynamic stability.17 This system enables measurements of highly reversible folding reactions solely for the monomeric forms of these ALS variants, in the absence of their self-association to dimeric species. Although the C6A/C111S and F50E/G51E mutations might themselves perturb the aggregation propensity of SOD1 (C6F, C6G and C111Y are ALS variants8), the introduction of ALS-inducing mutations into the same background allows for a direct comparison of their perturbations on the thermodynamic properties and populations of the SOD1 system.
Stabilities of disulfide-reduced apo-mAS-SOD1
The role of the disulfide bond in stabilizing mAS-SOD1Apo was determined by reducing the disulfide bond and monitoring the ellipticity at pH 7.2 and 20 °C as a function of the urea concentration. The choice of urea as the denaturant was necessitated by the expected low thermodynamic stability of these proteins. The far-UV circular dichroism (CD) spectra of the disulfide-reduced monomers can be seen in Figure 2A. WT and S134N exhibit a minimum at ∼208 nm and a slight shoulder at 230 nm, consistent with a significant degree of folding. In contrast, A4V, L38V, G93A and L106V exhibit unfolded-like CD spectra with the minimum shifted to ∼200 nm, and they lack the shoulder at 230 nm characteristic of the native fold.
Figure 2.

CD spectra and urea titrations reveal that A4V, L38V, G93A and L106V are partially unfolded. The CD spectra of WT (solid gray), A4V (dash-dot black), L38V (dotted black), G93A (dashed black), L106V (solid black), S134N (dashed gray) (A) and (B) at 20 °C and pH 7.2. Equilibrium unfolding curves at 20 °C and pH 7.2 for (C) and (D) variants and the calculated fraction unfolded plots for (E) and (F) variants measured by CD at 230 nm: WT (filled circle), A4V (triangle), L38V (hexagon), G93A (diamond), L106V (square), S134N (upside-down triangle). Best fit curves to a two-state U⇆M folding model are shown in solid lines. The thermodynamic data are presented in Table 1. The protein concentration for these experiments was 10 μM.
The equilibrium urea denaturation curves for variants are shown in Figure 2C. WT is stable to ∼1.2 M urea, where it undergoes a cooperative unfolding reaction that is complete by ∼2.6 M urea. The S134N variant is nearly coincident with WT with the exception of a slightly negatively sloped native baseline prior to the cooperative unfolding transition. The A4V, L38V, G93A and L106V variants are all completely denatured by 1 M urea and lack native baselines. These equilibrium data were fit to a two-state model, U⇆M, assuming a linear dependence of the apparent free energy of folding, ΔG°, on the denaturant concentration: ΔG° (urea) = ΔG° (H2O) − m[urea] (Table 1). All data were fit using Savuka 6.2, an in-house nonlinear least squares program using the Marquardt-Levenberg algorithm. The fits were performed by globally analyzing the ellipticity at 20 different wavelengths between 220 and 240 nm. Both WT and S134N exhibit well defined baselines and were fit without constraining the parameters. From the determined thermodynamic stability (Table 1), it is evident that both of these proteins are >99% folded under these conditions (Figure 2E). For the highly-destabilized ALS variants where no native baseline was observed, the m-value and the slope and intercept of the native baseline were fixed to the same values as WT. The constraint on several variables, required to fit these titrations, likely results in an underestimated error of the fit, suggesting caution in the accuracy of these estimates for stability. Nevertheless, the lack of native baselines and the reduced CD signal in the absence of denaturant (Figure 2B) make it clear that substantial fractions of A4V, L38V, G93A and L106V are unfolded at 20 °C and pH 7.2 (Figure 2E).
Table 1. Thermodynamic parameters, microscopic rate constants and kinetic m‡-values for monomeric variants.*.
| WT | A4V | L38V | G93A | L106V | S134N | ||
|---|---|---|---|---|---|---|---|
| Apo reduceda | |||||||
|
|
-3.95±0.1 | -1.09c | -0.22c | +0.12c | -0.40c | -3.97±0.16 | |
| m(U/M) | 2.23±0.11 | 2.23d | 2.23d | 2.23d | 2.23d | 2.08±0.08 | |
| Apo oxidizeda, e | |||||||
|
|
-4.48±0.07 | -2.83±0.07 | -2.32±0.15 | -1.87±0.07 | -2.52±0.08 | -4.54±0.10 | |
| m(U/M) | 1.65±0.02 | 1.88±0.04 | 1.91±0.08 | 1.89±0.04 | 2.09±0.05 | 1.73±0.03 | |
| Apo oxidizeda, e | |||||||
| kf | 0.056±0.002 | 0.15±0.02 | 0.033±0.006 | 0.029±0.004 | 0.030±0.003 | 0.039±0.004 | |
|
|
1.20±0.05 | 1.5±0.1 | 1.36±0.20 | 1.37±0.18 | 1.33±0.13 | 1.13±0.04 | |
| ku | (2.02±0.40)×10-5 | (1.0±0.1)×10-3 | (3.5±1.5)×10-4 | (1.2±0.2)×10-3 | (5.0±1.0)×10-4 | (1.2±0.1)×10-5 | |
|
|
-0.58±0.01 | -0.64±0.03 | -0.78±0.09 | -0.74±0.03 | -0.74±0.05 | -0.65±0.01 | |
|
|
-4.61±0.12 | -2.92±0.2 | -2.65±0.28 | -1.85±0.13 | -2.37±0.13 | -4.70±0.08 | |
| m(U/M)f | 1.79±0.05 | 2.14±0.1 | 2.14±0.22 | 2.11±0.18 | 2.07±0.14 | 1.78±0.04 | |
| Zn oxidizedg | |||||||
| kf | 0.74±0.01 | 0.87±0.15 | 0.27±0.05 | 0.19±0.05 | 0.34±0.05 | 0.64±0.01 | |
|
|
2.93±0.07 | 3.0±0.1 | 2.89±0.09 | 2.48±0.17 | 3.39±0.10 | 2.95±0.10 | |
| ku | (0.23±0.08)×10-7 | (1.77±0.39)×10-7 | (2.8±0.8)×10-7 | (4.1±1.2)×10-7 | (13.3±3.9)×10-7 | (2.8±1.2)×10-7 | |
|
|
-1.76±0.05 | -1.85±0.03 | -2.01±0.06 | -1.88±0.06 | -1.68±0.06 | -1.47±0.13 | |
|
|
-18.0±0.2 | -17.0±0.2 | -15.3±0.5 | -15.6±0.2 | -15.2±0.2 | -16.5±0.3 | |
| m(U/M)f | 4.69±0.09 | 4.85±0.1 | 4.90±0.11 | 4.36±0.18 | 5.07±0.12 | 4.42±0.13 | |
|
|
-13.4±0.2 | -14.1±0.2 | -12.7±0.5 | -13.8±0.2 | -12.8±0.2 | -11.8±0.3 | |
| Kdh | 100±39 pM | 31±15 pM | 360±350 pM | 55±23 pM | 270±110 pM | 1600±840 pM | |
Units for the U ⇆ M reaction: kf and ku, s-1; , and m(U/M), kcal mol-1 M-1; .
. kcal mol-1.
Denaturation was performed with urea.
Equilibrium data were fit to a two-state model, U⇆M, by globally analyzing the CD signal at 20 different wavelengths between 220 and 240 nm.
Due to the constraints required to fit these titrations, the error in the free energy of folding is approximated to be ±0.3 kcal mol-1.
Parameter was fixed to the WT value.
WT values adapted from Svensson et al.16
Calculated according to mtot = |m‡u| + |m‡f.|
Denaturation was performed with Gdn-HCl.
The free energy difference between U+Zn ⇆ N•Zn and the Zn affinity was calculated as described in Kayatekin et al. 30
The large fraction of unfolded protein for the A4V, L38V, G93A and L106V variants at 20 °C and the known endothermic unfolding reaction for SOD127 prompted a comparison of the urea denaturation reactions for WT and A4V over the temperature range from 4 – 37 °C to determine the extent of unfolding at physiological temperature, 37 °C. While the stability of WT protein can be measured reliably at 37 °C, A4V is too destabilized at these temperatures to allow accurate fitting of the data. For both A4V and WT, the thermodynamic stability was determined by an singular value decomposition (SVD) analysis of CD spectra from 220-240 nm.28 The SVD vectors were normalized to the unfolded baseline and fit by holding the slope and y-intercept of the native baseline fixed to the 4 °C data set. A van't Hoff plot of the results is shown in Figure 3. The data are well-described by a simple linear dependence whose calculated enthalpy changes, 23.8±2.8 kcal mol-1 for WT and 20.3±2.2 kcal mol-1 for A4V, allow for the prediction of the stabilities at 37 °C.
Figure 3.

The van't Hoff plot of WT (circle) and A4V (triangle) . The stability at each temperature was determined from a urea titration. Linear extrapolations to physiological temperature are shown by the solid black lines. The black, dashed, vertical line indicates 37 °C while the dashed line represents a folding free energy of zero, where the protein is half unfolded. The buffer used was 20 mM HEPES, 1 mM EDTA, 1 mM TCEP pH 7.2.
The stability of WT is 2.0 kcal mol-1 at pH 7.2 and 37 °C, meaning that ∼97% of the protein is folded under these conditions. By contrast, the stability of A4V , which could be measured reliably up to 27 °C, falls from 2.1 kcal mol-1 at 4 °C to 0.8 kcal mol-1 at 27 °C. Linear extrapolation to 37 °C reveals that the stability is reduced to ∼0 kcal mol-1, indicating that A4V is half unfolded under physiological conditions. These results are consistent with previous native-state hydrogen exchange experiments that showed WT retained some protection against amide hydrogen exchange at 37 °C while A4V did not.29 Urea titrations at 37 °C for all variants can be seen in Figure 4. For the highly destabilized variants, these titration curves entirely lack native baselines and display a very small portion of the unfolding transition, consistent with the conclusion that these proteins are largely unfolded at 37 °C. On the other hand, the S134N and WT variants are nearly indistinguishable and appear reasonably well folded under these conditions.
Figure 4.
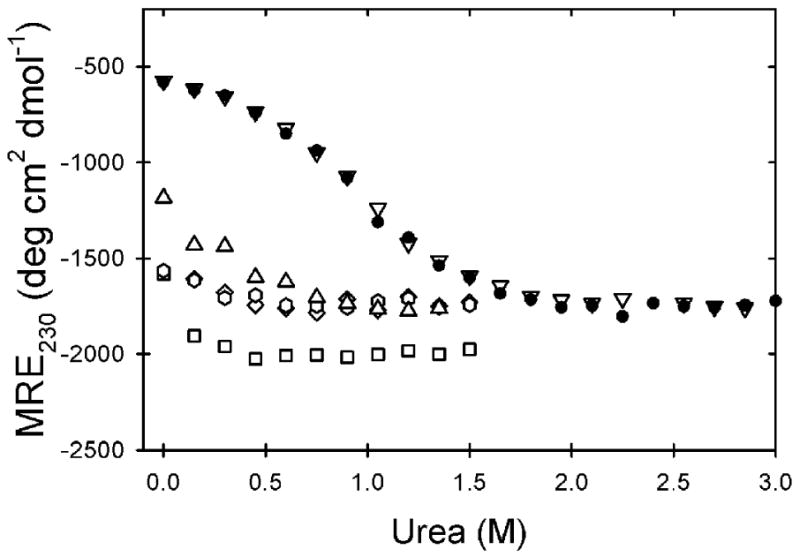
Equilibrium unfolding curves at 37 °C and pH 7.2 for WT and ALS variants as measured by CD at 230 nm: WT (filled circle), A4V (triangle), L38V (hexagon), G93A (diamond), L106V (square), S134N (upside-down triangle). The buffer used was 20 mM HEPES, 1 mM EDTA, 1 mM TCEP pH 7.2.
Stabilities of disulfide-oxidized apo-mAS-SOD1
The far-UV CD spectra of the disulfide-oxidized monomers can be seen in Figure 2B. In contrast with disulfide-reduced proteins, all variants exhibit well folded CD spectra similar to WT . Nevertheless, the characteristic shoulder at 230 nm is diminished for A4V, L38V, L106V and G93A, suggesting some structural differences may exist between these variants and WT .
The stabilities of the disulfide-oxidized and metal-free variants were measured by both kinetic and equilibrium methods. These complementary approaches towards stability measurement are possible because both kinetic and equilibrium mechanisms are well-described by two-state processes. The equilibrium urea denaturation curves for variants at 20 °C and pH 7.2, and the fraction unfolded plots for these titrations are shown in Figure 2D and Figure 2F, respectively. WT and S134N are stable to ∼1.8 M urea, where both proteins undergo a cooperative unfolding reaction that is complete by ∼3.8 M urea. The next most stable variant, A4V undergoes a cooperative unfolding reaction beginning around 0.8 M urea and is complete by 2.8M urea. L38V, G93A and L106V variants exhibit sloped native baselines and are completely unfolded by 2 M urea. These equilibrium data were fit to a two-state model as described for the titrations, except no were constraints imposed on the thermodynamic parameters or baselines and CD spectra between 215 and 245 nm were fit. From the calculated stabilities (Table 1) and the fraction unfolded plots (Figure 2F), it is evident that all disulfide-oxidized variants are at least 95% folded at 20 °C and pH 7.2.
The stabilities of the variants were also measured kinetically in order to facilitate an appropriate comparison with the Zn-bound data (see below). The kinetic unfolding and refolding reactions of the ALS-variants at 20 °C and pH 7.2 were monitored by far-UV CD at 230 nm. Both reactions were well described by single exponentials. Semi-log plots of the relaxation times as a function of the final denaturant concentration, i.e., chevron plots, are shown in Figure 5. The rate constants for the global unfolding and refolding reactions in the absence of denaturant were obtained by linear extrapolation of the exponentially-dependent regions of the chevron plots and calculated by the reciprocal of the relaxation times, k = 1/τ. The refolding rates only vary by a factor of 5 from the slowest, G93A, with a rate constant of 0.029 s-1, to the fastest, A4V, with a rate constant of 0.15 s-1. The largest perturbations are observed in the unfolding rates, ranging from 168-fold faster for G93A to 1.7-fold faster for S134N (Table 1), closely paralleling the perturbation in stability for each variant.
Figure 5.
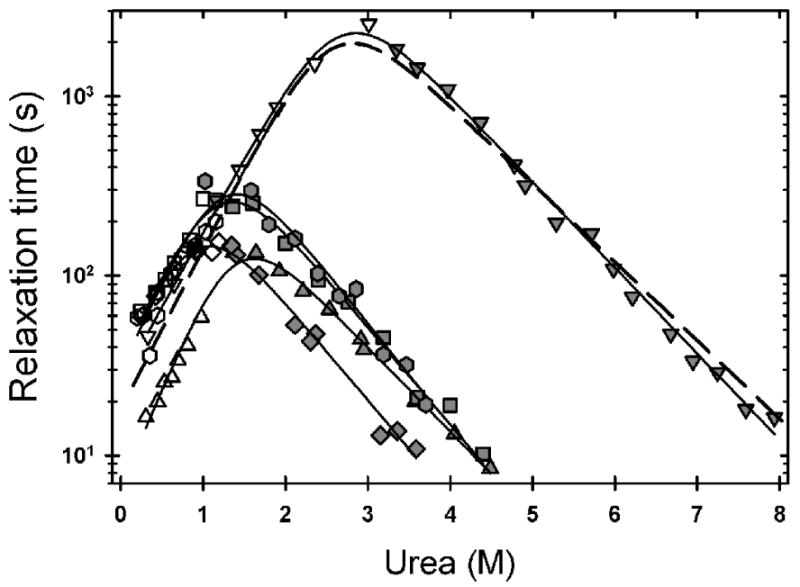
Observed refolding (open symbols) and unfolding (filled symbols) relaxation times of variants monitored by CD at 230 nm, at pH 7.2 and 20 °C, and plotted as a function of final denaturant concentration. The data for A4V (triangle), L38V (hexagon), G93A (diamond), L106V (square), S134N (upside-down triangle) are shown with the best fit line two a two-state folding reaction (solid lines). The fit to the disulfide-oxidized WT data (dashed-line) was adapted from Svensson et al.16 The buffer used was 10 mM KPi, 1 mM EDTA pH 7.2.
The free energy of folding for each variant, determined from the extrapolated rates, ΔG°=-RTln(kf/ku), is in very good agreement with the free energy of folding determined from the equilibrium titrations (Table 1). The observed change in stability range from no significant difference for S134N to as large as a 2.76 kcal mol-1 destabilization for G93A by the kinetic measurement (Table 1).
The effect of ALS mutations on the apparent Zn affinity of mAS-SOD1
The apparent Zn affinity of mAS-SOD1SS variants was determined from the difference in the free energies of their Zn-bound and Zn-free forms relative to the Zn-free unfolded state. Accurate estimates of the stability of the species cannot be obtained from equilibrium titrations because Zn appears to scramble to the Cu site in the unfolding transition zone.30 Alternatively, the equilibrium constant for the reversible unfolding reaction can be calculated from the ratio of the rate constants of unfolding and refolding reactions for zinc-bound unfolded state and the zinc-bound folded state. When combined with the affinity of Zn for the unfolded protein, estimated from a peptide model of the Zn-binding loop,30 the free energy of Zn binding (ΔΔG°Zn), and the corresponding apparent Zn affinity, can be obtained from the difference in the metal-bound (ΔG°Zn) and the metal-free stability(ΔG°Apo): ΔΔG°Zn = ΔG°Zn(Gdn-HCl) - ΔG °Apo(Urea), where ΔΔG°Zn=-RTln(Kd) (see Kayatekin et al.30 for a detailed explanation of the analysis).
The unfolding and refolding reactions of 10 μM protein loaded with stoichiometric Zn at 20 °C and pH 7.2 were monitored by far-UV CD at 230 nm. Guanidine hydrochloride (Gdn-HCl) rather than urea was used as a denaturant to sample the entire folding reaction coordinate because Zn greatly enhances the stability of SOD1 and requires a more potent denaturant to access the unfolded state. The presence of Zn also required a HEPES buffer because Zn3(PO4)2 precipitates at mM buffer concentrations. The requirement to change both the denaturant and the buffer to make this comparison means that the dissociation constants should be considered apparent and not absolute. However, because the WT and ALS-variants were subjected to the same procedures, the differences in the Zn binding free energies are expected to reflect the inherent differences in affinities induced by the mutations.
Above stoichiometric Zn:protein concentrations for all variants, the refolding reactions were well described by a single exponential phase, corresponding to the faster of the two observed relaxation times at stoichiometric or lower Zn concentrations. The slower reaction corresponds to the folding of .30 The unfolding reactions displayed a second, faster exponential in the transition zone that has previously been attributed to the scrambling of the Zn ion to the Cu site.30 The chevron plots for A4V, L38V and WT are shown in Figure 6A, and the chevron plots for G93A and L106V and S134N are shown in Figure 6B.
Figure 6.
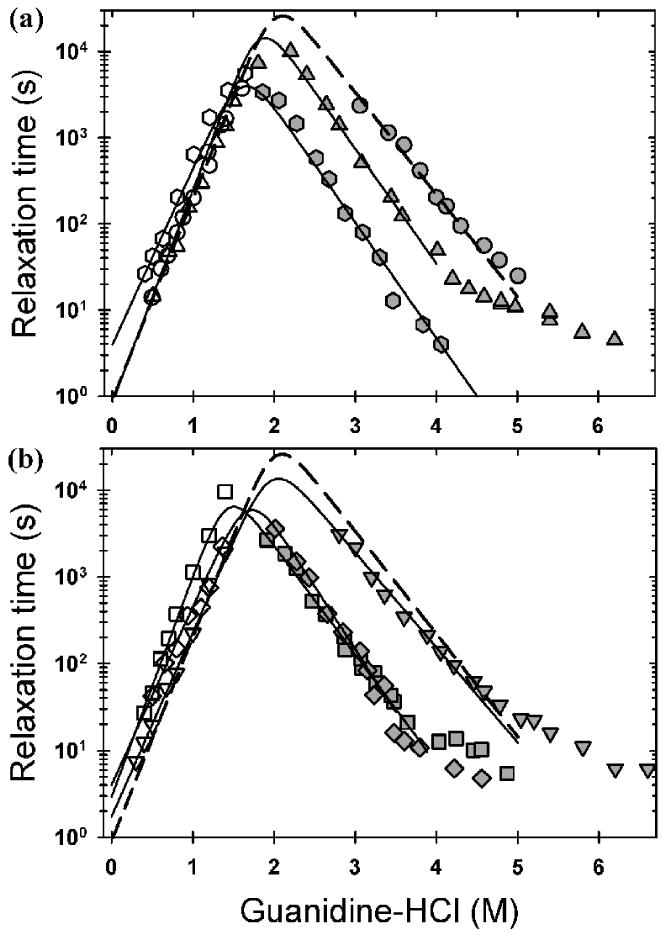
Observed refolding (open symbols) and unfolding (filled symbols) relaxation times monitored by CD at 230 nm, at pH 7.2 and 20 °C, and plotted as a function of final denaturant concentration. (A) The data for WT (circle), A4V (triangle), L38V (hexagon) and (B) the data for G93A (diamond), L106V (square) and S134N (upside-down triangle) are shown with the best fit line two a two-state folding reaction. The WT data were adapted from Kayatekin et al.30 The concentration of protein and Zn for all ALS variants was 10 μM and the buffer used was 20 mM HEPES pH 7.2.
The refolding relaxation times increase exponentially with increasing denaturant concentration to a maximum around 1.8-2.2 M Gdn-HCl. At higher denaturant concentrations the unfolding reaction is favored and the observed unfolding relaxation times decrease exponentially with increasing denaturant concentration. Above 4-5 M Gdn-HCl, the unfolding relaxation times in the presence of Zn for WT and all of the variants except for L38V rollover to become nearly independent of the denaturant concentration. The L38V variant may roll over at fast unfolding rates which are within the dead time of a manual-mixing experiment. These data suggest either that Zn has introduced a rate-limiting rearrangement reaction in the native conformation or that Zn has slowed a rearrangement reaction that was previously undetectable for the apoprotein with manual-mixing techniques. Because this reaction is not kinetically coupled to the major unfolding reaction that controls unfolding above 4-5 M Gdn-HCl, its presence does not interfere with the kinetic measurement of stability.
The refolding rate constants of all the proteins are within a factor of ∼5 of each other, ranging from 0.19±0.05 s-1 for G93A, to 0.74±0.01 s-1 for WT to 0.87±0.15 s-1 for A4V (Table 1). The effects of the mutations are substantially greater on the unfolding rate constants, which parallel the effects on stability. WT is the slowest to unfold, with a rate constant of 0.23±0.08×10-7 s-1, while L106V unfolds the most rapidly, roughly 60-fold faster, with a rate constant of 13.3±3.9×10-7 s-1.
These data can be compared with the stabilities of the Zn-free forms of these proteins to yield an apparent Zn affinity for the ALS variants (Table 1). A4V and G93A variants bind Zn slightly tighter than WT mAS-SOD1SS, while L38V, L106V and S134N bind Zn more weakly than the WT protein. The corresponding dissociation constants, Kd, for A4V, L38V, G93A and L106V are within a factor of three of the Kd for WT, 100 pM, and, with the exception of S134N, are nearly within the estimated errors of the measurement. The apparent Zn affinity of S134N is reduced by roughly an order-of-magnitude compared to WT. Contrasting with the dramatic effects on stability with the loss of the disulfide bond, Zn has little differential effect on the standard state stability of the A4V, L38V, G93A and L106V variants. Only the apparent Zn affinity of S134N is significantly reduced from WT mAS-SOD1SS.
The apparent Zn affinity of the disulfide-reduced protein is greatly decreased
The thermodynamic coupling between the disulfide bond and Zn binding was assessed by measuring the Zn affinity of WT and S134N mAS-SOD12SH with the kinetic assay described above for the mAS-SODSS species. The chevron plot of WT and S134N are shown in Figure 7A. In both refolding and unfolding, only a single exponential phase was observed. Neither the denaturant dependence of the refolding rate nor the unfolding rate is significantly affected by the loss of the disulfide-bond. Both variants exhibit an identical response to disulfide-bond reduction, where the refolding rates are slowed ∼3 fold compared to the disulfide-oxidized protein and the unfolding rate is accelerated ∼5 fold (Table 2).
Figure 7.

Observed refolding (open symbols) and unfolding (filled symbols) relaxation times of WT (circles) and S134N (upside-down triangle) (A) and (B) monitored by CD at 230 nm, at pH 7.2 and 20 °C, and plotted as a function of final denaturant concentration. The data are shown with the best fit line to a two-state folding reaction for WT(solid line) and S134N (dashed line). The protein and Zn concentrations were 10 μM and the buffer used was 20 mM HEPES, 1 mM TCEP pH 7.2.
Table 2. Thermodynamic parameters, microscopic rate constants and kinetic m ‡-values for disulfide-reduced WT and S134N.*.
| WTapoa | S134Napoa | WTZnb | S134NZnb | ||
|---|---|---|---|---|---|
| kf | 0.027±0.006 | 0.019±0.003 | 0.099±0.005 | 0.064±0.014 | |
|
|
1.43±0.23 | 1.19±0.12 | 3.6±0.1 | 2.87±0.17 | |
| ku | (1.0±0.3)×10-4 | (6.6±1.7)×10-5 | (2.7±0.5)×10-5 | (5.2±0.6)×10-6 | |
|
|
-0.57±0.03 | -0.65±0.03 | -0.92±0.03 | -1.04±0.02 | |
|
|
3.26±0.22 | 3.30±0.18 | 4.78±0.11 | 5.48±0.14 | |
| m(U/M)c | 2.00±0.23 | 1.84±0.12 | 4.52±0.1 | 3.91±0.17 | |
|
|
- | - | 9.52±0.25 | 10.2±0.2 | |
| Kdd | - | - | 75±33 nM | 25±10 nM | |
Units for the U ⇆ M reaction: kf and ku, s-1; , , and m(U/M), kcal mol-1 M-1; .
. kcal mol-1.
Denaturation was performed with urea.
Denaturation was performed with Gdn-HCl.
Calculated according to mtot = |m‡u| + |m‡f|
The free energy difference between U+Zn ⇆ N•Zn and the Zn affinity was calculated as described in Kayatekin et al. 30
When Zn was introduced, two phases were observed in refolding and unfolding, comparable to the disulfide-oxidized protein. The slower phase in refolding matched the refolding rate of the metal-free protein. The chevron plot of the slow exponential phase in unfolding and the fast phase in refolding at stoichiometric Zn concentrations for WT and S134N are shown in Figure 7B and the associated thermodynamic and kinetics parameters are reported in Table 2. Although the status of the disulfide bond does not alter the denaturant-dependence of the refolding reaction, the denaturant-dependence of the unfolding reaction is substantially decreased when the disulfide-bond is reduced. This change presumably reflects a less structured native state for compared to . The refolding rate in the absence of denaturant for WT is nearly 10-fold slower than for , while the unfolding rate constant is nearly 1000-fold faster.
Surprisingly, the disulfide-reduced S134N variant clearly exhibits an ∼3-fold slower unfolding rate than the WT protein in the presence of Zn, while the refolding rates for both proteins are comparable. This result suggests that the replacement of serine with asparagine in the electrostatic loop may actually lead to interactions which stabilize Zn binding compared to the WT protein when the disulfide-bond is reduced and the protein relaxes (see Discussion).
Discussion
Patients with the five ALS-variants of SOD1 examined in this study, A4V, L38V, G93A, L106V and S134N, exhibit an average life expectancy of less than three years after the onset of disease.31 Our approach towards testing the potential role of monomeric species in the aggregation hypothesis for toxicity in SOD1-mediated fALS has been to employ classical thermodynamic and kinetic folding experiments to define the folding free energy surface of stable monomeric versions of these variants. The coupling between mutations, disulfide-bond status and Zn binding can be related to the changes in the relative populations of the folded and unfolded states (Figure 8). Enhanced populations of marginally-soluble monomeric forms would be expected to increase the propensity for aggregation and, possibly, promote toxicity.
Figure 8.

The fraction of unfolded species populated at equilibrium for each variant, (black), (light gray), (dark gray) at 10 μM protein concentration, 20 °C and pH 7.2. The errors for A4V, L38V, G93A and L106V were approximated at 20%. All other errors were determined from the error in the ΔG°.
Disulfide-reduced ALS variants are very unstable
The reduction of the disulfide bond in the stable monomeric models of the A4V, L38V, G93A and L106V variants causes a significant reduction in their stability (Table 1) and a corresponding increase in the population of their unfolded states (Figure 8). For example G93A , the least stable variant, is only 5% unfolded at 20 °C. Upon reduction of the disulfide bond, the population of unfolded protein increases to 55%. The effect is exacerbated at 37 °C, where the unfolded populations of A4V, L38V, G93A and L106V exceed 50%, while only 3% of the WT protein is unfolded under these conditions. These results demonstrate that unfolded is prevalent in these ALS variants under physiological conditions and, therefore, could play a critical role in aggregation. A recent report has demonstrated that even WT-SOD1Apo can aggregate under conditions that enhance the unfolded state, such as the addition of reducing agent or the presence of 1M Gdn-HCl, when agitated at 37 °C.32 Given that the unfolded thermodynamic state represents a manifold of rapidly-interconverting conformations, it is also possible that a transient population of sub-structure might be more aggregation-prone and, therefore, responsible for the initiation of the aggregation reaction.
Implications for mitochondrial damage
The dramatic destabilization of the disulfide-reduced forms of the A4V, L38V, G93A and L106V variants may also play a role in mitochondrial damage observed in mouse models of ALS.33 The transport of SOD1 into yeast mitochondria is greatly favored for the reduced metal-free monomeric form,34 presumably reflecting the ease of access to an unstructured unfolded state required to traverse the outer membrane. Once inside the intermembrane space, a resident disulfide relay system oxidizes the intramolecular disulfide bond, which, along with Zn binding and dimer formation, would effectively trap SOD1 inside the mitochondria.35 The dominant population of the unfolded state for the disulfide-reduced form of the four WT-like variants, A4V, L38V, G93A and L106V, at pH 7.2 and 37 °C and the unusually slow folding reaction to the disulfide-competent folded state could provide an opportunity for the unfolded state to aggregate or to associate with other components in the intermembrane space. The observed increased localization of G93A and other ALS variants of SOD1 in mitochondria,36 especially in the spinal cord of transgenic mice,37 and their capacity to damage this essential organelle may, therefore, be a manifestation of their thermodynamic and kinetic folding properties.
Zn binding is marginally affected in WT-like ALS variants
NMR and crystallographic studies have shown that Zn binding has an important role in defining the structure of SOD1 by organizing the Zn binding and electrostatic loops of the WT protein.25,26,38 As shown in the present study, Zn binding to mAS-SOD1SS significantly stabilizes the folded state for all of the variants (Table 1). However, the apparent dissociation constants for the WT-like variants, A4V, L38V, G93A and L106V, vary by less than an order of magnitude from the 100 pM apparent Kd for the WT protein and are not consistently decreased. Despite the small differences in the measured Zn affinity, these results correlate well with the observed metal content of the mutant protein when isolated from insect cells.39
These results may appear to contradict the findings by Crow et al., 40 who found that the dimeric A4V and L38V SOD1 variants had a 30-fold and 18-fold weakening of the Zn affinity relative to WT protein in 2 M urea; the contradiction can be understood in terms of the increased population of the folded monomeric forms for these variants and their lower zinc affinity relative to the dimeric state.30 The 66 hours required to extract zinc from these variants in the Crow et al. study would provide ample opportunity for the monomeric form to serve as the conduit for zinc loss.
S134N, a metal-binding variant of SOD1, differs from the β-sandwich variants in that the apparent Zn affinity is significantly reduced. While the S134N variant is not destabilized significantly in its metal-free forms, the weakening in the apparent Kd for Zn binding from 100 pM for WT mAS-SOD1SS to 1.6 nM for S134N mAS-SOD1SS offers the possibility that aggregation of the monomeric forms may still be the source of toxicity. Although this variant can be loaded with Zn and Cu in vitro, 41,42 Hayward and coworkers43 have observed decreased Zn and Cu content in as-isolated samples of S134N SOD1 expressed in insect cells. The replacement of serine with asparagine in the electrostatic loop may similarly impede full metalation in motor neurons. The potential for aggregation might be further compounded by the spontaneous reduction of the disulfide bond of the metal-free species in the cytoplasm. Thus, even the small fraction of unfolded mAS-SOD12SH observed for S134N may be sufficient to cause aggregation. Because the predicted metal-free unfolded and folded populations for S134N are nearly identical to WT, the intriguing possibility arises that the loss of Zn even from the WT protein may be sufficient to trigger aggregation.
Zn binding is dramatically decreased by reduction of the disulfide bond
In contrast to the response to mutations, the reduction of the disulfide bond has a dramatic effect on the Zn affinity of mAS-SOD1, resulting in a ∼750-fold weakening in the apparent Kd from 100 pM to 75 nM for the WT protein and a 16-fold weakening in the apparent Kd for the S134N variant from 1.6nM to 25 nM. The 3-fold tighter Kd for disulfide-reduced S134N vs. WT protein contrasts with a 16-fold weaker apparent Kd in the presence of the disulfide bond. This behavior demonstrates thermodynamic coupling between the disulfide bond and Zn binding. Inspection of the crystal structure of SOD1 shows that the disulfide bond is proximal to the Cu site, which shares a ligand with the Zn site. Apparently, the constraints on the structure introduced by the disulfide bond are transmitted to the Zn binding site in a way that reduces the affinity of S134N for Zn. This may be due to the inability of the S134N variant to induce compactness in the electrostatic loop by the formation of the S134 - D125 hydrogen bond.41 When the protein relaxes in the absence of the disulfide bond, the electrostatic loop may be able to access conformers that form stabilizing electrostatic interactions absent in the WT protein.
These results suggest that disulfide bond oxidation may be the key step in the maturation of SOD1. Because would not compete effectively for free Zn at ∼1 nM concentration in vivo, Zn would be expected to bind after the oxidation of the disulfide bond when the apparent Kd decreases to 100 pM. This hypothesis is supported by the discovery that the presence of Zn does not accelerate the formation of the disulfide bond in SOD1 expressed in rabbit reticulocyte assays.44
Aggregation following synthesis on the ribosome?
The analysis presented above presumes that the populations of species in vivo dictated by the equilibrium free energy surface of SOD1 are those responsible for aggregation and toxicity. Alternately, the aggregation of SOD1 may occur soon after synthesis on the ribosome, prior to its several maturation steps including disulfide bond formation, dimerization and Zn and Cu loading.44 Disulfide-reduced SOD1 folds surprisingly slowly in vitro, requiring ∼40 seconds at 20 °C, nearly four orders of magnitude slower than predicted by its topology.45 Although the time required for folding is comparable to the length of time required to translate a 153 residue eukaryotic protein, ∼30-50 s,46 co-translational folding of SOD1 is unlikely since the rate-limiting step of folding involves the docking of the first four N-terminal β-strands with the β7 strand near the C-terminus.47 The observation that the four ALS variants in the β-sandwich structure, A4V, L38V, G93A and L106V, have substantially increased unfolded populations of their disulfide-reduced forms implies an enhanced opportunity for the aggregation of these species prior to their ultimate maturation to their native dimeric forms. The subsequent formation of the disulfide bond increases the stability of the folded monomeric form and drives the dimerization reaction.21 The high concentration of SOD1 in motor neurons, >10 μM,9 and its half-life of ∼1 day,48 requires a constant and significant rate of synthesis on ribosomes. Thus, the aggregation of SOD1 may involve both kinetic, i.e., following synthesis and prior to maturation, and thermodynamic, i.e., at steady-state after maturation,20 mechanisms.
Materials and Methods
Protein purification
Recombinant proteins were expressed in and purified from BL21-Gold(DE3) PLysS cells (Stratagene®, Inc. Cedar Creek, TX). The protein was purified using the procedure previously described in Kayatekin et al.30 Protein identity and purity was determined by measuring the molecular weight with LC-ESI mass spectrometry. The protein concentrations were calculated using an extinction coefficient of 5,400 M-1 cm-1.
Disulfide-bond reduction
All variants of mAS-SOD were reduced by 24 hour incubation in 1 mM TCEP. The reduction of the disulfide-bond and absence of reoxidation after the incubation in reducing agent was confirmed by kinetic unfolding or refolding experiments for mAS-SOD1Apo. Fully reduced protein exhibited a single exponential phase with a relaxation time 5-fold faster than the disulfide-oxidized protein in unfolding.
Equilibrium experiments
All circular dichroism spectroscopy was performed on a Jasco-810 spectropolarimeter (Jasco Inc., Easton, MD) equipped with a water-cooled Peltier temperature control system. The CD spectra of disulfide-reduced protein were an average of 6 spectra obtained using a 1 mm path length quartz cuvette, a scan rate of 20 nm min-1 and a response time of 4 s. The buffer used was 10 mM potassium phosphate, 1 mM ETDA, 1 mM TCEP, pH 7.2. The urea induced unfolding curves were monitored from 220-240 nm in a 0.5 cm path length quartz cuvette using a scan rate of 20 nm min-1 and a response time of 8 s. The buffer used was 20 mM HEPES, 1 mM EDTA, 1 mM TCEP for disulfide-reduced and 10 mM KPi, 1 mM EDTA for disulfide-oxidized samples, pH 7.2. Urea concentrations were determined by refractive index on a Leica Mark II refractometer. Titration samples were made from concentration matched stocks of folded protein in standard buffer and unfolded protein at 8 M urea and incubated at the experimental temperature overnight.
Kinetic experiments
The unfolding and refolding kinetics, initiated by manual mixing, were monitored by CD. For metal-free protein, 10 mM potassium phosphate, 1 mM EDTA, pH 7.2 was used as the buffer. For metal-bound protein the potassium phosphate was replaced with 20 mM HEPES and the 1 mM EDTA was omitted. Data were collected at 230 nm in a 1 cm2 cuvette under continuous mixing with a solution volume of 1.9 mL. Unfolding jumps were initiated from protein in 0 M urea or Gdn-HCl and refolding jumps were initiated from protein denatured in 4-6 M Gdn-HCl or 8 M urea to various final concentrations. For Zn-loaded samples, the protein was incubated with stoichiometric Zn prior to dilution in the unfolding/refolding buffer. The final concentration of Gdn-HCl was measured by index of refraction. Protein concentrations were 10 μM for all kinetic experiments.
Acknowledgments
We thank Dr. Osman Bilsel, Sagar V. Kathuria, Steven F. Trueman and Ian M. Love for many helpful discussions, Nicole Washington for purifying S134N mAS-SOD1 and Jessica Adefusika, Graham Dobereiner and Matt Samberg for creating the bacterial constructs. This work was supported by National Institutes of Health grant GM54836, and support for mass spectrometry was provided by NICHD IDDRC Core Grant HD04147.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- CD
circular dichroism
- fALS
familial amyotrophic lateral sclerosis
- Gdn-HCl
guanidine hydrochloride
- mAS-SOD1
F50E/G51E/C6A/C11S monomeric variant of SOD1
- SOD1
human Cu, Zn superoxide dismutase
- TCEP
(tris(2-carboxyethyl)phosphine)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Andersen PM, Sims KB, Xin WW, Kiely R, O'Neill G, Ravits J, Pioro E, Harati Y, Brower RD, Levine JS, Heinicke HU, Seltzer W, Boss M, Brown RH., Jr Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes. Amyotroph Lateral Scler Other Motor Neuron Disord. 2003;4:62–73. doi: 10.1080/14660820310011700. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–4. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 5.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–93. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 6.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 7.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- 8.Seetharaman SV, Prudencio M, Karch C, Holloway SP, Borchelt DR, Hart PJ. Immature Copper-zinc Superoxide Dismutase and Familial Amyotrophic Lateral Sclerosis. Exp Biol Med (Maywood) 2009 doi: 10.3181/0903-MR-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 10.Rakhit R, Robertson J, Vande Velde C, Horne P, Ruth DM, Griffin J, Cleveland DW, Cashman NR, Chakrabartty A. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–9. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 11.Hough MA, Grossmann JG, Antonyuk SV, Strange RW, Doucette PA, Rodriguez JA, Whitson LJ, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc Natl Acad Sci U S A. 2004;101:5976–81. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zetterstrom P, Stewart HG, Bergemalm D, Jonsson PA, Graffmo KS, Andersen PM, Brannstrom T, Oliveberg M, Marklund SL. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci U S A. 2007;104:14157–62. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–98. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 14.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 15.Teilum K, Smith MH, Schulz E, Christensen LC, Solomentsev G, Oliveberg M, Akke M. Transient structural distortion of metal-free Cu/Zn superoxide dismutase triggers aberrant oligomerization. Proc Natl Acad Sci U S A. 2009;106:18273–8. doi: 10.1073/pnas.0907387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. J Mol Biol. 2006;364:1084–102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci U S A. 2004;101:15893–8. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindberg MJ, Tibell L, Oliveberg M. Common denominator of Cu/ Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc Natl Acad Sci U S A. 2002;99:16607–12. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassall KA, Stathopulos PB, Rumfeldt JA, Lepock JR, Meiering EM. Equilibrium thermodynamic analysis of amyotrophic lateral sclerosis-associated mutant apo Cu,Zn superoxide dismutases. Biochemistry. 2006;45:7366–79. doi: 10.1021/bi0600953. [DOI] [PubMed] [Google Scholar]
- 20.Svensson AK, Bilsel O, Kayatekin C, Adefusika JA, Zitzewitz JA, Matthews CR. Metal-free ALS-Variants of Dimeric Human Cu,Zn-Superoxide Dismutase Have Enhanced Populations of Monomeric Species. PloS One. 2010 doi: 10.1371/journal.pone.0010064. In Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnesano F, Banci L, Bertini I, Martinelli M, Furukawa Y, O'Halloran TV. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–8003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa Y, O'Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280:17266–74. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 23.Bertini I, Piccioli M, Viezzoli MS, Chiu CY, Mullenbach GT. A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase. Eur Biophys J. 1994;23:167–76. doi: 10.1007/BF01007608. [DOI] [PubMed] [Google Scholar]
- 24.Hallewell RA, Imlay KC, Lee P, Fong NM, Gallegos C, Getzoff ED, Tainer JA, Cabelli DE, Tekamp-Olson P, Mullenbach GT, et al. Thermostabilization of recombinant human and bovine CuZn superoxide dismutases by replacement of free cysteines. Biochem Biophys Res Commun. 1991;181:474–80. doi: 10.1016/s0006-291x(05)81443-3. [DOI] [PubMed] [Google Scholar]
- 25.Banci L, Bertini I, Cantini F, D'Onofrio M, Viezzoli MS. Structure and dynamics of copper-free SOD: The protein before binding copper. Protein Sci. 2002;11:2479–2492. doi: 10.1110/ps.0210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strange RW, Antonyuk S, Hough MA, Doucette PA, Rodriguez JA, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328:877–91. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 27.Stathopulos PB, Rumfeldt JA, Karbassi F, Siddall CA, Lepock JR, Meiering EM. Calorimetric analysis of thermodynamic stability and aggregation for apo and holo amyotrophic lateral sclerosis-associated Gly-93 mutants of superoxide dismutase. J Biol Chem. 2006;281:6184–93. doi: 10.1074/jbc.M509496200. [DOI] [PubMed] [Google Scholar]
- 28.Ionescu RM, Smith VF, O'Neill JC, Jr, Matthews CR. Multistate equilibrium unfolding of Escherichia coli dihydrofolate reductase: thermodynamic and spectroscopic description of the native, intermediate, and unfolded ensembles. Biochemistry. 2000;39:9540–50. doi: 10.1021/bi000511y. [DOI] [PubMed] [Google Scholar]
- 29.Shaw BF, Durazo A, Nersissian AM, Whitelegge JP, Faull KF, Valentine JS. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–76. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- 30.Kayatekin C, Zitzewitz JA, Matthews CR. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J Mol Biol. 2008;384:540–55. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chattopadhyay M, Durazo A, Sohn SH, Strong CD, Gralla EB, Whitelegge JP, Valentine JS. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc Natl Acad Sci U S A. 2008;105:18663–8. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hervias I, Beal MF, Manfredi G. Mitochondrial dysfunction and amyotrophic lateral sclerosis. Muscle Nerve. 2006;33:598–608. doi: 10.1002/mus.20489. [DOI] [PubMed] [Google Scholar]
- 34.Field LS, Furukawa Y, O'Halloran TV, Culotta VC. Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J Biol Chem. 2003;278:28052–9. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 35.Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta. 2008;1783:601–9. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Kawamata H, Manfredi G. Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria. Hum Mol Genet. 2008;17:3303–17. doi: 10.1093/hmg/ddn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, Brannstrom T, Gredal O, Wong PC, Williams DS, Cleveland DW. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Banci L, Bertini I, Cramaro F, DelConte R, Viezzoli MS. Solution Structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez JA, Valentine JS, Eggers DK, Roe JA, Tiwari A, Brown RH, Jr, Hayward LJ. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J Biol Chem. 2002;277:15932–7. doi: 10.1074/jbc.M112088200. [DOI] [PubMed] [Google Scholar]
- 40.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–44. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 41.Banci L, Bertini I, D'Amelio N, Gaggelli E, Libralesso E, Matecko I, Turano P, Valentine JS. Fully metallated S134N Cu,Zn-superoxide dismutase displays abnormal mobility and intermolecular contacts in solution. J Biol Chem. 2005;280:35815–21. doi: 10.1074/jbc.M506637200. [DOI] [PubMed] [Google Scholar]
- 42.Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, Hart PJ. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–7. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 43.Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, Valentine JS, Brown RH., Jr Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–31. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 44.Bruns CK, Kopito RR. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. Embo J. 2007;26:855–66. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–94. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 46.Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–55. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 47.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc Natl Acad Sci U S A. 2006;103:10218–23. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci U S A. 1994;91:8292–6. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]


