Abstract
Tumorigenesis is in many respects a process of dysregulated cellular evolution that drives malignant cells to acquire six phenotypic hallmarks of cancer, including their ability to proliferate and replicate autonomously, to resist cytostatic and apoptotic signals, and to induce tissue invasion, metastasis, and angiogenesis. Transforming growth factor-β (TGF-β) is a potent pleiotropic cytokine that functions as a formidable barrier to the development of cancer hallmarks in normal cells and tissues. Paradoxically, tumorigenesis counteracts the tumor suppressing activities of TGF-β, thus enabling TGF-β to stimulate cancer invasion and metastasis. Fundamental gaps exist in our knowledge of how malignant cells overcome the cytostatic actions of TGF-β, and of how TGF-β stimulates the acquisition of cancer hallmarks by developing and progressing human cancers. Here we review the molecular and cellular mechanisms that underlie the ability of TGF-β to mediate tumor suppression in normal cells, and conversely, to facilitate cancer progression and disease dissemination in malignant cells.
Keywords: Angiogenesis, Apoptosis, Epithelial-mesenchymal Transition, Immunosurveillance, Invasion, Metastasis, Signaling Transduction, Tumor Suppressor
1. Introduction
Since the inception of the National Cancer Act of 1971, science and medicine have waged an intense battle aimed at conquering cancer. Although considerable progress has been achieved in terms of our understanding of the molecular mechanisms that underlie cancer development and progression, cancer itself remains a significant health concern and burden in the United States. Indeed, 1 in 4 deaths in the United States results from cancer, which also is the leading cause of death in individuals younger than 65 years of age. Despite these grim statistics, overall cancer incidence and mortality rates have begun to decline over the last decade [1]. Continuing along this positive trend will require the development of new diagnostic and chemotherapeutic regimens, as well as the elucidation of new knowledge of how cancer cells acquire the six essential phenotypes, or hallmarks, necessary to become malignant. Included in this phenotypic list is the ability of cancer cells to (i) grow autonomously; (ii) disregard cytostatic signals; (iii) ignore apoptotic signals; (iv) stimulate angiogenesis; (v) invade and metastasize; and (vi) become immortal [2]. Failure by developing neoplasms to acquire each of these phenotypes prevents their conversion to aggressive states, suggesting that these cancer hallmarks represent various rate-limiting steps during malignant development. As such, pharmacological targeting of cancer hallmarks, both singly and in combination, may offer new inroads to effectively treat the development and dissemination of human malignancies.
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that regulates mammalian development, differentiation, and homeostasis. It is also a potent anticancer agent that prohibits the uncontrolled proliferation of epithelial, endothelial, and hematopoietic cells. Aberrations in the TGF-β signaling pathway bring about resistance to TGF-β-mediated growth arrest, and thus give rise to human malignances [3–5]. Paradoxically, these genetic and epigenetic aberrations conspire to convert TGF-β from a suppressor of tumor formation to a promoter of their growth, survival, and metastasis. Although the molecular details underlying the oncogenic activities of TGF-β remain to be fully elucidated, recent evidence implicates TGF- β as a principle player involved in regulating the acquisition of cancer hallmarks by malignant cells [3–5]. This review focuses on the complex roles played by TGF-β during cancer progression, particularly its ability to regulate the development and progression of malignant cells through each of the individual hallmarks of cancer (Figure 1).
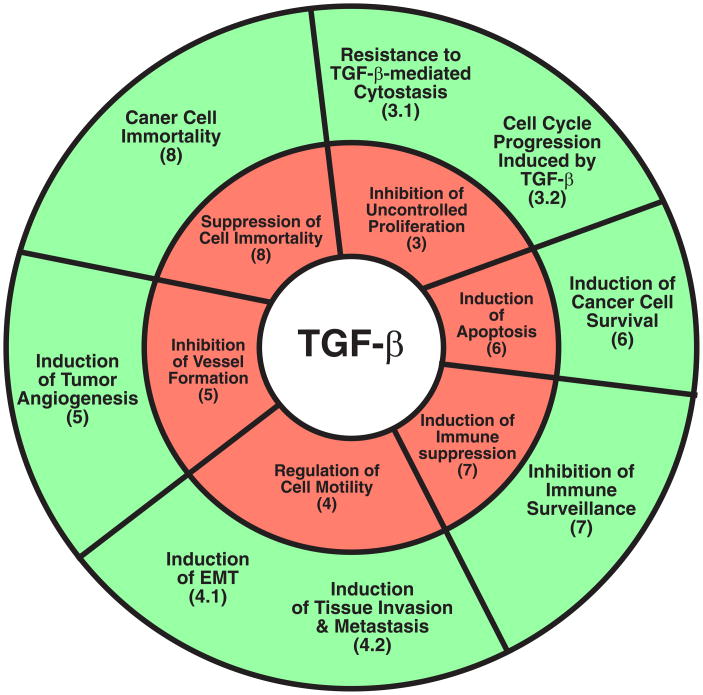
Fig. 1.
TGF-β and the hallmarks of cancer. Tumorigenesis converts TGF-β from a powerful tumor suppressor to a lethal tumor promoter that enables evolving cancer cells to acquire the 6 phenotypic traits or hallmarks of cancer. In normal epithelial, endothelial, and hematopoietic cells, TGF-β suppresses the formation of cancer hallmarks (red wheel). However, genetic or epigenetic events conspire to inactivate the tumor suppressing functions of TGF-β, thereby conferring oncogenic behaviors upon this multifunctional cytokine and its ability to stimulate the development of cancer hallmarks (green wheel). Bracketed numbers correspond to individual chapter subheadings that describe the indicated functions of TGF-β.
2. TGF-β signaling system
2.1. Canonical TGF-β signaling
The diverse biological activities of TGF-β are mediated through its stimulation of a deceptively simple signaling system that at its core is comprised of three TGF-β receptors, types I (TβR-I), II (TβR-II), and III (TβR-III), and three latent transcription factors, Smads 2, 3, and 4 (Figure 2; [4–8]). The initiation of transmembrane signaling by TGF-β takes place upon its binding to TβR-III, which then presents TGF-β to TβR-II [9]. This ligand presentation mechanism is especially important for TGF-β2, which only interacts with TβR-II when bound to TβR-III [10]. It should be noted that the requirement for TβR-III in propagating messages by TGF-β2 is not absolute, particularly in cells that express a TβR-II variant that binds TGF-β2 independently of TβR-II expression [11]. In contrast, TGF-β1 and TGF-β3 both readily bind to TβR-II and induce intracellular signaling in the absence or presence of TβR-III. The differential requirements of individual TGF-β isoforms for TβR-III, coupled with the spatiotemporal differences observed in their expression and activation patterns [12], likely underlies the more than 30 distinct phenotypes observed in TGF-β1-, TGF-β2-, and TGF-β3-deficient mice [13]. Regardless of its mode of activation, ligand-bound TβR-II subsequently associates with and binds to TβR-I. Both TβR-I and TβR-II house intrinsic Ser/Thr protein kinase activity in their cytoplasmic domains, and the conversion of these ligand:receptor ternary complexes from their inactive to active states requires TβR-II to transphosphorylate TβR-I, thereby stimulating its protein kinase activity [14]. Activated TβR-I in turn stimulates Smads 2 and 3 by phosphorylating these latent transcriptional factors at their C-terminal SXS motif. Phosphorylated Smads 2 and 3 undergo a rapid conformational change that facilitates their association with common Smad, Smad4. This conformational change also unveils cryptic nuclear localization sequences in Smads 2, 3, and 4 that promotes heterocomplex accumulation in the nucleus [4–8]. Access to the nucleus enables Smad2/3/4 complexes to interact with an expanding list of transcriptional co-activators and -repressors that collectively alter cell fates through the coordinated induction and repression of TGF-β-responsive genes in a cell- and context-specific manner [4–8].
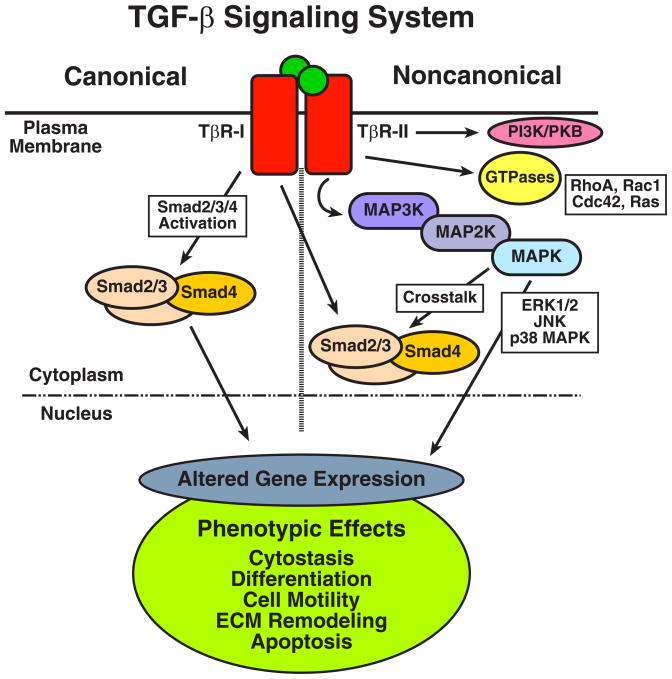
Fig. 2.
Canonical and noncanonical TGF-β signaling. TGF-β stimulates responsive cells by binding and activating two transmembrane Ser/Thr protein kinase receptors termed, TGF-β type I (TβR-I) and type II (TβR-II). Activation of these ligand:receptor ternary complexes requires TβR-II to transphosphorylate TβR-I, which phosphorylates and activates Smad2/3. Once activated, Smad2/3 form heterocomplexes with Smad4, which collectively translocate to the nucleus to mediate canonical signaling events by TGF-β (left panel). Noncanonical TGF-β signaling takes place through the ability of TGF-β to stimulate MAP kinases, small GTPases, and PI3K/AKT, and to inhibit NF-κB. Altered coupling of TGF-β to its canonical and noncanonical effector pathways contributes to the development of oncogenic signaling by TGF-β. See text for specific details of TGF-β signaling in normal and malignant cells.
Signaling through this canonical TGF-β pathway is regulated and fine-tuned via multiple mechanisms that span all cellular compartments. For instance, several adapter/anchoring proteins such as SARA [15], Hgs [16], and Dab2 [17] bind Smad2/3 and facilitate their phosphorylation and activation by TβR-I. This phosphotransferase reaction can be negated by expression of the inhibitory Smad, Smad7, which (i) interacts physically with TβR-I and occludes its access to Smad2/3 [18–20], and (ii) recruits the E3 ligase Smurf1/2 to facilitate TGF-β receptor ubiquitination and degradation [21, 22]. Moreover, the ability of Smad7 to inhibit TGF-β signaling is augmented by its interaction with STRAP [23], and conversely, is attenuated by its association with either AMSH2 [24] or Arkadia [25–27].
The cellular response to TGF-β also is fine-tuned by the continual nucleocytoplasmic shuttling of Smad2/3, which facilitates their ability to sense and respond rapidly to fluctuations in TGF-β receptor activity [28, 29]. Moreover, regulated phosphorylation of Smad2/3 linker regions is mediated by a variety of protein kinases, including MAP kinases (i.e., ERK1/2 [30], JNK [31], p38 MAPK [32, 33]), Ca++-calmodulin kinase II [34], casein kinase I-ε [35, 36], and CDKs 2 and 4 [37], and is readily reversed by SCP1/2/3-mediated dephosphorylation of Smad2/3 linker regions [38]. At present, the precise role of linker phosphorylation in regulating the biology and pathology of TGF-β action remains to be fully clarified. Similar ambiguity exists concerning the role of sumoylation in regulating the function of Smad4, whose transcriptional activity can be strengthened or weakened by sumoylation in a promoter-dependent manner [39–43]. Finally, termination of Smad2/3/4 signaling takes place primarily through three distinct mechanisms. First, Smad2/3 are rapidly dephosphorylated and inactivated by the nuclear phosphatase PPM1A [44]. Second, Smad2/3/4 undergo polyubiquitination by the E3 ligases Smurf1, Smurf2, and SCF/Roc1 [45–49], leading to their proteosomal degradation. And third, Smad4 activity is suppressed by Ecto/Tifγ, which monoubiquitinates Smad4 on Lys519 and prevents its association with phosphorylated Smad2 [50]. This inhibitory event is readily reversed by FAM/USP9x, which deubiquitinates Smad4 and restores its responsiveness to TGF-β [50].
2.2. Noncanonical TGF-β signaling
Besides its ability to stimulate the canonical Smad2/3 pathway, TGF-β also alters cell behavior through its activation of Smad2/3-independent signaling systems. Included in this growing list of noncanonical effector molecules stimulated by TGF-β are (i) the MAP kinases ERK1/ERK2, p38 MAPK, and JNK; (ii) the growth and survival kinases PI3K, AKT/PKB, and mTOR; and the small GTP-binding proteins Ras, RhoA, Rac1, and Cdc42 [51–56]. Additionally, activation of the NF-κB signaling system typically is repressed by TGF-β in normal epithelial cells [57]. Recent evidence also implicates TGF-β in mediating the activation of a number of protein tyrosine kinases, including FAK [58, 59], Src [60–63], and Abl [60–62], particularly in mesenchymal or dedifferentiated epithelial cells. Importantly, amplified activation of these noncanonical TGF-β signaling components has been shown to override the normal cellular homeostatic mechanisms governed by Smad2/3 in a manner that figures prominently in the development of human cancers. Precisely how TGF-β couples to the activation of these noncanonical effector systems remains unknown, as does the manner in which their activities becomes dysregulated in response to TGF-β in developing and progressing human cancers. Recent studies linking the inappropriate and/or aberrant activation of alterative TGF-β signaling systems to the acquisition of various cancer hallmarks in developing neoplasms will be presented in the following sections.
2.3. Mutated TGF-β signaling components and cancer
Initial studies aimed at establishing how tumorigenesis negates the cytostatic and tumor suppressing activities of TGF-β were focused primarily on monitoring the expression, mutation, and/or activation status of various TGF-β signaling components. Indeed, considering the fact that TGF-β is a principle player operant in preventing the uncontrolled growth of epithelial cells, and that ~90% of all human malignancies derive from epithelial cell origins [64], it should come as no surprise to learn that components of the TGF-β signaling system are indeed subjected to frequent mutational inactivation. Moreover, many of these genetic defects are heritable and predispose affected individuals to develop cancer [65, 66]. This scenario is especially evident in colon cancers that exhibit microsatellite instability, which produces frameshift mutations in TβR-II and the synthesis of nonfunctional TβR-II proteins that elicit complete cellular insensitivity to TGF-β [67–69]. Clinically, patients with defective TβR-II proteins present with significantly more polyps and preneoplastic lesions than do their TGF-β-responsive counterparts [70]. However, in an interesting twist of fate, patients housing these TβR-II mutants possess a better overall survival rate due to their failure to progress to aggressive, metastatic disease [71]. As will be discussed later, these findings point to an essential role for TGF-β signaling in promoting cancer progression and the acquisition of metastatic phenotypes, particularly in late stage cancers that garner a selective advantage by maintaining their ability to respond to TGF-β. In addition, TβR-II defects also are observed in cancers of the stomach, prostate, breast, lung, liver, and pancreas, and in some hematological malignancies (see [3, 4, 72]).
Likewise, diminished expression and/or activity of TβR-I has been correlated with the development of TGF-β resistance in cancers of the colon, pancreas, ovary, breast, cervix, head and neck, and in T and B cell leukemias [3, 72–75]. As a group, TβR-I mutations tend to partition into two general classes, namely those that target its signal sequence [75–77, 78], and those that target its protein kinase domain [79, 80]. Interestingly, an inactivating Ser387Tyr mutation in TβR-I was observed to be more prevalent in metastatic lesions as compared to their corresponding primary tumors, suggesting that this mutation occurs predominantly in late stage cancers where it may play a role in promoting metastasis. Along these lines, cancers of the breast, ovary, prostate, lung, and pancreas frequently lose expression of TβR-III [81–85], an event that enhances the ability of cancer cells to undergo EMT and, consequently, to acquire invasive and metastatic phenotypes. Thus, while TβR-III-deficiency resulting from a loss-of-heterozygousity at or epigenetic silencing of the TβR-III locus clearly drives progression of tumors from indolent to aggressive states, the molecular mechanisms underlying the ability of TβR-III to normally suppress these tumorigenic processes remains to be fully elucidated.
Given the importance of Smads 2, 3, and 4 in mediating the tumor suppressing activities of TGF-β, it is not surprisingly to learn that these latent transcription factors also manifest a variety of missense mutations during tumorigenesis. Structurally, Smad2/3/4 are comprised of three distinct domains: (i) a globular N-terminal Mad-homology 1 (MH1) domain, which binds TGF-β-regulated promoters at stimulatory Smad-binding elements (SBE; GTCTCAGA, CAGA, or AGAC), or at inhibitory repressive SBE (RSBE; GGCGGG) or TGF-β inhibitory elements (TIE; GNNTTGGtGa) [8, 86-90]; (ii) a central Pro-rich linker region subject to regulated phosphorylation by a variety of protein kinases (see above); and (iii) a globular C-terminal MH2 domain, which mediates transactivation through its ability to interact physically with Smad partners, and with various components of the nuclear pore and transcriptional machinery [4, 6, 8]. Most missense mutations identified to date in Smads 2 and 4 localize to the oligomerization sequences within their MH2 domains [91], and to a lesser extent, to the DNA-binding sequences within their MH1 domains [92]. In either scenario, the ability of TGF-β to mediate cytostasis is compromised severely, which enables malignant cells to progress unabated through the cell cycle even in the continued presence of cytokine. This situation occurs frequently in cancers of the gastrointestinal tract, where Smad4 mutations are observed in 50% of pancreatic cancers [66], in 30% of colorectal cancers [3], and in 25% of patients with juvenile polyposis syndrome [3]. Targeted deletion of Smad4 in mouse keratinocytes [93] or T cells [94], or global Smad4 heterozygousity in combination with APC inactivation [95] further confirmed the essential function of Smad4 in suppressing tumor formation, particularly that in the gastrointestinal track. Although the prevalence of Smad2 mutations is considerably less than that observed for Smad4, mutations in Smad2 are detected in 11% of colorectal cancers, and in 7% of lung cancers [3]. Lastly, despite the fact that Smad3 mutations have thus far remained undetected in human cancers, its expression is lost in 37% of human gastric cancers [96], and in essentially all childhood cases of acute T cell ALL [97]. Furthermore, unlike mice deficient in either Smad2 or Smad4, mice lacking Smad3 are viable and develop colorectal cancers only in response to Helicobacter infection and its accompanying inflammatory reaction [98]. More importantly, Smad3-deficiency protects mice from developing chemically-induced skin carcinomas [99]. Collectively, these findings indicate Smad2 and Smad3 mediate distinct aspects of the biology and pathology of TGF-β; they also suggest that retaining selective features of the TGF-β signaling system functions in promoting oncogenic signaling by TGF-β. The molecular mechanisms that underlie oncogenic signaling by TGF-β, as well as their role in achieving the hallmarks of cancers are discussed below.
3. TGF-β and dysregulated cell proliferation
3.1. Resistance to TGF-β-mediated cytostasis
TGF-β is a principle player involved in suppressing autonomous growth by epithelial, endothelial, and hematopoietic cell lineages, doing so primarily through its ability to induce cell cycle arrest, but also through its stimulation of cell differentiation or apoptosis in a cell- and context-specific manner. Importantly, the ability to preneoplastic cells to acquire resistance to growth arrest governed by TGF-β represents a major hallmark in the development of numerous human cancers. The ability of TGF-β to mediate cytostasis takes place late in the G1 phase of the cell cycle and occurs primarily through the initiation of two synchronized events. First, TGF-β downregulates the expression of the growth-promoting transcription factors, c-Myc and Ids 1–3 [4, 6, 100, 101]. In doing so, TGF-β inhibits Myc transcription by inducing the binding of Smad3 to E2F4/5 and p107 at RSBE sites housed in the Myc promoter [87, 89]. Smad3 also mediates Id1 repression by TGF-β through a “self-enabled” signaling system that first requires Smad3 to induce the expression of ATF3, which subsequently interacts with Smad3 to inhibit Id1 promoter activation [102]. Loss of Id2 expression requires TGF-β to induce the expression of the Myc antagonists, Mad2 and Mad4, which form heterodimers with Max and prevent Id2 transcription [101].
The second major pathway whereby TGF-β mediates cytostasis occurs through its production of the cyclin-dependent protein kinase (CDK) inhibitors, p15INK4b and p21CIP [4, 6, 100]. The ability of p15 to prevent cell cycle progression takes place by its binding to CDK4/6, which occludes their access to and activation by cyclin D. TGF-β induces p15 expression via a bimodal mechanism involving (i) Smad3-mediated repression of Myc, which together with Miz-1 binds and inactivates p15 transcription; and (ii) the formation of Sp1:Smad3:Miz-1 complexes that stimulate p15 transcription [103, 104]. Whereas p15 preferentially inactivates CDK4/6, p21 preferentially targets and antagonizes cyclin E:CDK2 complexes [100]. In doing so, TGF-β stimulates the formation of Sp1:Smad2/3:FoxO complexes that transactivate the p21 promoter [105, 106].
Developing and progressing neoplasms have evolved several mechanisms to override the cytostatic activities of TGF-β. For example, dysregulated Myc expression negates cell cycle arrest induced by TGF-β by (i) promoting the formation of Myc:Miz-1 complexes, which inhibit p15 and p21 transcription induced by Smad3:Miz-1 complexes [103, 104]; and (ii) recruiting the DNA methyltransferase, Dnmt3a, to Myc:Miz-1 complexes, which methylates and inactivates p21 transcription [107]. In addition, human cancers frequently exhibit hyperactivation of the PI3K/AKT pathway [108], which inactivates the cytostatic activities of TGF-β by (i) phosphorylating FoxO, which subsequently is exported from the nucleus and sequestered in the cytoplasm by 14-3-3, and as such, is unavailable to induce p21 expression with Smad3 [105, 109]; and (ii) binding and sequestering inactive Smad3 to prevent its stimulation by active TGF-β receptors [110, 111]. Moreover, in conjunction with activated AKT, FoxG1 interacts physically with Smad3:FoxO complexes to inhibit their induction of p21 in glioblastomas [105]. Whether FoxG1 or other Forkhead family members function similarly to inactive p15 and p21 expression in other human cancers remains to be determined definitively.
Cancer cells also evade the cytostatic activities of TGF-β via cancer cell-specific alternative splicing of C/EBPβ into transcriptionally active LAP or inactive LIP variants [112]. Under normal circumstances, LAP C/EBPβ variants participate in TGF-β-mediated cytostasis by cooperating with Smad3:FoxO complexes to induce p15 expression, and with Smad3:E2F4/5 complexes to repress Myc expression [112]. Cancer cells, particularly those of the breast, elevate their expression of inactive LIP C/EBPβ variants, which uncouples TGF-β from regulation Myc and p15 expression and correlates with metastatic disease development [112]. Along these lines, ELAC2 recently was identified as a transcriptional co-factor that mediates p21 expression in conjunction with Smad2:FAST-1 complexes in prostate epithelial cells [113]. Importantly, loss of ELAC2 in developing and progressing prostate cancers inactivates their ability to undergo growth arrest in response to TGF-β [114]. Finally, overexpression or oncogenic activation of the Ski and SnoN induces cellular transformation in part via their ability to bind Smad2/3/4 complexes and recruit transcriptional inactivation machinery, including N-Cor, mSin3A, and HDAC, which collectively repress gene induction stimulated by TGF-β [115, 116].
3.2. Cell cycle progression induced by TGF-β
In addition to developing resistance to TGF-β-mediated growth arrest, cancer cells routinely acquire the ability to undergo enhanced proliferation when stimulated by TGF-β. The precise mechanisms underlying this phenomenon has yet to be established, but likely reflects a combination of the inactivation of cytostasis mediated by TGF-β coupled to its ability to induce the expression of cytokines and growth factors or their cognate receptors. For instance, TGF-β stimulates the synthesis of IL-1 [117], CTGF [118], bFGF [119], PDGF [120], and TGF-α [121], and of the receptors for PDGF [122] and EGF [123]. Moreover, a common feature of mitogenic signaling systems is their coupling to activation of the Ras/MAP kinase pathways, which results in ERK1/2-mediated phosphorylation of the linker region of Smad2/3 linker and their exclusion from the nucleus, perhaps via Smurf-mediated sequestration of Smad2/3 from the nuclear pore machinery [124]. Hyperactivation of the Ras/MAP kinase pathway also promotes ERK1/2-mediated phosphorylation of TGIF, leading to its stabilization and nuclear localization where it functions as a transcriptional co-repressor by recruiting Smad2:HDAC complexes to the p15 promoter [125]. More recently, activation of RTK and Ras/MAP kinase signaling was shown to stimulate CK1ε/δ phosphorylation of p53, which interacted physically with Smad2/3 to promote the initiation of the cytostatic program by TGF-β. Importantly, restoring p53 function to cancer cells devoid of p53 expression or activity reinstated the ability of TGF-β to induce p21 expression and, consequently, cell cycle arrest [36]. Finally, besides their ability to activate the Ras/MAP kinase pathway, mitogens also activate CDKs when promoting cell cycle progression. In doing so, the cytostatic function of TGF-β can be subverted by CDK2- and CDK4-mediated phosphorylation of the linker region of Smad2, which inactivates its ability to induce the expression of p15 and p21, and to repress the expression of Myc [37].
4. TGF-β and cancer cell motility
4.1. TGF-β and epithelial-mesenchymal transition
The process whereby immotile, polarized epithelial cells to transdifferentiate into highly motile, apolar mesenchymal cells is known as epithelial-mesenchymal transition (EMT), was described initially as a phenomenon that took place during chick embryonic development nearly 100 years ago by Frank Lillie [126], and which was first documented as a discrete physiological event in 1982 by Greenburg and Hay [127]. Today, EMT is recognized as a normal and essential physiological process operant in mediating embryonic development, wound healing, and tissue morphogenesis, remodeling, and repair. EMT itself compromises several distinct features, including (i) the loss of cell polarity due to downregulated expression of epithelial cell markers (e.g., E-cadherin, zona occluden-1, and β4 integrin); (ii) cytoskeletal architecture reorganization and intracellular organelle redistribution; (iii) upregulated expression of fibroblast markers (e.g., vimentin, N-cadherin, α-smooth muscle actin); and (iv) elevated cell invasion and migration [126, 128–131]. Besides its role in mediating normal tissue morphogenesis and repair, inappropriate initiation of EMT also underlies the development of several human pathologies, such as chronic inflammation, rheumatoid arthritis, and chronic fibrotic degenerative disorders of the lung, liver, and kidney [126, 128–132]. In addition, the acquisition of metastatic phenotypes by dedifferentiated tumors is critically dependent on EMT and its ability to actively remodel tumor microenvironments in a manner that promotes the evolution and selection of metastatic cells [126, 128–131].
The essential role of TGF-β in mediating EMT was first described by Miettinen et al [133]who showed that TGF-β stimulation of normal mammary epithelial cells readily induced their transdifferentiation into fibroblastoid-like cells. Since that time, TGF-β has been identified as a master regulator both of physiological and pathophysiological EMT in a variety of epithelial cell types and tissues. For instance, TGF-β3-deficiency in mice elicits defective palatogenesis and cleft palate formation due to faulty initiation of EMT [134]. Along these lines, neutralization of TGF-β2 function impairs chick heart endocardial cushion development by inhibiting Slug expression and, consequently, its induction of EMT [135]. Finally, targeted-deletion of Smad3 in mice prevents their development of EMT-driven retinal [136, 137] and renal [138] fibrosis. Aberrant EMT also is an essential facet of the oncogenic activities of TGF-β, particularly its ability to stimulate the progression and metastasis of late stage cancers. The molecular mechanisms underlying the ability of TGF-β to induce cancer cell EMT and metastasis are described below.
4.2. TGF-β, invasion, and metastasis
The activities of fibroblasts and their associated stromal components play important roles in determining whether TGF-β either suppresses or promotes tumor formation [139, 140]. Indeed, the ability of TGF-β to inhibit tumor formation occurs not only through its actions in epithelial cells, but also through its stimulation of adjacent fibroblasts, which synthesize and secrete a variety of cytokines, growth factors, and extracellular matrix (ECM) proteins that mediate tissue homeostasis and suppress cancer development. Importantly, genetic or epigenetic events that inactivate paracrine TGF-β signaling between adjacent epithelial and stromal compartments can lead to the formation of neoplastic cells, as well as to their selection and expansion by promoting their growth, survival, and motility [141, 142]. For instance, conditional inactivation of TβR-II in mouse fibroblasts elicits the formation of prostate intraepithelial neoplasias and invasive carcinoma of the forestomach [143]. Similar targeted inactivation of TβR-II in mouse mammary fibroblasts inhibits mammary gland ductal morphogenesis, as well as the formation of terminal end buds [144]. Importantly, grafting mammary carcinoma cells with TβR-II-deficient fibroblasts, not their normal counterparts, significantly enhanced the growth and invasion of breast cancer cells in subrenal capsules [144]. Mechanistically, inactivation of TGF-β signaling in fibroblasts promotes tumorigenesis by upregulating the expression and signaling of TGF-α, MSP, and HGF within cell microenvironments [139, 140, 143, 144]. Furthermore, inactivating TGF-β signaling in fibroblasts also promotes breast cancer metastasis by activating two chemokine receptor axes, namely SDF-1:CXCR4 and CXCL5:CXCR2, which recruit immature GR1+CD11b+ myeloid cells that (i) suppress host tumor immunosurveillance, and (ii) induce MMP expression within tumor microenvironments that promotes the dissemination of breast cancer cells [145].
In addition to its metastatic-promoting activities initiated in fibroblasts and tumor microenvironments, TGF-β also induces EMT and metastasis by directly effecting the activities and behaviors of developing and progressing malignant cells. For instance, TGF-β stimulates human MDA-MB-231 breast cancer cells to metastasize specifically to bone by inducing their expression of PTHrP, IL-11, and CTGF [146–148]. Interestingly, although TGF-β1 had no effect on the growth of mammary tumors induced by transgenic polyomavirus middle T antigen expression, conditional TGF-β1 expression significantly enhanced the ability of these same mammary tumors to metastasize to the lungs [149]. In addition, TGF-β stimulation of EMT in breast cancer cells has been linked to the selection and expansion of cancer initiating/stem cells [150–153], which may underlie breast cancer resistance to neoadjuvant chemotherapies, as well as disease progression and recurrence. Thus, following inactivation of its cytostatic function, TGF-β preferentially promotes the acquisition of metastatic phenotypes in previously nonmetastatic malignant cells.
Accumulating evidence indicates that TGF-β stimulates cancer cell EMT and metastasis through a combination of Smad2/3-dependent and -independent signaling systems. Indeed, engineering metastatic human MCF10ACA1a breast cancer cells to express a dominant-negative Smad3 [154] or a TβR-I mutant incapable of activating Smad2/3 (i.e., L45 mutant) [154] both significantly reduced the ability of MCF10ACA1a cells to colonize the lung. Along these lines, Smad4-deficiency elicited by RNA interference inhibited the ability of MDA-MB-231 xenografts to metastasize to bone in part via diminished expression of IL-11 and CTGF in response to TGF-β [146, 155]. As above, the extent of tumor formation and their rate of growth was unaffected by altering the response of these breast cancer cells to TGF-β [146, 155], which again points to the importance of maintaining the fidelity of the TGF-β signaling system during metastasis development. Accordingly, whereas Smad4-deficiency conspires with oncogenic K-Ras to promote pancreatic cancer initiation and development, maintaining intact Smad4 signaling is necessary to induce EMT and TGF-β-dependent growth of advanced pancreatic ductal adenocarcinomas [156]. In addition, overexpression of Smad7, which inhibits TGF-β stimulation of Smad2/3 [18–20], prevents the invasion of breast [157] and head and neck cancers [158, 159], as well as impairs the ability of melanoma cells to metastasize to bone [160]. Recently, TGF-β was shown to cooperate with Ras to promote the formation of mutant p53/p63 complexes that are bridged by Smad2/3, and that result the inactivation of p63 and its ability to downregulate the expression of metastasis suppressor genes, Sharp-1 and cyclin G2 [161]. Along these lines, Smad2 has been observed to promote EMT in mammary epithelial cells by stimulating the DNA binding activity of the DNA methyltransferase, DNMT1, leading to the chronic epigenetic silencing of epithelial-associated genes, including CDH1, CGN, CLDN4, and KLK10 [162]. Collectively, these findings demonstrate the importance of Smad2/3/4 signaling in mediating metastasis stimulated by TGF-β; they also suggest that the development and implementation of novel Smad2/3 antagonists may improve the clinical course of metastatic cancer patients whose tumors house functional Smad2/3 signaling systems.
Equally important to the ability of TGF-β to stimulate cancer cell EMT and metastasis is its coupling to activation of noncanonical signaling systems (Fig. 2; [7]). Indeed, activation of Ras/MAP kinase [51, 163–167], PI3K/AKT [52], Rho/ROCK [53]; NF-κB [168], Jagged/Notch [169], Wnt/β-catenin [170], and MDM2/p53 [171] pathways by TGF-β all play essential roles in mediating its induction of cancer cell EMT, invasion, and metastasis. In particular, Ras/MAP kinase signaling cooperates synergistically with TGF-β to elicit EMT and metastasis during the progression of skin cancers from squamous to spindle cell phenotypes [172–174]. In addition, TGF-β stimulation of NF-κB promotes EMT in and lung colonization by breast cancer cells that house oncogenic Ras [168]. Furthermore, activation of the Ras effector, c-Raf-1, induces autocrine TGF-β expression and activation that ultimately results in its stimulation of EMT and invasion [175]. Finally, TGF-β-induces elevated MDM2 expression that leads to the destabilization of p53, a key component of EMT [171].
Integrins also have been implicated in regulating oncogenic signaling by TGF-β, particularly its ability to induce EMT, invasion, and metastasis. For instance, administering neutralizing antibodies against β1 integrin prevented TGF-β stimulation of p38 MAPK and EMT in mammary epithelial cells [163]. In addition, TGF-β stimulates the binding of the adapter protein Dab2 [17] to β1 integrin, leading to FAK activation and the induction of EMT in mammary epithelial cells [176]. Similarly, work in our laboratory led to the discovery of a novel αvβ3 integrin:Src:phospho-Y284-TβR-II:Grb2:p38 MAPK signaling axis whose activation mediates oncogenic signaling – i.e., EMT, invasion, and metastasis – by TGF-β in normal and malignant mammary epithelial cells [60–62]. Importantly, the ability of TGF-β to stimulate the growth and pulmonary metastasis of breast cancers in mice absolutely requires activation of this oncogenic signaling complex [62]. In addition to its role in mediating pulmonary metastasis of breast cancers, αvβ3 integrin also mediates breast cancer metastasis to bone [177, 178] in a TGF-β-dependent manner. Lastly, new insights into the role of TGF-β in regulating tight junction dissolution during EMT recently was elucidated by Wrana and colleagues [179] who observed that the tight-junction assembly protein, PAR-6, interacts physically with TβR-I. Activation of TGF-β receptors by ligand results in TβR-II-mediated phosphorylation of PAR-6, which binds Smurf1 and coordinates its ubiquitination and degradation of RhoA. The net effect of these TGF- β-dependent events results in the dissolution of epithelial cell tight junctions and the disassembly of their actin cytoskeleton, leading to the induction of EMT [179]. Future studies need to more thoroughly examine the role of additional phosphorylation events in potentially regulating PAR-6 function, as well as the role of integrins and other adhesion-regulated signaling systems to collaborate with TGF-β in mediating EMT and tight junction dissolution. Similar analyses aimed at determining the importance of these events in regulating metastasis development stimulated by TGF-β also is warranted.
5. TGF-β and tumor angiogenesis
Angiogenesis is a normal physiological process whereby new blood vessels develop from preexisting vessels, and which is an essential component of embryonic development, wound healing, and the female reproductive cycle [180, 181]. Pathological activation of angiogenesis also figures prominently in mediating the development of a number of human diseases, including rheumatoid arthritis, diabetic retinopathy, and age-related macular degeneration [181, 182]. Indeed, inappropriate initiation of angiogenesis perhaps is best known and characterized for its role in mediating the development and progression of tumors, whose growth is limited by the extent of nutrient diffusion into tumor microenvironments. Tumor angiogenesis circumvents this limitation by providing developing neoplasias with an efficient supply of nutrients, and ultimately, with a route for their metastatic spread. Angiogenesis is comprised of two distinct and sequential phases referred to as angiogenesis activation and angiogenesis resolution [180–182]. Angiogenesis activation encompasses the initiation and development of new blood vessels, and is characterized by (i) increased endothelial cell (EC) permeability, proliferation, migration, and invasion, and (ii) reduced EC adhesion and basement membrane integrity. In contrast, angiogenesis resolution encompasses the maturation of newly formed vessels and entails (i) increased EC adhesion and basement membrane deposition, (ii) decreased EC proliferation and motility, and (iii) the recruitment of perivascular cells, namely pericytes, vascular smooth muscle cells, or mural cells, that function in regulating vessel stability and hemodynamics. Importantly, normal endothelium in adult tissues exists in a quiescent, inactive state, a fact that underlies the continued enthusiasm to selectively target tumor vascular when treating human malignancies.
Consistent with its multifunctional nature, TGF-β has been reported to regulate the activation and resolution phases of angiogenesis [183–186]. In addition, TGF-β also affects ECM production and remodeling in EC microenvironments, and as such, impacts the interactions and communications between ECs and their supporting mesenchymal cells [187]. The ability of TGF- β to regulate angiogenesis was discovered by analyzing the vasculature of TGF-β1-deficient mice, which exhibit severe defects in hematopoiesis and EC differentiation that results in the production of weak, aberrantly formed capillaries [188]. Additional studies demonstrating a crucial role of TGF-β signaling in regulating angiogenesis were gleaned from analyses of mice lacking TβR-I [189], TβRII [190, 191], TβR-III [192, 193], ALK1 [194, 195], endoglin [196], Smad1 [197], or Smad5 [198], all of which exhibited vascular and endothelial defects. Clinically, the development of hereditary hemorrhagic telangiectasia type 1 (HTT1) results from the loss or inactivation of the gene for endoglin [199, 200], while the loss or inactivation of the gene for ALK1 elicits HHT2 [201, 202]. Moreover, HHT1 and HHT2 both are phenocopied in knockout mice lacking either endoglin [196, 203] or ALK1 [204], respectively. Thus, aberrant TGF-β signaling clearly underlies the development of multiple vascular disorders [205].
Genetic inactivation of ALK1 [195] and ALK5 [189] elicits embryonic lethality at E11.5 and E10.5, respectively, demonstrating the indispensable function of these genes in mediating normal embryonic angiogenesis and vasculogenesis. Quite surprisingly, recent studies have established that TGF-β differentially activates TβR-I/ALK5 versus ALK1, which elicits dramatically different angiogenic responses by ECs. For instance, TGF-β stimulation of TβR-I/ALK5 activates Smad2/3 and their transcription of the ECM proteins, plasminogen activator inhibitor type 1 (PAI-1) and fibronectin, which mediate angiostasis and vessel maturation [194, 206, 207]. In addition, Miyazono and colleagues [208] showed that TβR-I/ALK5 mediates TGF-β stimulation of EC gene expression profiles characteristic of angiostasis and vessel maturation, and of periendothelial cell differentiation. Thus, activation of TβR-I/ALK5 by TGF-β functions preferentially in regulating angiogenesis resolution [185, 208]. In contrast, activation of ALK1 by TGF-β stimulates Smad1/5/8 transcriptional activity and the expression of angiogenic genes, such as Id1 and interleukin 1 receptor-like 1 [194, 206–208]. Thus, activation of ALK1 by TGF-β functions preferentially in regulating angiogenesis activation. Interestingly, the decision by TGF-β to couple to either ALK1 or ALK5 likely depends on the balance between TGF-β and angiogenic cytokines within tumor microenvironments. Indeed, low TGF-β concentrations enhance the ability of bFGF and VEGF to stimulate EC proliferation and angiogenic sprouting, while high TGF-β concentrations inhibit these angiogenic activities [185, 186].
In addition to regulating angiogenesis via its activation of ALK1 and ALK5, TGF-β also controls vessel development via its actions on the co-receptors, TβR-III/betaglycan and endoglin. Several recent studies have shown the importance of epithelial cell expression of TβR-III in suppressing the growth and metastasis of breast [81], lung [82], pancreatic [83], ovarian [84], and prostate [85]cancers. However, the ability of soluble TβR-III to bind and sequester TGF-β has been used successfully to antagonize tumor angiogenesis and, consequently, to inhibit the growth and progression of human tumors produced in mice [209–211]. In contrast, endoglin is expressed predominantly on proliferating ECs, and its expression also can be upregulated by ALK1 [208]. When expressed in human umbilical vein endothelial cells, endoglin inhibits ALK5 signaling and promotes EC proliferation, migration, and tubulogenesis in conjunction with stimulation of ALK1 [212, 213]. Moreover, the redundant clinical symptoms of HHT1 (i.e., endoglin defects) and HHT2 (i.e., ALK1 defects) in humans, together with the overlapping phenotypes observed between ALK1- and endoglin-deficient mice, suggest that ALK1 and endoglin both function as negative regulators of TGF-β/ALK5 signaling.
Collectively, these studies highlight the complexities associated with the ability of TGF-β to regulate EC activities coupled to angiogenesis. Future studies clearly need to (i) better define the precise mechanisms that enable TGF-β and its downstream effectors to govern the induction of angiogenic or angiostatic gene expression profiles; (ii) establish the impact of EC and perivascular cell differentiation states to influence the angiogenic response to TGF-β; and (iii) identify the microenvironmental cues and signals the cooperate with TGF-β in mediating angiogenesis activation and resolution.
6. TGF-β and cancer cell survival
Although cancer typically is viewed as a disease that arises from uncontrolled cell proliferation, it also is a disease of dysregulated apoptosis that enhances cancer cell survival by conferring developing neoplasms resistance to stimuli that would normally induce their programmed cell death [2]. Under normal physiological conditions, TGF-β inhibits autonomous cell growth by inducing cell cycle arrest or by stimulating cellular differentiation [3, 4, 214]; however, TGF-β also guards against the development of dysregulated cell growth through its ability to promote programmed cell death, particularly in lymphocytes and hepatocytes [3, 4, 214, 215]. Importantly, TGF-β stimulation of apoptosis occurs independently of its regulation of EMT and cell motility [216, 217], and is mediated through its regulation of a variety of proapoptotic proteins (Table 1).
Table 1.
Expression of Proapoptotic Genes Induced by TGF-β
| Gene | Mechanism of Action | Reference |
|---|---|---|
| SHIP | SH2-containing lipid phosphatase that inhibits cell survival mediated by PI3K | [268] |
| DAPK | Ca++/calmodulin-dependent protein kinase that regulates mitochondrial-, caspase-, and autophagic cell death | [269, 270] |
| TIEG1 | Zinc-finger transcription factor that represses Bcl-2 expression | [271, 272] |
| Daxx | TβR-II-adaptor protein that stimulates JNK | [54] |
| ARTS | Mitochondrial septin-like protein that inactivates XIAP to elicit caspase activation | [273, 274] |
| TAK1 | MAP kinase kinase kinase that stimulates JNK and p38 MAPK | [275–278] |
One of the hallmarks of cancer is the ability of malignant cells to acquire resistance to apoptotic stimuli [2]. As mentioned previously, tumorigenesis frequently subverts the tumor suppressing activity of TGF-β, thereby enabling TGF-β to promote oncogenesis by stimulating the growth, invasion, metastasis, and angiogenesis of developing tumors (see above). Along these lines, alterations within the TGF-β signaling system also conspire to convert TGF-β from activator of apoptosis to a promoter of cancer cell survival. This altered cellular response to TGF-β is especially evident through its ability to protect and promote the recovery of cancer cells following their targeting by radiation and chemotherapy treatments [218, 219]. Thus, the continued reliance on radiation and chemotherapy regimens to treat cancer patients necessitates that science and medicine fully elucidate the molecular mechanisms that affords TGF-β to protect developing neoplasms from apoptotic stimuli.
Numerous studies have established that dysregulated activation of the NF-κB and PI3K/AKT pathways by TGF-β functions in promoting cancer cell survival during tumorigenesis [220–223]. Interestingly, TGF-β routinely suppresses the activation of NF-κB in normal epithelial cells by inducing their expression of IκBα, a phenomenon that is inactivated during oncogenic progression [157, 215]. Quite intriguingly, TGF-β stimulation of late stage cancer of the breast, prostate, and liver results in NF-κB activation, leading to the induction of pro-survival and -tumorigenic gene expression profiles [220, 224–226]. The aberrant activation of NF-κB by TGF-β has been attributed its stimulation of TAK1, which induces IKK activation [220, 224, 225]. Indeed, recent work in our laboratory defined a novel TAB1:TAK1:IKKβ:NF-κB signaling axis that forms aberrantly in breast cancer cells, and in normal mammary epithelial cells following their induction of EMT by TGF-β [225]. Once formed, this signaling axis enables oncogenic signaling by TGF-β in part via activation of NF-κB and its consequential production of proinflammatory cytokines, and of Cox-2 [227], which engages a PGE2:EP2 signaling axis coupled to breast cancer development and progression [228]. In addition, TAK1-deficiency abrogated the ability of TGF-β to induce the invasion of breast cancer cells, as well as restored their ability to undergo cytostasis in response to TGF-β. More importantly, expression of a dominant-negative TAB1 mutant dramatically reduced the growth of mammary tumors in normal mice, as well as in their immunocompromised counterparts, suggesting a potentially important role of NF-κB in regulating innate immunity by TGF-β [225]. Recently, TGF-β was shown to induce xIAP expression in hepatic and endometrial carcinomas, which enhanced their survival and invasiveness via xIAP-mediated regulation of PK3K/AKT, TAK1, PKC, JNK, and MMP-9 [229, 230]. Along these lines, work in our laboratory has identified xIAP as an essential mediator underlying the formation of TAB1:TAK1:IKKβ complexes and, consequently, their activation of NF-κB solely in cancer cells [231]. Thus, pharmacological targeting of xIAP may represent a novel approach to antagonize the oncogenic activities of TGF-β in developing and progressing human malignancies.
Altered coupling of TGF-β that results in its stimulation of AKT [232] and p38 MAPK [233] may also serve in potentiating the activation of IKKβ and NF-κB. Furthermore, activated IKK [234] complexes and AKT [223] both directly phosphorylate FoxO, which subsequently is inactivated and sequestered in the cytoplasm by its binding to 14-3-3 proteins. In doing so, phosphorylated FoxO transcription factors are no longer available to participate in Smad2/3-mediated expression of apoptotic and cytostatic genes, thereby enhancing cancer cell survival. Finally, activated AKT has been shown to bind and sequester inactive, cytoplasmic Smad3, which prevents TGF-β stimulation of programmed cell death [110, 111].
In addition to its anti-apoptotic function, NF-κB also plays an important role in mediating TGF-β stimulation of EMT and metastasis [168], and of pro-inflammatory gene expression [226]. For instance, inhibiting NF-κB activity in Ras-transformed mammary epithelial cells blocked the ability of TGF-β to (i) induce MMP-13 and MCP1 expression, and (ii) repress E-cadherin expression. Moreover, TGF-β stimulation of EMT is sufficient in eliciting its ability to stimulate, not inhibit, NF-κB activity in mammary epithelial cells [225]. When stimulated with TGF-β, cancer cells synthesize and secrete an array of pro-inflammatory genes following NF-κB activation. Included in this list of proinflammatory mediators regulated by TGF-β and its activation of NF-κB are Cox-2, GM-CSF, TNF-α, and interleukins 1, 6, and 8 [225, 226, 235, 236], which collectively drive oncogenesis by stimulating tumor angiogenesis and metastasis, as well as by inhibiting host immunosurveillance. Recently, aberrant expression of Cox-2 was shown to negate the cytostatic activities of TGF-β [237], and to promote its ability to induce breast cancer metastasis to bone [235]. Thus, pharmacological targeting of Cox-2 may enhance the tumor suppressing activities of TGF-β. Finally, TGF-β also enhances cancer cell survival through its ability to modulate and suppress immunosurveillance mediated by the adaptive immune system [238, 239]. The role of TGF-β in regulating immune system function during tumorigenesis is discussed below.
7. TGF-β and dysregulated immunosurveillance
It has been appreciated from many years now that cancer initiation, promotion, and progression all are linked to aberrant and/or persistent inflammation within tumor microenvironments [240]. Indeed, the balance between host immunosurveillance and proinflammatory activity within the tumor milieu plays an essential role in determining whether tumor development and progression is induced or inhibited [240]. An implicit factor in regulating this delicate balancing act is TGF-β, which governs the activities and behaviors of cancer cells and their associated stromal components [239]. Indeed, while many cytokines and chemokines participate in the inflammatory process, the prominent and essential role of TGF-β in maintaining proper immune system function and homeostasis is underscored by the finding that TGF-β1-deficient mice exhibit lethal multifocal inflammatory disease [241, 242], while mice lacking Smad3 exhibit defects in the responsiveness and chemotaxis of their neutrophils, and their T and B cells [243]. In addition, a characteristic feature of cancer cells is their capacity to increase the production and secretion of TGF-β into tumor microenvironments, and into the general circulation of cancer patients [244–246]. Elevated concentrations of active TGF-β also are detected within the tumor milieu due to enhanced ECM degradation mediated by resident and recruited leukocytes –i.e., monocytes/macrophages, dendritic cells, granulocytes, mast cells, T cells, and natural killer (NK) cells – that either promote or suppress tumor development in a context-specific manner [240]. It is interesting to note that the recruitment of leukocytes to developing neoplasms in many respects reflects the processes underlying their normal recruitment to regions of wounding, inflammation, or infection. This observation, coupled with the fact that cancer cells house genetic or epigenetic alterations that elicit persistent tumor inflammation and chronic leukocyte infiltration undoubtedly led to Dvorak to liken “tumors to wounds that never heal” [247].
In general, high levels of TGF-β inactivate host anti-tumor immunosurveillance systems, which confers immune privilege to developing neoplasms and ensures for their continued progression. Mechanistically, TGF-β inhibits the proliferation and differentiation of NK and T cells, as well as their production of cytotoxic effector molecules. Moreover, the ability of TGF-β to affect the behaviors of cytotoxic CD8+ T cells occurs in a manner that reflects their differentiation status. For instance, TGF-β is a potent inhibitor of naïve CD8+ T cell populations, but elicits little to no response in their fully differentiated and activated counterparts, which downregulate expression of TβR-II. The insensitivity of differentiated CD8+ T cells to TGF-β can be overcome by the production of either IL-1o or IL-2, or by the expression of the co-stimulatory molecule CD28 [238, 239, 248]. Interestingly, TGF-β stimulation of T cells expressing CD28 ultimately promotes the survival of memory/effector phenotypes in thymic and peripheral T cell populations.
The lymphocyte defects observed in Smad3-deficient mice suggest an important role for this latent transcription factor in mediating immunosuppression by TGF-β. Accordingly, activation of Smad3 by TGF-β prevents mitogenesis in CD8+ T cells by (i) inhibiting their production of IL-2; (ii) repressing their expression of c-Myc, cyclin D2, and cyclin E; and (iii) stimulating the expression of the CDKIs p15, p21, and p27 [239, 248, 249]. In addition, TGF-β also represses the expression and production of cytotoxic effector molecules, including IFN-γ, lymphotoxin-α (LT- α), perforin/granzyme, and Fas ligand [239, 248, 249]. Although TGF-β has no effect on proliferation of CD4+ T cells, it does inhibit CD4+ T cell differentiation into T helper 1(Th1) and Th2 cell lineages, which readily secrete IFN-γ and LT-α (Th1) or IL-4, IL-5, and IL-13 (Th2), respectively. The inhibitory activities of TGF-β in CD4+ T cells result from its ability to downregulate T cell receptor expression, to diminish intracellular Ca++ signaling, and to reduce the expression and activation of transcription factors [239, 248, 249], which collectively serve to alleviate host immunosurveillance. Accordingly, inactivation of TGF-β in CD8+ or CD4+ T cells results in T cell-mediated eradication of skin [250] and prostate [251] in mice. The ability of TGF-β to suppress the activities of tumor-infiltrating T cells also has been associated with its activation of Tregs, which represent a subset of CD4+ and CD8+ peripheral T cells that also express CD25 and Fox3P, and which routinely localize to tumor microenvironments where they promote immune privilege by inhibiting tumor targeting T and NK cells [248].
In addition to regulating the adaptive immune system, TGF-β also figures prominently in governing the behavior and activation of the innate immune system. Indeed, TGF-β is a potent inhibitor of NK cell cytolytic activity, presumably by attenuating NKp30 and NKG2D receptor activation. Neutralizing TGF-β not only prevents NKG2D downregulation, but also restores NK cell anti-tumor reactivity [252]. In addition, NK cell production of IFN-γ is repressed by TGF-β through cellular mechanisms that remain incompletely understood. Dendritic cells are professional antigen presenting cells whose expression of MHC class II, of the co-stimulatory molecules CD40, CD80, and CD86, and of cytokines TNF-α, IL-12, and CCL5/Rantes is inactivated following their stimulation with TGF-β [239, 248, 249, 253]. In addition, the increased production of TGF-β within tumor microenvironments serves as a chemoattractant for mast cells that enhance tumor progression through their synthesis and release of a variety of factors, including histamine, proteases, and cytokines (e.g., VEGF and TGF-β) [249, 254]. Lastly, TGF-β stimulation of resting monocytes (i.e., non-phagocytic) promotes their chemotaxis to and infiltration into tumor microenvironments where they (i) enhance oncogenesis by stimulating ECM degradation, and by inducing tumor angiogenesis, invasion, and metastasis, and (ii) contribute to immunosuppression through upregulated production and release of TGF-β into tumor microenvironments [255, 256]. Furthermore, activation of differentiated macrophages by TGF-β inhibits their expression of the scavenger receptors, CD36 and SR-A, of the opsonizing IgG receptors, FcγRI and FcγRIII, and of the antigen presenting receptor, MHC class II, which collectively function to enhance the progression and survival of developing neoplasms. In addition to macrophages, TGF-β also induces the infiltration of tumor-associated neutrophils (TAN) that bear a tumor promoting phenotype, and as such, antagonizing TGF-β function facilitates the recruitment of TANs that possess an anti-tumor phenotype [257].
8. TGF-β and cancer cell immortality
An important and widely recognized hallmark of cancer lies in the ability of malignant cells to become immortal, a process achieved through the reactivation of human telomerase reverse transcriptase (hTERT) and its consequential lengthening of chromosomal telomeres. Structurally, telomeres are complex nucleoproteins comprised of single- and double-stranded DNA and associated proteins that function in safeguarding against chromosomal end-to-end fusions. With the exception of germ cells, hTERT expression is silenced and undetectable in normal somatic cells. Thus, in the absence of hTERT and its ability to maintain telomere integrity, each successive cell division results in telomere erosion and, ultimately, in cell senescence, crisis, and apoptosis [258]. Collectively, these events conspire to suppress dysregulated cell growth and survival, and as such, to prevent tumor formation.
In contrast to their normal counterparts, cancer cells readily overcome the negative constraints that repress hTERT expression, resulting in the reinitiation and activation of telomerase activity and the acquisition of cellular immortalization. Human telomerase consists of a telomerase RNA template (hTER) and hTERT, both of which function in coordinating the synthesis of telomere repeats that mediate chromosomal integrity and prolong cellular replication [258, 259]. Given the importance of dysregulated hTERT expression and activity to promoting cancer development, it is fitting that TGF-β may in fact be the principle cytokine operant in silencing hTERT expression in normal cells [260]. Indeed, the extent of TGF-β signaling in responsive cells displays an inverse relationship with the levels of hTERT expression observed in cancers of the colon and breast [260–263]. Moreover, elevating hTERT expression in epithelial cells elicits resistance to the cytostatic activities of TGF-β [264].
Current evidence indicates that TGF-β represses hTERT expression via several distinct mechanisms. For instance, cellular depletion of SIP1/ZEB2, a zinc-finger transcription factor more commonly associated with induction of EMT, significantly attenuates the ability of TGF-β to repress hTERT expression in human osteosarcoma cells [265]. Moreover as mentioned above, TAK1 plays a major role in promoting the activation of NF-κB by TGF-β in breast cancer cells, thereby enhancing their tumorigenicity in mice [225]. Quite surprisingly, activation of TAK1 by TGF-β also has been linked to the repression of hTERT expression in human lung cancer cells. In doing so, TAK1 suppresses Sp1 transcriptional activity via recruitment of HDAC to the hTERT promoter [266]. Future studies clearly are warranted to delineate how TAK1 participates in mediating tumor suppression (i.e., repressing hTERT expression) and tumor promotion (i.e., NF-κB activation and proinflammatory gene expression) in response to TGF-β. Along these lines, upregulated expression of the negative regulator of TGF-β signaling, Smurf2, contradicts the immortalization activities of hTERT in a manner independent of the ability of Smurf2 to ubiquitinate target proteins and inhibit TGF-β signaling [267]. Thus, the interplay between Smurf2, hTERT, and TGF-β awaits further clarification. Finally, two recent studies have shown that TGF-β represses hTERT transcription by inducing the assembly of Myc:Smad3 complexes on E-box and SBE sites in the hTERT promoter [262]. Thus, dysregulated Myc expression or aberrant Smad3 signaling can promote upregulated telomerase expression and activity.
9. Concluding remarks
Defining the molecular mechanisms that underlie the initiation of the TGF-β paradox in human malignancies remains the most important and unanswered question concerning the biology and pathology of this multifunctional cytokine. In their seminal paper, Hanahan and Weinberg [2] proposed that all neoplasms must evolve 6 physiological traits during the course of their malignant progression. Importantly, the acquisition of each trait – i.e., autonomous cell growth, resistance to cytostatic, apoptotic, and morality signals, and the induction of angiogenesis and invasion/metastasis – represents an essential rate-limiting step operant in mediating cancer development and progression. The studies reviewed herein show that TGF-β plays a major role, both directly and indirectly, in regulating the acquisition by cancer cells of each of these cancer hallmarks, and as such, the development of novel TGF-β chemotherapeutics capable of targeting these cancer hallmarks offers new inroads into alleviating the devastating effects of TGF-β in promoting metastatic disease development in humans.
Acknowledgments
We thank members of the Schiemann Laboratory for critical comments and reading of the manuscript. W.P.S. was supported by grants from the National Institutes of Health (CA129359), the Department of Defense (BC084651), the Komen Foundation (BCTR0706967), and the Case Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. CA Cancer J Clin. 2007;57(1):43. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Cell. 2000;100(1):57. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Blobe GC, Schiemann WP, Lodish HF. N Engl J Med. 2000;342(18):1350. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 4.Galliher AJ, Neil JR, Schiemann WP. Future Oncology. 2006;2(6):743. doi: 10.2217/14796694.2.6.743. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Massague J. Cell. 2003;113(6):685. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 6.Massague J, Gomis RR. FEBS Lett. 2006;580(12):2811. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Moustakas A, Heldin CH. J Cell Sci. 2005;118(Pt 16):3573. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 8.Feng XH, Derynck R. Annu Rev Cell Dev Biol. 2005;21:659. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Casillas F, Wrana JL, Massague J. Cell. 1993;73(7):1435. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 10.Sankar S, Mahooti-Brooks N, Centrella M, McCarthy TL, Madri JA. Journal of Biological Chemistry. 1995;270(22):13567. doi: 10.1074/jbc.270.22.13567. [DOI] [PubMed] [Google Scholar]
- 11.Rotzer D, Roth M, Lutz M, Lindemann D, Sebald W, Knaus P. Embo J. 2001;20(3):480. doi: 10.1093/emboj/20.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massague J. Annu Rev Biochem. 1998;67:753. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 13.Chang H, Brown CW, Matzuk MM. Endocr Rev. 2002;23(6):787. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 14.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Nature. 1994;370(6488):341. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 15.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. Cell. 1998;95(6):779. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 16.Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K. Mol Cell Biol. 2000;20(24):9346. doi: 10.1128/mcb.20.24.9346-9355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hocevar BA, Smine A, Xu XX, Howe PH. Embo J. 2001;20(11):2789. doi: 10.1093/emboj/20.11.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MAJ, Wrana JL, Falb D. Cell. 1997;89:1165. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 19.Nakao A, Afrakht M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Nature. 1997;389:631. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 20.Souchelnytskyi S, Nakayama T, Nakao A, Moren A, Heldin CH, Christian JL, ten Dijke P. J Biol Chem. 1998;273:25364. doi: 10.1074/jbc.273.39.25364. [DOI] [PubMed] [Google Scholar]
- 21.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. J Biol Chem. 2001;276(16):12477. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 22.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Mol Cell. 2000;6(6):1365. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 23.Datta PK, Moses HL. Mol Cell Biol. 2000;20(9):3157. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibarrola N, Kratchmarova I, Nakajima D, Schiemann WP, Moustakas A, Pandey A, Mann M. BMC Cell Biol. 2004;5:2. doi: 10.1186/1471-2121-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, Miyazono K. Embo J. 2003;22(24):6458. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu FY, Li XZ, Peng YM, Liu H, Liu YH. Am J Nephrol. 2007;27(2):176. doi: 10.1159/000100518. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, Ye Z, Zhang S, Chan SC, Chen YG, Han J, Lin SC. Embo J. 2006;25(8):1646. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inman GJ, Nicolas FJ, Hill CS. Mol Cell. 2002;10(2):283. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 29.Xu L, Massague J. Nat Rev Mol Cell Biol. 2004;5(3):209. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- 30.Kretzschmar M, Doody J, Timokhina I, Massaguâe J. Genes & development. 1999;13(7):804. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel ME, McDonnell MA, Law BK, Moses HL. J Biol Chem. 1999;274(52):37413. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa F, Matsuzaki K, Mori S, Tahashi Y, Yoshida K, Sugano Y, Yamagata H, Matsushita M, Seki T, Inagaki Y, Nishizawa M, Fujisawa J, Inoue K. Hepatology. 2003;38(4):879. doi: 10.1053/jhep.2003.50384. [DOI] [PubMed] [Google Scholar]
- 33.Kamaraju AK, Roberts AB. J Biol Chem. 2005;280(2):1024. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- 34.Wicks SJ, Lui S, Abdel-Wahab N, Mason RM, Chantry A. Mol Cell Biol. 2000;20(21):8103. doi: 10.1128/mcb.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waddell DS, Liberati NT, Guo X, Frederick JP, Wang XF. J Biol Chem. 2004;279(28):29236. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- 36.Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. Science. 2007;315(5813):840. doi: 10.1126/science.1135961. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Nature. 2004;430(6996):226. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 38.Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. J Biol Chem. 2006;281(52):40412. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- 39.Lee PS, Chang C, Liu D, Derynck R. J Biol Chem. 2003;278(30):27853. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 40.Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. J Biol Chem. 2003;278(33):31043. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 41.Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. J Biol Chem. 2003;278(21):18714. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- 42.Long J, Wang G, He D, Liu F. Biochem J. 2004;379(Pt 1):23. doi: 10.1042/BJ20031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohshima T, Shimotohno K. J Biol Chem. 2003;278(51):50833. doi: 10.1074/jbc.M307533200. [DOI] [PubMed] [Google Scholar]
- 44.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. Cell. 2006;125(5):915. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K. Mol Biol Cell. 2001;12(5):1431. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izzi L, Attisano L. Oncogene. 2004;23(11):2071. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 47.Lin X, Liang M, Feng XH. J Biol Chem. 2000;275(47):36818. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 48.Lo RS, Massague J. Nat Cell Biol. 1999;1(8):472. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. Nature. 1999;400(6745):687. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 50.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. Cell. 2009;136(1):123. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 51.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. Journal of cell science. 2002;115(Pt 15):3193. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 52.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. The Journal of biological chemistry. 2000;275(47):36803. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 53.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Mol Biol Cell. 2001;12(1):27. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perlman R, Schiemann WP, Brooks MW, Lodish HF, Weinberg RA. Nat Cell Biol. 2001;3(8):708. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- 55.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP. Proc Natl Acad Sci U S A. 2001;98(12):6686. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamouille S, Derynck R. J Cell Biol. 2007;178(3):437. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azuma M, Motegi K, Aota K, Yamashita T, Yoshida H, Sato M. Exp Cell Res. 1999;250(1):213. doi: 10.1006/excr.1999.4503. [DOI] [PubMed] [Google Scholar]
- 58.Horowitz JC, Rogers DS, Sharma V, Vittal R, White ES, Cui Z, Thannickal VJ. Cell Signal. 2007;19(4):761. doi: 10.1016/j.cellsig.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thannickal VJ, Lee DY, White ES, Cui Z, Larios JM, Chacon R, Horowitz JC, Day RM, Thomas PE. J Biol Chem. 2003;278(14):12384. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- 60.Galliher AJ, Schiemann WP. Breast Cancer Res. 2006;8(4):R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galliher AJ, Schiemann WP. Cancer Res. 2007;67(8):3752. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 62.Galliher-Beckley AJ, Schiemann WP. Carcinogenesis. 2008;29:244. doi: 10.1093/carcin/bgm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park SS, Eom YW, Kim EH, Lee JH, Min DS, Kim S, Kim SJ, Choi KS. Oncogene. 2004;23(37):6272. doi: 10.1038/sj.onc.1207856. [DOI] [PubMed] [Google Scholar]
- 64.Elenbaas B, Weinberg RA. Exp Cell Res. 2001;264(1):169. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 65.Myeroff LL, Parsons R, Kim S-J, Hedrick L, Cho KR, Orth K, Mathis M, Kinzler KW, Lutterbaugh J, Park K, Bang Y-J, Lee HY, Park J-G, Lynch HT, Roberts AB, Vogelstein B, Markowitz SD. Cancer Res. 1995;55:5545. [PubMed] [Google Scholar]
- 66.Waite KA, Eng C. Nat Rev Genet. 2003;4(10):763. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 67.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Cancer Research. 1999;59(2):320. [PubMed] [Google Scholar]
- 68.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Willson JKV. Science. 1995;268(5215):1336. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 69.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Cancer Research. 1995;55(23):5548. [PubMed] [Google Scholar]
- 70.Grady W, Willis J, Trobridge P, Romero-Gallo J, Munoz N, Olechnowicz J, Ferguson K, Gautam S, Markowitz S. Int J Cancer. 2006;118(3):600. doi: 10.1002/ijc.21399. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB, 3rd, Hamilton SR. N Engl J Med. 2001;344(16):1196. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elliott RL, Blobe GC. J Clin Oncol. 2005;23(9):2078. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 73.Seoane J. Carcinogenesis. 2006 doi: 10.1093/carcin/bgl068. [DOI] [PubMed] [Google Scholar]
- 74.Schiemann WP, Pfeifer WM, Levi E, Kadin ME, Lodish HF. Blood. 1999;94(8):2854. [PubMed] [Google Scholar]
- 75.Schiemann WP, Rotzer D, Pfeifer WM, Levi E, Rai KR, Knaus P, Kadin ME. Cancer Detect Prev. 2004;28(1):57. doi: 10.1016/j.cdp.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Bian Y, Caldes T, Wijnen J, Franken P, Vasen H, Kaklamani V, Nafa K, Peterlongo P, Ellis N, Baron JA, Burn J, Moeslein G, Morrison PJ, Chen Y, Ahsan H, Watson P, Lynch HT, de la Chapelle A, Fodde R, Pasche B. J Clin Oncol. 2005;23(13):3074. doi: 10.1200/JCO.2005.00.281. [DOI] [PubMed] [Google Scholar]
- 77.Kaklamani VG, Baddi L, Liu J, Rosman D, Phukan S, Bradley C, Hegarty C, McDaniel B, Rademaker A, Oddoux C, Ostrer H, Michel LS, Huang H, Chen Y, Ahsan H, Offit K, Pasche B. Cancer Res. 2005;65(8):3454. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- 78.Pasche B, Knobloch TJ, Bian Y, Liu J, Phukan S, Rosman D, Kaklamani V, Baddi L, Siddiqui FS, Frankel W, Prior TW, Schuller DE, Agrawal A, Lang J, Dolan ME, Vokes EE, Lane WS, Huang CC, Caldes T, Di Cristofano A, Hampel H, Nilsson I, von Heijne G, Fodde R, Murty VV, de la Chapelle A, Weghorst CM. Jama. 2005;294(13):1634. doi: 10.1001/jama.294.13.1634. [DOI] [PubMed] [Google Scholar]
- 79.Chen T, Carter D, Garrigue-Antar L, Reiss M. Cancer Res. 1998;58(21):4805. [PubMed] [Google Scholar]
- 80.Chen T, Yan W, Wells RG, Rimm DL, McNiff J, Leffell D, Reiss M. Int J Cancer. 2001;93(5):653. doi: 10.1002/ijc.1381. [DOI] [PubMed] [Google Scholar]
- 81.Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC. J Clin Invest. 2007;117(1):206. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finger EC, Turley RS, Dong M, How T, Fields TA, Blobe GC. Carcinogenesis. 2008 doi: 10.1093/carcin/bgm289. [DOI] [PubMed] [Google Scholar]
- 83.Gordon KJ, Dong M, Chislock EM, Fields TA, Blobe GC. Carcinogenesis. 2007 doi: 10.1093/carcin/bgm249. [DOI] [PubMed] [Google Scholar]
- 84.Hempel N, How T, Dong M, Murphy SK, Fields TA, Blobe GC. Cancer Res. 2007;67(11):5231. doi: 10.1158/0008-5472.CAN-07-0035. [DOI] [PubMed] [Google Scholar]
- 85.Turley RS, Finger EC, Hempel N, How T, Fields TA, Blobe GC. Cancer Res. 2007;67(3):1090. doi: 10.1158/0008-5472.CAN-06-3117. [DOI] [PubMed] [Google Scholar]
- 86.Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. J Biol Chem. 1998;273(33):21145. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 87.Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Mol Cell Biol. 2004;24(6):2546. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Massague J, Seoane J, Wotton D. Genes Dev. 2005;19(23):2783. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 89.Chen CR, Kang Y, Siegel PM, Massague J. Cell. 2002;110(1):19. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 90.White LA, Mitchell TI, Brinckerhoff CE. Biochim Biophys Acta. 2000;1490(3):259. doi: 10.1016/s0167-4781(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 91.Heldin C-H, Miyazono K, ten Dijke P. Nature. 1997;390:465. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 92.Friedl W, Uhlhaas S, Schulmann K, Stolte M, Loff S, Back W, Mangold E, Stern M, Knaebel HP, Sutter C, Weber RG, Pistorius S, Burger B, Propping P. Hum Genet. 2002;111(1):108. doi: 10.1007/s00439-002-0748-9. [DOI] [PubMed] [Google Scholar]
- 93.Yang L, Mao C, Teng Y, Li W, Zhang J, Cheng X, Li X, Han X, Xia Z, Deng H, Yang X. Cancer Res. 2005;65(19):8671. doi: 10.1158/0008-5472.CAN-05-0800. [DOI] [PubMed] [Google Scholar]
- 94.Kim BG, Li C, Qiao W, Mamura M, Kasperczak B, Anver M, Wolfraim L, Hong S, Mushinski E, Potter M, Kim SJ, Fu XY, Deng C, Letterio JJ. Nature. 2006;441(7096):1015. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 95.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Cell. 1998;92(5):645. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 96.Han SU, Kim HT, Seong do H, Kim YS, Park YS, Bang YJ, Yang HK, Kim SJ. Oncogene. 2004;23(7):1333. doi: 10.1038/sj.onc.1207259. [DOI] [PubMed] [Google Scholar]
- 97.Wolfraim LA, Fernandez TM, Mamura M, Fuller WL, Kumar R, Cole DE, Byfield S, Felici A, Flanders KC, Walz TM, Roberts AB, Aplan PD, Balis FM, Letterio JJ. N Engl J Med. 2004;351(6):552. doi: 10.1056/NEJMoa031197. [DOI] [PubMed] [Google Scholar]
- 98.Maggio-Price L, Treuting P, Zeng W, Tsang M, Bielefeldt-Ohmann H, Iritani BM. Cancer Res. 2006;66(2):828. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li AG, Lu SL, Zhang MX, Deng C, Wang XJ. Cancer Res. 2004;64(21):7836. doi: 10.1158/0008-5472.CAN-04-1331. [DOI] [PubMed] [Google Scholar]
- 100.Siegel PM, Massague J. Nat Rev Cancer. 2003;3(11):807. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 101.Siegel PM, Shu W, Massague J. J Biol Chem. 2003;278(37):35444. doi: 10.1074/jbc.M301413200. [DOI] [PubMed] [Google Scholar]
- 102.Kang Y, Chen CR, Massague J. Mol Cell. 2003;11(4):915. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 103.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Oncogene. 2003;22(3):351. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 104.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. Nat Cell Biol. 2001;3(4):400. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 105.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Cell. 2004;117(2):211. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 106.Moustakas A, Kardassis D. Proc Natl Acad Sci U S A. 1998;95(12):6733. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, Pelicci PG, Amati B, Kouzarides T, de Launoit Y, Di Croce L, Fuks F. Embo J. 2005;24(2):336. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo J, Manning BD, Cantley LC. Cancer Cell. 2003;4(4):257. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 109.Atfi A, Abecassis L, Bourgeade MF. EMBO Rep. 2005 doi: 10.1038/sj.embor.7400501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Conery AR, Cao Y, Thompson EA, Townsend CM, Ko TC, Luo K. Nat Cell Biol. 2004;6:366. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- 111.Remy I, Montmarquette A, Michnick SW. Nat Cell Biol. 2004;6:358. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 112.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. Cancer Cell. 2006;10(3):203. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 113.Noda D, Itoh S, Watanabe Y, Inamitsu M, Dennler S, Itoh F, Koike S, Danielpour D, Ten Dijke P, Kato M. Oncogene. 2006 doi: 10.1038/sj.onc.1209571. [DOI] [PubMed] [Google Scholar]
- 114.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA. Nat Genet. 2001;27(2):172. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 115.Luo K. Curr Opin Genet Dev. 2004;14(1):65. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 116.Liu X, Sun Y, Weinberg RA, Lodish HF. Cytokine Growth Factor Rev. 2001;12(1):1. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 117.Karakas B, Weeraratna A, Abukhdeir A, Blair BG, Konishi H, Arena S, Becker K, Wood W, 3rd, Argani P, De Marzo AM, Bachman KE, Park BH. Oncogene. 2006;25(40):5561. doi: 10.1038/sj.onc.1209540. [DOI] [PubMed] [Google Scholar]
- 118.Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Mol Biol Cell. 1993;4(6):637. doi: 10.1091/mbc.4.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Strutz F, Zeisberg M, Renziehausen A, Raschke B, Becker V, van Kooten C, Muller G. Kidney Int. 2001;59(2):579. doi: 10.1046/j.1523-1755.2001.059002579.x. [DOI] [PubMed] [Google Scholar]
- 120.Seifert RA, Coats SA, Raines EW, Ross R, Bowen-Pope DF. Journal of Biological Chemistry. 1994;269(19):13951. [PubMed] [Google Scholar]
- 121.Chantry D, Turner M, Abney E, Feldmann M. J Immunol. 1989;142(12):4295. [PubMed] [Google Scholar]
- 122.Ishikawa O, LeRoy EC, Trojanowska M. J Cell Physiol. 1990;145(1):181. doi: 10.1002/jcp.1041450124. [DOI] [PubMed] [Google Scholar]
- 123.Feng P, Catt KJ, Knecht M. J Biol Chem. 1986;261(30):14167. [PubMed] [Google Scholar]
- 124.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Mol Cell. 2007;25(3):441. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 125.Lo RS, Wotton D, Massague J. Embo J. 2001;20(1–2):128. doi: 10.1093/emboj/20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thiery JP. Nat Rev Cancer. 2002;2(6):442. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 127.Greenburg G, Hay ED. J Cell Biol. 1982;95(1):333. doi: 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thiery JP. Curr Opin Cell Biol. 2003;15(6):740. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 129.Zavadil J, Bottinger EP. Oncogene. 2005;24(37):5764. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 130.Moustakas A, Heldin CH. Cancer Sci. 2007;98(10):1512. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Savagner P. Bioessays. 2001;23(10):912. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 132.Willis BC, Borok Z. Am J Physiol Lung Cell Mol Physiol. 2007;293(3):L525. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 133.Miettinen PJ, Ebner R, Lopez AR, Derynck R. Journal of Cell Biology. 1994;127(6 Pt 2):2021. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Nature Genetics. 1995;11(4):415. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 135.Romano LA, Runyan RB. Dev Biol. 2000;223(1):91. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- 136.Saika S, Kono-Saika S, Ohnishi Y, Sato M, Muragaki Y, Ooshima A, Flanders KC, Yoo J, Anzano M, Liu CY, Kao WW, Roberts AB. Am J Pathol. 2004;164(2):651. doi: 10.1016/S0002-9440(10)63153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saika S, Kono-Saika S, Tanaka T, Yamanaka O, Ohnishi Y, Sato M, Muragaki Y, Ooshima A, Yoo J, Flanders KC, Roberts AB. Lab Invest. 2004;84(10):1245. doi: 10.1038/labinvest.3700156. [DOI] [PubMed] [Google Scholar]
- 138.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. J Clin Invest. 2003;112(10):1486. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bhowmick NA, Moses HL. Curr Opin Genet Dev. 2005;15(1):97. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bhowmick NA, Neilson EG, Moses HL. Nature. 2004;432(7015):332. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fidler IJ. Semin Cancer Biol. 2002;12(2):89. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- 142.Fidler IJ. Differentiation. 2002;70(9–10):498. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- 143.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, Washington MK, Neilson EG, Moses HL. Science. 2004;303(5659):848. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 144.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R, Arteaga CL, Neilson EG, Hayward SW, Moses HL. Oncogene. 2005;24(32):5053. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, Moses HL. Cancer Cell. 2008;13(1):23. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, Manova-Todorova K, Blasberg R, Gerald WL, Massague J. Proc Natl Acad Sci U S A. 2005;102(39):13909. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. Cancer Cell. 2003;3(6):537. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 148.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. J Clin Invest. 1999;103(2):197. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Muraoka-Cook RS, Kurokawa H, Koh Y, Forbes JT, Roebuck LR, Barcellos-Hoff MH, Moody SE, Chodosh LA, Arteaga CL. Cancer Res. 2004;64(24):9002. doi: 10.1158/0008-5472.CAN-04-2111. [DOI] [PubMed] [Google Scholar]
- 150.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. Cell. 2008;133(4):704. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Cancer Cell. 2007;11(3):259. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 152.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. Nat Genet. 2008;40(5):499. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. PLoS ONE. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tian F, DaCosta Byfield S, Parks WT, Yoo S, Felici A, Tang B, Piek E, Wakefield LM, Roberts AB. Cancer Res. 2003;63(23):8284. [PubMed] [Google Scholar]
- 155.Deckers M, van Dinther M, Buijs J, Que I, Lowik C, van der Pluijm G, ten Dijke P. Cancer Res. 2006;66(4):2202. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 156.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Genes Dev. 2006;20(22):3130. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Azuma H, Ehata S, Miyazaki H, Watabe T, Maruyama O, Imamura T, Sakamoto T, Kiyama S, Kiyama Y, Ubai T, Inamoto T, Takahara S, Itoh Y, Otsuki Y, Katsuoka Y, Miyazono K, Horie S. J Natl Cancer Inst. 2005;97(23):1734. doi: 10.1093/jnci/dji399. [DOI] [PubMed] [Google Scholar]
- 158.Leivonen SK, Ala-Aho R, Koli K, Grenman R, Peltonen J, Kahari VM. Oncogene. 2006;25(18):2588. doi: 10.1038/sj.onc.1209291. [DOI] [PubMed] [Google Scholar]
- 159.Leivonen SK, Kahari VM. Int J Cancer. 2007;121(10):2119. doi: 10.1002/ijc.23113. [DOI] [PubMed] [Google Scholar]
- 160.Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Niewolna M, Andre J, Delmas V, Larue L, Guise TA, Mauviel A. Cancer Res. 2007;67(5):2317. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 161.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. Cell. 2009;137(1):87. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 162.Papageorgis P, Lambert AW, Ozturk S, Gao F, Pan H, Manne U, Alekseyev YO, Thiagalingam A, Abdolmaleky HM, Lenburg M, Thiagalingam S. Cancer Res. 2010;70(3):968. doi: 10.1158/0008-5472.CAN-09-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bhowmick NA, Zent R, Ghiassi M, McDonnell M, Moses HL. J Biol Chem. 2001;276(50):46707. doi: 10.1074/jbc.M106176200. [DOI] [PubMed] [Google Scholar]
- 164.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. J Cell Biochem. 2005;95(5):918. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 165.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, Adler G, Gress TM. Cancer Res. 2001;61(10):4222. [PubMed] [Google Scholar]
- 166.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. J Cell Biol. 2002;156(2):299. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Neoplasia. 2004;6(5):603. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. J Clin Invest. 2004;114(4):569. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Embo J. 2004;23(5):1155. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim K, Lu Z, Hay ED. Cell Biol Int. 2002;26(5):463. doi: 10.1006/cbir.2002.0901. [DOI] [PubMed] [Google Scholar]
- 171.Araki S, Eitel JA, Batuello CN, Bijangi-Vishehsaraei K, Xie XJ, Danielpour D, Pollok KE, Boothman DA, Mayo LD. J Clin Invest. 2010;120(1):290. doi: 10.1172/JCI39194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Oft M, Akhurst RJ, Balmain A. Nat Cell Biol. 2002;4(7):487. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 173.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. Cell. 1996;86(4):531. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 174.Fowlis DJ, Cui W, Johnson SA, Balmain A, Akhurst RJ. Cell Growth & Differentiation. 1996;7(5):679. [PubMed] [Google Scholar]
- 175.Lehmann K, Janda E, Pierreux CE, Rytèomaa M, Schulze A, McMahon M, Hill CS, Beug H, Downward J. Genes & development. 2000;14(20):2610. doi: 10.1101/gad.181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prunier C, Howe PH. J Biol Chem. 2005;280(17):17540. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 177.Sloan EK, Pouliot N, Stanley KL, Chia J, Moseley JM, Hards DK, Anderson RL. Breast Cancer Res. 2006;8(2):R20. doi: 10.1186/bcr1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bandyopadhyay A, Agyin JK, Wang L, Tang Y, Lei X, Story BM, Cornell JE, Pollock BH, Mundy GR, Sun LZ. Cancer Res. 2006;66(13):6714. doi: 10.1158/0008-5472.CAN-05-3565. [DOI] [PubMed] [Google Scholar]
- 179.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Science. 2005;307(5715):1603. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 180.Carmeliet P. Nat Med. 2000;6(4):389. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 181.Carmeliet P. Nat Med. 2003;9(6):653. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 182.Carmeliet P, Jain RK. Nature. 2000;407(6801):249. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 183.Bertolino P, Deckers M, Lebrin F, ten Dijke P. Chest. 2005;128(6 Suppl):585S. doi: 10.1378/chest.128.6_suppl.585S. [DOI] [PubMed] [Google Scholar]
- 184.Lebrin F, Deckers M, Bertolino P, Ten Dijke P. Cardiovasc Res. 2005;65(3):599. doi: 10.1016/j.cardiores.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 185.Pepper MS. Cytokine Growth Factor Rev. 1997;8(1):21. doi: 10.1016/s1359-6101(96)00048-2. [DOI] [PubMed] [Google Scholar]
- 186.Pepper MS, Vassalli JD, Orci L, Montesano R. Exp Cell Res. 1993;204(2):356. doi: 10.1006/excr.1993.1043. [DOI] [PubMed] [Google Scholar]
- 187.Wolff RA, Malinowski RL, Heaton NS, Hullett DA, Hoch JR. J Vasc Surg. 2006;43(5):1028. doi: 10.1016/j.jvs.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 188.Dickson MC, Martin JS, Cousins FM, Kulkarni AB, Karlsson S, Akhurst RJ. Development. 1995;121(6):1845. doi: 10.1242/dev.121.6.1845. [DOI] [PubMed] [Google Scholar]
- 189.Larsson J, Goumans MJ, Sjostrand LJ, van Rooijen MA, Ward D, Leveen P, Xu X, ten Dijke P, Mummery CL, Karlsson S. Embo J. 2001;20(7):1663. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goumans MJ, Zwijsen A, van Rooijen MA, Huylebroeck D, Roelen BA, Mummery CL. Development. 1999;126(16):3473. doi: 10.1242/dev.126.16.3473. [DOI] [PubMed] [Google Scholar]
- 191.Oshima M, Oshima H, Taketo MM. Dev Biol. 1996;179(1):297. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- 192.Brown CB, Boyer AS, Runyan RB, Barnett JV. Science. 1999;283(5410):2080. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 193.Compton LA, Potash DA, Brown CB, Barnett JV. Circ Res. 2007;101(8):784. doi: 10.1161/CIRCRESAHA.107.152082. [DOI] [PubMed] [Google Scholar]
- 194.Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, Karlsson S, ten Dijke P. Mol Cell. 2003;12(4):817. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 195.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, ten Dijke P, Kim S, Li E. Proc Natl Acad Sci U S A. 2000;97(6):2626. doi: 10.1073/pnas.97.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Arthur HM, Ure J, Smith AJ, Renforth G, Wilson DI, Torsney E, Charlton R, Parums DV, Jowett T, Marchuk DA, Burn J, Diamond AG. Dev Biol. 2000;217(1):42. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
- 197.Lechleider RJ, Ryan JL, Garrett L, Eng C, Deng C, Wynshaw-Boris A, Roberts AB. Dev Biol. 2001;240(1):157. doi: 10.1006/dbio.2001.0469. [DOI] [PubMed] [Google Scholar]
- 198.Chang H, Huylebroeck D, Verschueren K, Guo Q, Matzuk MM, Zwijsen A. Development. 1999;126(8):1631. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- 199.Shovlin CL, Hughes JM, Scott J, Seidman CE, Seidman JG. Am J Hum Genet. 1997;61(1):68. doi: 10.1086/513906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J, et al. Nat Genet. 1994;8(4):345. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 201.Berg JN, Gallione CJ, Stenzel TT, Johnson DW, Allen WP, Schwartz CE, Jackson CE, Porteous ME, Marchuk DA. Am J Hum Genet. 1997;61(1):60. doi: 10.1086/513903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A, Guttmacher AE, Jackson CE, Attisano L, Kucherlapati R, Porteous ME, Marchuk DA. Nat Genet. 1996;13(2):189. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 203.Bourdeau A, Dumont DJ, Letarte M. J Clin Invest. 1999;104(10):1343. doi: 10.1172/JCI8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Srinivasan S, Hanes MA, Dickens T, Porteous ME, Oh SP, Hale LP, Marchuk DA. Hum Mol Genet. 2003;12(5):473. doi: 10.1093/hmg/ddg050. [DOI] [PubMed] [Google Scholar]
- 205.Goumans MJ, Lebrin F, Valdimarsdottir G. Trends Cardiovasc Med. 2003;13(7):301. doi: 10.1016/s1050-1738(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 206.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Embo J. 2002;21(7):1743. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu X, Ma J, Han JD, Wang N, Chen YG. Microvasc Res. 2006;71(1):12. doi: 10.1016/j.mvr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 208.Ota T, Fujii M, Sugizaki T, Ishii M, Miyazawa K, Aburatani H, Miyazono K. J Cell Physiol. 2002;193(3):299. doi: 10.1002/jcp.10170. [DOI] [PubMed] [Google Scholar]
- 209.Bandyopadhyay A, Lopez-Casillas F, Malik SN, Montiel JL, Mendoza V, Yang J, Sun LZ. Cancer Res. 2002;62(16):4690. [PubMed] [Google Scholar]
- 210.Bandyopadhyay A, Zhu Y, Cibull ML, Bao L, Chen C, Sun L. Cancer Res. 1999;59(19):5041. [PubMed] [Google Scholar]
- 211.Bandyopadhyay A, Zhu Y, Malik SN, Kreisberg J, Brattain MG, Sprague EA, Luo J, Lopez-Casillas F, Sun LZ. Oncogene. 2002;21(22):3541. doi: 10.1038/sj.onc.1205439. [DOI] [PubMed] [Google Scholar]
- 212.Lebrin F, Goumans MJ, Jonker L, Carvalho RL, Valdimarsdottir G, Thorikay M, Mummery C, Arthur HM, ten Dijke P. Embo J. 2004;23(20):4018. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li C, Hampson IN, Hampson L, Kumar P, Bernabeu C, Kumar S. Faseb J. 2000;14(1):55. doi: 10.1096/fasebj.14.1.55. [DOI] [PubMed] [Google Scholar]
- 214.Massague J, Blain SW, Lo RS. Cell. 2000;103(2):295. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 215.Sanchez-Capelo A. Cytokine Growth Factor Rv. 2005;16:15. doi: 10.1016/j.cytogfr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 216.Docherty NG, O'Sullivan OE, Healy DA, Murphy M, O'Neill AJ, Fitzpatrick JM, Watson RW. Am J Physiol Renal Physiol. 2006;290:F1202. doi: 10.1152/ajprenal.00406.2005. [DOI] [PubMed] [Google Scholar]
- 217.Yang Y, Pan X, Lei WJ, Song J. Oncogene. 2006;25:7235. doi: 10.1038/sj.onc.1209712. [DOI] [PubMed] [Google Scholar]
- 218.Biswas S, Guix M, Rinehart C, Dugger TC, Chytil A, Moses HL, Freeman ML, Arteaga CL. J Clin Invest. 2007;117(5):1305. doi: 10.1172/JCI30740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lei X, Bandyopadhyay A, Le T, Sun L. Oncogene. 2002;21:7514. doi: 10.1038/sj.onc.1205966. [DOI] [PubMed] [Google Scholar]
- 220.Arsura M, Panta GR, Bilyeu JD, Cavin LG, Sovak MA, Oliver AA, Factor V, Heuchel R, Mercurio F, Thorgeirsson SS, Sonenshein GE. Oncogene. 2003;22:412. doi: 10.1038/sj.onc.1206132. [DOI] [PubMed] [Google Scholar]
- 221.Lin SJ, Chang C, Ng AK, Wang SH, Li JJ, Hu CP. Apoptosis. 2007;12:1659. doi: 10.1007/s10495-007-0085-5. [DOI] [PubMed] [Google Scholar]
- 222.Rayet B, Gelinas C. Oncogene. 1999;18(49):6938. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 223.Shin I, Bakin AV, Rodeck U, Brunet A, Arteaga CL. Mol Biol Cell. 2001;12(11):3328. doi: 10.1091/mbc.12.11.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cao Y, Karin M. J Mammary Gland Biol Neoplasia. 2003;8(2):215. doi: 10.1023/a:1025905008934. [DOI] [PubMed] [Google Scholar]
- 225.Neil JR, Schiemann WP. Cancer Research. 2008;68:1462. doi: 10.1158/0008-5472.CAN-07-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Park J-I, Lee M-G, Cho K, Park B-J, Chae K-S, Byun D-S, Ryu B-K, Park Y-K, Chi S-G. Oncogene. 2003;22:4314. doi: 10.1038/sj.onc.1206478. [DOI] [PubMed] [Google Scholar]
- 227.Neil JR, Johnson KM, Nemenoff RA, Schiemann WP. Carcinogenesis. 2008;29(11):2227. doi: 10.1093/carcin/bgn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tian M, Schiemann WP. FASEB J. 2009;24:1105. doi: 10.1096/fj.09-141341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kaur S, Wang F, Venkatraman M, Arsura M. J Biol Chem. 2005;280:38599. doi: 10.1074/jbc.M505671200. [DOI] [PubMed] [Google Scholar]
- 230.Van Themsche C, Mathieu I, Parent S, Asselin E. J Biol Chem. 2007;282(7):4794. doi: 10.1074/jbc.M608497200. [DOI] [PubMed] [Google Scholar]
- 231.Neil JR, Tian M, Schiemann WP. J Biol Chem. 2009;284(32):21209. doi: 10.1074/jbc.M109.018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Madrid LV, Wang CY, Guttridge DC, Schottelius AJ, Baldwin AS, Jr, Mayo MW. Mol Cell Biol. 2000;20(5):1626. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr J Biol Chem. 2001;276:18934. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 234.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. Cell. 2004;117(2):225. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 235.Hiraga T, Myoui A, Choi ME, Yoshikawa H, Yoneda T. Cancer Res. 2006;66:2067. doi: 10.1158/0008-5472.CAN-05-2012. [DOI] [PubMed] [Google Scholar]
- 236.Lu S, Dong Z. Prostate. 2006;66(9):996. doi: 10.1002/pros.20424. [DOI] [PubMed] [Google Scholar]
- 237.Enders GA. Br J Cancer. 2007;97(10):1388. doi: 10.1038/sj.bjc.6604048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gorelik L, Flavell RA. Nat Rev Immunol. 2002;2:46. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 239.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Annu Rev Immunol. 2006;24:99. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 240.Lin WW, Karin M. J Clin Invest. 2007;117:1175. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gorelik L, Flavell RA. Immunity. 2000;12(2):171. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 242.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Proc Natl Acad Sci. 1993;90:770. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C. EMBO J. 1999;18(5):1280. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dalal BI, Keown PA, Greenberg AH. Am J Pathol. 1993;143(2):381. [PMC free article] [PubMed] [Google Scholar]
- 245.Gorsch SM, Memoli VA, Stukel TA, Gold LI, Arrick BA. Cancer Res. 1992;52:6949. [PubMed] [Google Scholar]
- 246.Ivanovic V, Todorovic-Rakovic N, Demajo M, Neskovic-Konstantinovic Z, Subota V, Ivanisevic-Milovanovic O, Nikolic-Vukosavljevic D. Eur J Cancer. 2003;39(4):454. doi: 10.1016/s0959-8049(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 247.Dvorak HF. N Engl J Med. 1986;315(26):1650. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 248.Teicher BA. Clin Cancer Res. 2007;13(21):6247. doi: 10.1158/1078-0432.CCR-07-1654. [DOI] [PubMed] [Google Scholar]
- 249.Wrzesinski SH, Wan YY, Flavell RA. Clin Cancer Res. 2007;13(18):5262. doi: 10.1158/1078-0432.CCR-07-1157. [DOI] [PubMed] [Google Scholar]
- 250.Gorelik L, Flavell RA. Nat Med. 2001;7(10):1118. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 251.Zhang Q, Yang X, Pins M, Javonovic B, Kuzel T, Kim SJ, Parijs LV, Greenberg NM, Liu V, Guo Y, Lee C. Cancer Res. 2005;65(5):1761. doi: 10.1158/0008-5472.CAN-04-3169. [DOI] [PubMed] [Google Scholar]
- 252.Kopp HG, Placke T, Salih HR. Cancer Res. 2009;69(19):7775. doi: 10.1158/0008-5472.CAN-09-2123. [DOI] [PubMed] [Google Scholar]
- 253.Larmonier N, Cathelin D, Larmonier C, Nicolas A, Merino D, Janikashvili N, Audia S, Bateman A, Thompson J, Kottke T, Hartung T, Katsanis E, Vile R, Bonnotte B. Exp Cell Res. 2007;313(11):2345. doi: 10.1016/j.yexcr.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 254.Conti P, Castellani MI, Kempuraj D, Salini V, Vecchiet J, Tete S, Mastrangelo F, Perrella A, De Lutiis MA, Tagen M, Theoharides TC. Ann Clin Lab Sci. 2007;37(4):315. [PubMed] [Google Scholar]
- 255.Lin EY, Li JF, Gnatovskiy L, DY, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW. Cancer Res. 2006;66(23):11238. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 256.Pollard JW. Nat Rev Cancer. 2004;4(1):71. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 257.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM. Cancer Cell. 2009;16(3):183. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Stewart SA, Weinberg RA. Annu Rev Cell Dev Biol. 2006;22:531. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 259.Ducrest AL, Szutorisz H, Lingner J, Nabholz M. Oncogene. 2002;21(4):541. doi: 10.1038/sj.onc.1205081. [DOI] [PubMed] [Google Scholar]
- 260.Li H, Xu D, Toh BH, Liu JP. Cell Res. 2006;16(2):169. doi: 10.1038/sj.cr.7310023. [DOI] [PubMed] [Google Scholar]
- 261.Elkak AE, Newbold RF, Thomas V, Mokbel K. Cancer Cell Int. 2003;3(1):9. doi: 10.1186/1475-2867-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Li H, Xu D, Li J, Berndt MC, Liu JP. J Biol Chem. 2006;281(35):25588. doi: 10.1074/jbc.M602381200. [DOI] [PubMed] [Google Scholar]
- 263.Yang H, Kyo S, Takatura M, Sun L. Cell Growth Differ. 2001;12(2):119. [PubMed] [Google Scholar]
- 264.Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Proc Natl Acad Sci U S A. 2001;98(8):4498. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lin SY, Elledge SJ. Cell. 2003;113(7):881. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 266.Fujiki T, Miura T, Maura M, Shiraishi H, Nishimura S, Imada Y, Uehara N, Tashiro K, Shirahata S, Katakura Y. Oncogene. 2007;26(36):5258. doi: 10.1038/sj.onc.1210331. [DOI] [PubMed] [Google Scholar]
- 267.Zhang H, Cohen SN. Genes Dev. 2004;18(24):3028. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Valderrama-Carvajal H, Cocolakis E, Lacerte A, Lee EH, Krystal G, Ali S, Lebrun JJ. Nat Cell Biol. 2002;4:936. doi: 10.1038/ncb885. [DOI] [PubMed] [Google Scholar]
- 269.Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. Nat Cell Biol. 2002;4:51. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 270.Kondo Y, Kanzawa T, Sawaya R, Kondo S. Nat Rev Cancer. 2005;5:726. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 271.Chalaux E, Lopez-Rovira T, Rosa Jl, Pons G, Boxer LM, Bartrons R, Ventura F. FEBS Lett. 1999;457:478. doi: 10.1016/s0014-5793(99)01051-0. [DOI] [PubMed] [Google Scholar]
- 272.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. J Clin Invest. 1997;99:2365. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, Book-Melamed N, Timberg R, Duckett CS, Lechleider RJ, Steller H, Orly J, Kim SJ, Roberts AB. Nat Cell Biol. 2000;2:915. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- 274.Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S. EMBO J. 2004;23:1627. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Thiefes A, Wolter S, Mushinski JF, Hoffmann E, Dittrich-Breiholz O, Graue N, Dorrie A, Schneider H, Wirth D, Luckow B, Resch K, Kracht M. J Biol Chem. 2005;18:18. doi: 10.1074/jbc.M411657200. [DOI] [PubMed] [Google Scholar]
- 276.Edlund S, Bu S, Schuster N, Aspenstrom P, Heuchel R, Heldin NE, ten Dijke P, Heldin CH, Landstrom M. Mol Biol Cell. 2003;14:529. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, Nishida E, Hagiwara M. J Biol Chem. 1996;271(23):13675. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 278.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Science. 1995;270(5244):2008. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]