Abstract
Chloroplasts fulfill important functions in cellular metabolism. The majority of plastid genome-encoded genes is involved in either photosynthesis or chloroplast gene expression. Whether or not plastid genes also can determine extraplastidic functions has remained controversial. We demonstrate here an essential role of plastid protein synthesis in tobacco leaf development. By using chloroplast transformation, we have developed an experimental system that produces recombination-based knockouts of chloroplast translation in a cell-line-specific manner. The resulting plants are chimeric and, in the presence of translational inhibitors, exhibit severe developmental abnormalities. In the absence of active plastid protein synthesis, leaf blade development is abolished because of an apparent arrest of cell division. This effect appears to be cell-autonomous in that adjacent sectors of cells with translating plastids are phenotypically normal but cannot complement for the absence of plastid translation in mutant sectors. Developmental abnormalities also are seen in flower morphology, indicating that the defects are not caused by inhibited expression of plastid photosynthesis genes. Taken together, our data point to an unexpected essential role of plastid genes and gene expression in plant development and cell division.
Plastids arose through endosymbiosis and stem from cyanobacterial ancestors. The gradual conversion of the endosymbiont into a cell organelle was accompanied by a dramatic reduction in genome size. Whereas the cyanobacterium Synechocystis has >3,100 genes (and ORFs) in a genome of 3.57 Mbp (1–3), higher plant plastid genomes harbor only ≈130 genes in ≈150 kbp (4). Three processes have contributed to chloroplast genome shrinking on an evolutionary time scale: (i) the loss of dispensable genetic information (e.g., genes for cyanobacterial cell-wall biosynthesis), (ii) the elimination of redundant genetic information that was present in both the host's and the endosymbiont's genomes (e.g., pathways for carbohydrate, amino acid, and lipid biosyntheses), and (iii) the massive translocation of genes from the endosymbiont's DNA to the host cell's nuclear genome. Most of the residual 130 genes on the genome of present-day chloroplasts either are photosynthesis-related genes or encode components of the plastid gene expression machinery (e.g., rRNAs, a complete set of tRNAs, some ribosomal proteins; ref. 4). It is generally assumed that plastid-encoded gene products function exclusively inside the plastid, although metabolic signals may leave the chloroplast and influence processes outside the plastid (5, 6). Collectively, such signals have been termed “plastid factors.” They were initially identified by their capacity to promote transcription of defined subsets of nuclear genes. At least two different plastid factors seem to operate in plant cells: redox signals (7, 8) and intermediates in chlorophyll biosynthesis (9–11).
In contrast to this well established exchange of metabolic signals between the chloroplast and the nucleus, a role of chloroplast genes and gene functions in developmental processes has remained speculative. Early genetic work involving reciprocal crosses between two sexually compatible evening primrose species, Oenothera odorata and Oenothera berteriana, provided evidence for some influence of the plastid genome type on leaf margin shape (intensity of toothing; ref. 12). Similarly, flower morphology (e.g., hypanthium length) seemed to be influenced to a limited extent by the plastid genome type present in the hybrid progeny (12, 13). Unfortunately, this early genetic work was discontinued, the results were never reproduced, and their interpretation has remained controversial (14, 15).
To address the role of chloroplast genes and their expression during plant development, we have set up a transgenic chloroplast system that allows us to monitor the effects of inactivated plastid gene expression on the development of tobacco plants in a cell-line-dependent manner. We show here that plastid protein synthesis absolutely is required for development of the leaf blade and also for flower development. These findings point to an unexpected critical role of plastid genes and their expression in plant development.
Materials and Methods
Plant Material. Tobacco plants (Nicotiana tabacum cv. Petit Havana) were grown under aseptic conditions on agar-solidified Murashige and Skoog medium containing 30 g/l sucrose. Transplastomic lines were rooted and propagated on the same medium in the presence or absence of plastid translational inhibitors.
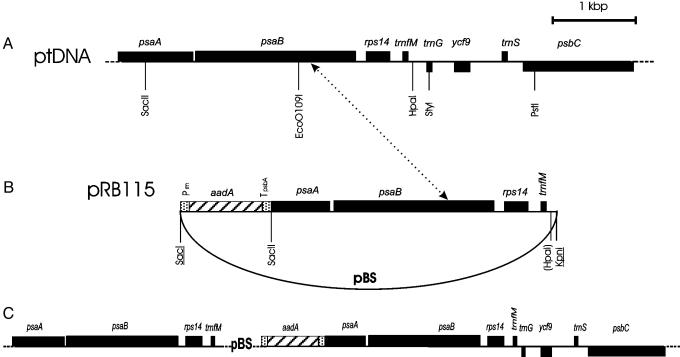
Vector Construction and Cloning Procedures. Plastid transformation vector pRB115 (Fig. 1) was derived from the psaA/B region in a cloned tobacco ptDNA fragment. An HpaI/SacII restriction fragment was subcloned into a pBluescript vector (Stratagene) cut with EcoRV and SacII, yielding plasmid pRB114. A chimeric aadA gene (16) subsequently was inserted as a blunt-end Ecl136II/DraI fragment into the SacI site of the pRB114 polylinker (after linearization with the blunt-end cutting isoschizomer Ecl136II). A clone was selected that contained the plastid selectable marker gene aadA in the same transcriptional orientation as the psaA/B fragment (Fig. 1). This clone was designated pRB115 and used as chloroplast transformation vector.
Fig. 1.
Recombination-induced knockout of plastid translation (RIKT). (A) Region of the tobacco plastid genome from which the chloroplast transformation vector pRB115 was derived. (B) Physical map of the plastid targeting region in transformation vector pRB115 (cloned as SacI/KpnI fragment into pBluescript). Note that the selectable marker gene aadA is flanked by only one region of homology with the plastid genome. Restriction sites lost because of ligation of heterologous ends are shown in parentheses. Sites from the pBluescript (pBS) polylinker are underlined. Prrn, promoter derived from the plastid rRNA operon; TpsbA, 3′ UTR derived from the psbA gene. (C) Cointegrate produced by plastid transformation with pRB115. A single homologous recombination event (indicated by the dotted arrow between the two maps in A and B) results in incorporation of the entire plasmid vector into the plastid genome. The cointegrate is unstable, and homologous recombination in the directly repeated psaA/psaB/rps14/trnfM region will recreate the wild-type genome and eliminate the aadA-resistance gene. In the presence of the translational inhibitor spectinomycin, cell lines that have completely lost the aadA as the result of such recombination events will no longer produce plastid-encoded proteins. pBS, pBluescript backbone (not drawn to scale).
Plastid Transformation and Selection of Transplastomic Tobacco Lines. Young leaves were harvested from sterile tobacco plants and bombarded with plasmid DNA-coated gold particles with the BioRad PDS1000He biolistic gun. Spectinomycin-resistant cell lines were selected on RMOP regeneration medium containing 500 mg/l spectinomycin dihydrochloride (17). Primary plastid transformants were identified by a PCR strategy employing aadA-specific primers. Three independently generated transplastomic lines (designated Nt-pRB115–4, Nt-pRB115–10, and Nt-pRB115–12) were subjected to four additional rounds of regeneration on RMOP/spectinomycin to enrich the transplastome and eliminate wild-type genome copies. Shoots resulting from the final round of regeneration were rooted on phytohormone-free medium, transferred to the soil, and grown under greenhouse conditions.
Crosses, Inheritance Tests, and Phenotypic Assays. Seeds from Nt-pRB115 plants were germinated on Murashige and Skoog medium containing spectinomycin (500 mg/liter). Green and variegated seedlings were propagated further on the same medium. For comparative phenotypic analyses, cuttings from individual plants were transferred to Murashige and Skoog medium without antibiotic or with spectinomycin or streptomycin (500 mg/liter). Alternatively, plants were transferred to the soil, and leaf phenotypes were monitored in the absence of antibiotic selection.
Isolation of Nucleic Acids and Analysis of Recombination Products. Total plant nucleic acids were isolated from leaf tissue samples by a cetyltrimethylammoniumbromide (CTAB)-based method (18). Presence of plasmids in planta was assayed by direct transformation of extracted total DNA into the Escherichia coli strain XL-1 blue. Plasmids were analyzed for their identity with pRB115 by diagnostic restriction digests.
Blotting and Hybridization Techniques. Total cellular DNA was digested with restriction enzymes, separated by gel electrophoresis on 0.8% agarose gels, and transferred onto Hybond nylon membranes (Amersham Pharmacia) by capillary blotting. Blots for the analysis of recombination products were probed with a radiolabeled plasmid probe (pRB115 linearized with SalI). The presence of aadA was assayed for with a labeled cloned aadA fragment covering the entire coding region (NcoI/XbaI fragment cloned into pUC120), and the plastid psbA gene was detected with a labeled PCR product (generated by amplification with primers P5psbA: 5′-GATGGTATTCGTGAACCT-3′ and P3psbA: 5′-TGATCAAACTAGAAGTTACC-3′). Labeling of probes with [32P]dATP was carried out by random priming (Amersham Pharmacia). Hybridization was performed at 65°C in Rapid-Hyb buffer (Amersham Pharmacia) following the manufacturer's protocol.
Results and Discussion
A Strategy for Cell-Line-Specific Knockout of Plastid Translation. Most genes encoded in the tobacco plastid genome specify photosynthetic proteins and constituents of the plastid gene expression machinery. Photosynthesis is optional under heterotrophic growth conditions, and all photosynthesis-related genes can be easily deleted from the chloroplast genome without affecting cell viability and plant development as long as sucrose is provided as carbon source (e.g., refs. 19–21). If the production of photosynthesis-related proteins were the only function of the plastid genome, then all plastid gene functions should be dispensable under heterotrophic culture conditions. Although this appears to be the case in some plant species (like Zea mays and Brassica napus; refs. 22 and 23), it is clearly not the case in others (like N. tabacum and Chlamydomonas reinhardtii; refs. 22, 24, and 25). We recently have established that two large ORFs in the tobacco plastid genome are indispensable genes (24) providing indirect evidence for plastid gene functions being essential. Our attempts to construct knockout plants for the chloroplast ribosomal protein gene rps14 confirmed that plastid protein biosynthesis is indispensable: transplastomic plants were obtained, but homoplasmic rps14 knockout lines could not be purified (unpublished results). To devise an experimental system suitable to study the effects of the loss of plastid translation on plant development, we constructed a transplastomic line in which homologous recombination-induced deletion of a spectinomycin-resistance gene produces translation-deficient tobacco cells (Fig. 1). We subsequently will refer to this strategy as “recombination-induced knockout of translation” (RIKT).
RIKT plants were constructed by plastid transformation with a vector harboring only one plastid targeting region (Fig. 1 A and B). In conventional plastid transformation experiments, two regions of homology with the plastid genome flank the foreign sequences resulting in transgene incorporation by two homologous recombination events (16, 26, 27). In contrast, vector pRB115 (Fig. 1B) contains only one block of homologous sequence and, hence, will be incorporated into the plastid genome by a single homologous recombination event producing a cointegrate (Fig. 1C). This cointegrate is likely to be highly unstable and can be expected to be resolved frequently by homologous recombination in the directly repeated psaA/psaB/rps14/trnfM region (Fig. 1C). These resolution events will recreate the wild-type genome and eliminate the aadA-resistance gene. In the presence of the plastid translational inhibitor spectinomycin, cell lines that have lost the aadA as the result of such recombination events will no longer produce plastidencoded proteins. In this way, we can construct chimeras whose aadA-containing cell lines should be phenotypically normal, whereas the RIKT cell lines can be analyzed for the phenotypic effects of turned-off plastid protein biosynthesis.
Generation of RIKT Plants. Plastid transformation experiments with vector pRB115 (Fig. 1B) produced numerous transplastomic tobacco lines, three of which were studied in detail (referred to as Nt-pRB115–4, Nt-pRB115–10, and Nt-pRB115–12). Primary transformants were subjected to four additional rounds of regeneration on medium with spectinomycin to enrich the transplastome and reduce the number of wild-type genome copies (16, 26). Subsequently, regenerating shoots were rooted and transferred to the greenhouse. We suspected the high copy number of the plastid genome in somatic leaf cells (with up to 10,000 copies being present in a single cell; ref. 28) to largely cover up possible phenotypic consequences of RIKT in primary plastid transformants. Because only a few copies of the plastid genome are transmitted on sexual reproduction, we reasoned that germinating seedlings should show recombination-induced loss of the aadA-resistance gene at high frequency. We therefore obtained seeds from the transplastomic plants and germinated seed samples in the presence of spectinomycin (Fig. 2A). Frequent appearance of white and variegated seedlings (Fig. 2 A and B) indicated that elimination of the aadA-resistance gene by RIKT may indeed work with high efficiency.
Fig. 2.
Phenotype of tobacco plants with recombination-induced knockout of translation. (A) Segregating progeny of transplastomic RIKT plants. Variegated seedlings consist of both cells with translating and cells with nontranslating plastids whereas white seedlings have completely lost the spectinomycin-resistance gene by recombination and thus bleach out in the presence of the drug. (B) Close-up showing three seedlings with different degrees of variegation. (C) Phenotype of a RIKT plant on spectinomycin-containing synthetic medium. The arrow points to a leaf with half of the leaf blade missing. (D) Wild-type (left) and two RIKT (right) plants transferred from spectinomycin-containing synthetic medium to the soil. (E) The same plants after 3 weeks of growth in the soil. Note appearance of normal leaves in the RIKT plants in the absence of spectinomycin. (F) A RIKT plant transferred from spectinomycin-containing medium to drug-free medium. The lower leaves developed in the presence of the antibiotic (marked with filled arrows) whereas the upper leaves grew in the absence of spectinomycin (empty arrows) and gradually became more wild-type-like. (G) Reexposure of the same plant to spectinomycin-containing medium. Bleaching of leaf sectors in those leaves that developed in the absence of spectinomycin suggests that the these cell lines would have ceased to divide in the presence of the antibiotic and caused the phenotypic aberrations seen in leaves of RIKT plants. (H) Abnormal flower morphology in RIKT plants. The arrows point to missing sectors in the petals.
RIKT Leads to Severe Defects in Leaf Development. When we selected variegated seedlings and continued to grow them on spectinomycin-containing medium, we observed severe morphological alterations in leaf shape (Figs. 2C and 3). Leaf phenotypes ranged from nearly normal leaves (which, however, were only rarely seen) over highly asymmetric leaves lacking parts of the leaf blade to needle-like leaves almost entirely lacking the leaf blade (Fig. 3). The appearance of deformed leaves was not specific to spectinomycin but also occurred when the plants grew in the presence of streptomycin, another antibiotic of the aminoglycoside type against which the aadA gene confers resistance (16).
Fig. 3.
Spectrum of leaf phenotypes in RIKT plants. (Scale bar: 2 cm.) Upper left corner shows a wild-type leaf.
Spectinomycin and streptomycin are antibiotics that, in plant cells, specifically inhibit plastid protein biosynthesis by binding to plastid 70S ribosomes. Single point mutations at specific sites in the plastid 16S rRNA are known to be sufficient to prevent antibiotic binding to plastid ribosomes, resulting in spontaneous spectinomycin-resistant (or streptomycin-resistant) plants (29). These plants are phenotypically indistinguishable from the wild type and develop normally even in the presence of high concentrations of the antibiotic, unambiguously demonstrating that the action of spectinomycin and streptomycin is confined to chloroplast protein biosynthesis. This finding excludes the possibility that these aminoglycoside antibiotics exert some side effects outside the chloroplast that indirectly could have cause the observed phenotype in leaf development.
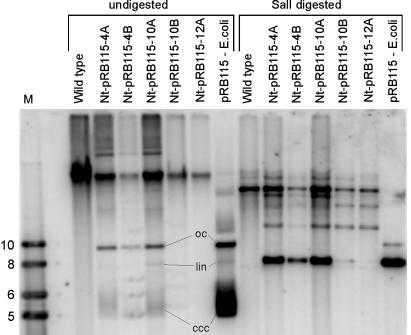
A reasonable explanation for the frequent formation of misshapen leaves is that cell lines in which RIKT has occurred and no aadA-containing plastid genome molecules are left cease to divide. This explanation would imply that leaf development is arrested in the absence of plastid translation. To test whether high-frequency excision of the aadA by homologous recombination is indeed occurring in RIKT plants, we assayed for the presence of recombined-out pRB115 plasmid in planta. DNA gel blot analyses with undigested or restriction-enzyme-digested DNA from individual Nt-pRB115 plants established that plasmid pRB115 is detected at high copy number in all those Nt-pRB115 plants that display a strong phenotype (Fig. 4). To ultimately confirm that the plasmid detected in planta is identical with the original transformation vector pRB115, we transformed E. coli with total plant DNA extracted from Nt-pRB115 plants followed by selection for ampicillin-resistant and spectinomycin-resistant clones. As expected, these experiments produced antibiotic-resistant clones at high frequency. Restriction mapping of the plasmids isolated from these bacterial clones confirmed their identity with pRB115 (data not shown).
Fig. 4.
Southern blot confirming active excision of pRB115 in vivo. Total cellular DNA from the wild type and five independently generated transplastomic plants was electrophoresed in a 0.7% agarose gel with or without prior digestion with SalI and probed with the radiolabeled insert from pRB115. Analysis of the undigested samples reveals presence of the identical conformations of pRB115 in planta as in E. coli (oc, open circular conformation; lin, linear conformation; ccc, covalently closed circular conformation = supercoiled plasmid). Digestion with SalI produces a prominent band of 7.6 kb as expected for linearized pRB115. Note that little or no excision by recombination has occurred in the Nt-pRB115–10B (faint 7.6-kb band) and Nt-pRB115–12A plants (virtual absence of the 7.6-kb band), which correlates with the lack of a strong phenotype in these plants. The strongly hybridizing fragment of 23 kb present in both the wild-type and transplastomic lines originates from hybridization to the psaA/B region of the chloroplast genome. Minor bands are likely to originate from plasmid multimers (31) and/or promiscuous plastid DNA present in the nuclear genome (21, 32). M, molecular weight marker (sizes indicated in kb).
Interestingly, plasmid pRB115 seems to adopt identical conformations in plastids and in E. coli with prominent bands for supercoiled, linear, and open circular conformations being readily detectable (Fig. 4). This finding may indicate that a similar set of topoisomerases acts in plastids and in bacterial cells.
Defects in Leaf Morphology Are Caused by the Absence of Plastid Translation. If the observed defects in leaf development were indeed caused by inhibited plastid translation, then shift of the plants to growth without spectinomycin should restore normal leaf development. Therefore, we transferred plants from antibiotic-containing medium to the soil and tested whether the newly formed leaves lack phenotypic abnormalities (Fig. 2 D and E). This was indeed the case: newly appearing leaves were phenotypically normal, demonstrating that, in the absence of spectinomycin, wild-type cells and RIKT cells are equally competent in leaf development (Fig. 2 D and E). Gradual phenotypic normalization was also seen in in vitro culture when RIKT plants were transferred to antibiotic-free medium (Fig. 2F).
Until now, the analysis of tobacco plant development in the absence of plastid gene expression had not been possible. Seed germination in the presence of inhibitors of plastid gene expression produces white seedlings that do not grow beyond the cotyledon stage and eventually die (see, e.g., figure 3 in ref. 26). Interestingly, white sectors seen in the cotyledons of variegated RIKT seedlings (Fig. 2 A and B) are not seen on newly developing leaves, which, instead of showing white sectors, lack parts of their leaf blades (Figs. 2C and 3). This finding is most likely explained with the development of the cotyledons taking place already during embryogenesis and seed maturation and, hence, in the absence of exposure to spectinomycin. In contrast, all true leaves develop after germination on spectinomycin-containing medium, suggesting that white sectors in the cotyledons are equivalent to missing sectors in true leaves. If this were the case, then phenotypically normal leaves developed in the absence of spectinomycin (Fig. 2F) should show sectorial bleaching on reexposure to spectinomycin. We therefore transferred the plants from drug-free medium (Fig. 2F) back to antibiotic-containing medium (Fig. 2G). As hypothesized, the phenotypically normal leaves displayed sectorial bleaching, and all newly developing leaves showed deformations with sectors of the leaf blade missing (Fig. 2G). This finding strongly suggests that the bleached sectors are equivalent to missing sectors in leaves developed under exposure to spectinomycin (in that the cell lines that give rise to white sectors would have ceased to divide in the presence of the antibiotic and caused the phenotypic aberrations seen in leaves of RIKT plants).
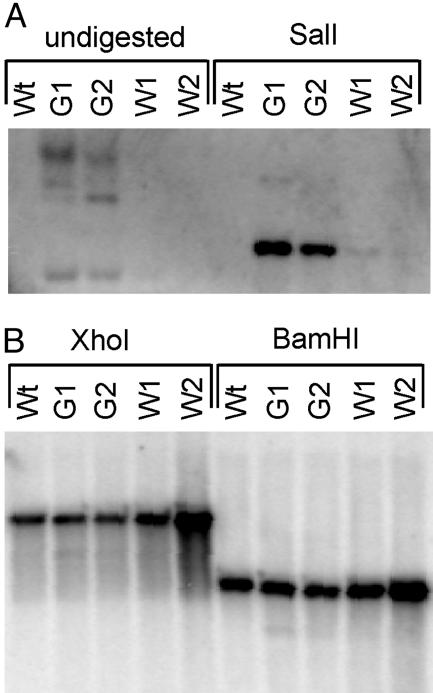
To ultimately confirm that the bleached sectors originate from recombinational loss of the aadA-resistance gene, we comparatively analyzed green and white sectors for the presence of the aadA by DNA gel blots. As expected, no aadA sequences were detectable in white sectors, whereas green sectors yielded strong hybridization signals (Fig. 5A). As a control, we assayed the same DNA preparations for the plastid psbA gene. This hybridization produced strong signals in all samples and detected restriction fragments of identical size as in the wild-type control (Fig. 5B). This experiment also excludes the possibility that the bleached sectors originate from recombination between the psbA-derived 3′ end of the chimeric aadA gene and the endogenous psbA locus. [Although such unwanted recombination events have never been observed with the short version of the psbA 3′ end used in this study, they were occasionally seen in plastid transformants produced with aadA gene versions carrying long psbA-derived 3′ ends (16).]
Fig. 5.
Recombinational loss of the aadA-resistance gene in bleached leaf sectors. Two green (G1 and G2) and two white (W1 and W2) leaf sectors from plants treated as shown in Fig. 2G were comparatively analyzed by DNA gel blot analyses. (A) Hybridization of undigested and SalI-digested DNA samples to an aadA-specific probe detects recombined-out pRB115 plasmids as well as aadA copies integrated into the plastid genome (compare with Fig. 4). (B) After digestion with XhoI or BamHI, hybridization to a psbA-specific probe detects identical fragments in green sectors, white sectors, and wild-type leaves (Wt). XhoI digestion produces a fragment of 8.5 kb; BamHI digestion generates a 4.8-kb fragment.
Cells in the bleached sectors were not dead as leaf explants taken from the white parts of the leaves regreened and produced normal shoots on antibiotic-free plant regeneration medium (data not shown).
The chimeric leaf structure revealed by occurrence of somatic variegation in our spectinomycin reexposure experiments (Fig. 2G) also demonstrates that RIKT acts cell-autonomously. The function of nontranslating plastids cannot be complemented by neighboring cells with translating plastids, making it unlikely that lack of synthesis of a transportable metabolite causes the mutant phenotype.
RIKT also Affects Development of Nonphotosynthetic Organs. Apart from a few ORFs of unknown function, most plastid gene products are involved in either photosynthesis or gene expression. Because photosynthesis is the main function of the plastid genome, we wanted to know whether the phenotypic abnormalities in RIKT plants were somehow linked to inhibited expression of plastid photosynthesis genes. We therefore analyzed flower morphology in RIKT plants grown on synthetic medium with spectinomycin. Phenotypic abnormalities were observed at high frequency (Fig. 2H) and, similar to the defects in leaf blade development, petal shape was severely altered with multiple sectors missing, demonstrating that, also in nonphotosynthetic organs, plastid protein biosynthesis is essential for development and cell division.
It seems conceivable that by influencing the rate of cell division, more subtle changes in plastid gene expression may have caused the plastome-dependent variation in leaf margin shape and hypanthium length of Oenothera flowers measured by Schwemmle more than 60 years ago (12, 13). Disturbed plastid gene expression also may explain altered leaf morphology seen in a heteroplasmic chloroplast protease subunit knockout (25).
In sum, we have provided evidence for an important extraplastidic function of plastid genes and gene expression in tobacco. It will be interesting to learn which plastid gene(s) and gene product(s) are directly involved in cell line survival during organ development. Requirement for a protein product encoded by a plastid gene seems to be the most plausible explanation for the observed plastid translation-dependent cell line survival. Systematic reverse genetics analyses of plastid-encoded genes and ORFs (e.g., refs. 20, 21, and 24 and reviewed in ref. 30) should ultimately identify these proteins and pave the way to studying the signaling mechanisms that link plastid gene expression to cell division and plant development.
Acknowledgments
We thank Dr. Jörg Kudla for helpful suggestions and critical reading of the manuscript. This research was supported by grants from the Deutsche Forschungsgemeinschaft to R.B.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: RIKT, recombination-induced knockout of translation.
References
- 1.Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., et al. (1996) DNA Res. 3, 109–136. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko, T. & Tabata, S. (1997) Plant Cell Physiol. 38, 1171–1176. [DOI] [PubMed] [Google Scholar]
- 3.Kotani, H. & Tabata, S. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 151–171. [DOI] [PubMed] [Google Scholar]
- 4.Wakasugi, T., Tsudzuki, T. & Sugiura, M. (2001) Photosynthesis Res. 70, 107–118. [DOI] [PubMed] [Google Scholar]
- 5.Jarvis, P. (2001) Curr. Biol. 11, 307–310. [DOI] [PubMed] [Google Scholar]
- 6.Rodermel, S. (2001) Trends Plant Sci. 6, 471–477. [DOI] [PubMed] [Google Scholar]
- 7.Escoubas, J. M., Lomas, M., LaRoche, J. & Falkowski, P. G. (1995) Proc. Natl. Acad. Sci. USA 92, 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfannschmidt, T. (2003) Trends Plant Sci. 8, 33–41. [DOI] [PubMed] [Google Scholar]
- 9.Kropat, J., Oster, U., Rüdiger, W. & Beck, C. F. (1997) Proc. Natl. Acad. Sci. USA 94, 14168–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strand, A., Asami, T., Alonso, J., Ecker, J. R. & Chory, J. (2003) Nature 421, 79–82. [DOI] [PubMed] [Google Scholar]
- 11.Larkin, R. M., Alonso, J. M., Ecker, J. R. & Chory, J. (2003) Science 299, 902–906. [DOI] [PubMed] [Google Scholar]
- 12.Schwemmle, J. (1941) Z. Indukt. Abstammungs-u. Vererbungslehre 79, 321–333. [Google Scholar]
- 13.Schwemmle, J. (1943) Flora 137, 61–72. [Google Scholar]
- 14.Rhoades, M. M. (1955) in Encyclopedia of Plant Physiology, ed. Ruhland, W. (Springer, Berlin), Vol. 1, pp. 19–57. [Google Scholar]
- 15.Hagemann, R. (1964) in Genetics Today: Proceedings of the XI International Congress of Genetics, The Hague, The Netherlands, ed. Geerts, S. J. (Pergamon, Oxford), pp. 613–625.
- 16.Svab, Z. & Maliga, P. (1993) Proc. Natl. Acad. Sci. USA 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svab, Z., Hajdukiewicz, P. & Maliga, P. (1990) Proc. Natl. Acad. Sci. USA 87, 8526–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle, J. J. & Doyle, J. L. (1990) Focus 12, 13–15. [Google Scholar]
- 19.Kanevski, I. & Maliga, P. (1994) Proc. Natl. Acad. Sci. USA 91, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruf, S., Kössel, H. & Bock, R. (1997) J. Cell Biol. 139, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hager, M., Biehler, K., Illerhaus, J., Ruf, S. & Bock, R. (1999) EMBO J. 18, 5834–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess, W. R., Hoch, B., Zeltz, P., Hübschmann, T., Kössel, H. & Börner, T. (1994) Plant Cell 6, 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zubko, M. K. & Day, A. (1998) Plant J. 15, 265–271. [DOI] [PubMed] [Google Scholar]
- 24.Drescher, A., Ruf, S., Calsa, T., Jr., Carrer, H. & Bock, R. (2000) Plant J. 22, 97–104. [DOI] [PubMed] [Google Scholar]
- 25.Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T. & Hashimoto, T. (2001) Plant Cell Physiol. 42, 264–273. [DOI] [PubMed] [Google Scholar]
- 26.Bock, R. (2001) J. Mol. Biol. 312, 425–438. [DOI] [PubMed] [Google Scholar]
- 27.Maliga, P. (2003) Trends Biotechnol. 21, 20–28. [DOI] [PubMed] [Google Scholar]
- 28.Bendich, A. J. (1987) BioEssays 6, 279–282. [DOI] [PubMed] [Google Scholar]
- 29.Svab, Z. & Maliga, P. (1991) Mol. Gen. Genet. 228, 316–319. [DOI] [PubMed] [Google Scholar]
- 30.Bock, R. & Hippler, M. (2002) Prog. Bot. 63, 106–131. [Google Scholar]
- 31.Staub, J. M. & Maliga, P. (1994) Proc. Natl. Acad. Sci. USA 91, 7468–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayliffe, M. A. & Timmis, J. N. (1992) Theor. Appl. Genet. 85, 229–238. [DOI] [PubMed] [Google Scholar]