Abstract
Hereditary hemochromatosis (HH) is an autosomal recessive disease characterized by iron accumulation in several organs, followed by organ damage and failure. The C282Y mutation in the HFE gene explains 80-90% of all diagnosed cases of HH in populations of northwestern European ancestry. Targeted disruption of the mouse Hfe gene (or introduction of the murine mutation analogous to the C282Y human mutation) produces a murine model of HH. Another mutation in the HFE gene, H63D, is more prevalent than C282Y. However, the physiological consequences of the H63D mutation (as well as C282Y/H63D compound heterozygosity) on iron homeostasis are less well established. To evaluate the phenotypic consequences of the C282Y/H63D and H63D/H63D genotypes, we produced H67D (corresponding to H63D in humans) and C294Y (corresponding to C282Y in humans) knock-in mice. H67D homozygous mice, C294Y homozygous mice, and H67D/C294Y compound heterozygous mice each demonstrated hepatic iron loading. Even on a standard diet, by 10 weeks of age, hepatic iron levels in mice of these three genotypes were significantly higher than those of wild-type littermates. The relative severity of hepatic iron loading was C294Y/C294Y > C294Y/H67D > H67D/H67D. We conclude that the H67D allele, when homozygous or combined with a more consequential mutation like C294Y, leads to hepatic iron loading. These observations indicate that the H67D mutation leads to partial loss of Hfe function and can contribute to murine HH.
Keywords: knock-in mice, C282Y mutation, H63D mutation, Cre-loxP
Hereditary hemochromatosis (HH) is an autosomal recessive disorder characterized by elevated transferrin saturations, hepatic iron overload, and variable clinical manifestations including liver cirrhosis, hypogonadism, heart failure, and arthropathy. Prevalence of mutations in the gene responsible for HH is estimated to be 2-5 per 1,000 in Caucasians. The gene mutated in HH, the hemochromatosis gene (HFE), encodes an MHC class I-like protein (1) that heterodimerizes with β2-microglobulin (β2M). Most cases of HH arise from a founder mutation in HFE (845G to -A) that results in the amino acid substitution C282Y and prevents the association of HFE with β2M (1-3). Functional significance of HFE-β2M complex in iron homeostasis was supported from the observations that HFE-β2M is physically associated with transferrin receptor 1 (TfR1) in human placenta (4), in crypt enterocytes of duodenum (5), and in cultured human cell lines (6, 7). The effect of HFE on transferrin-mediated iron uptake seems to depend on the particular cell type studied as well as the magnitude of expression of β2M (8).
Although the most common HFE mutation associated with HH is C282Y, the most common HFE mutation overall is H63D. Among Caucasian populations, the allele frequency of C282Y is 2-5% and that of H63D is 15-20% (9, 10). The H63D allele frequency has been reported to be as high as 30% in the Basque population of Spain (11). The H63D mutation seems to have evolutionarily predated C282Y and to have arisen on multiple haplotypes (12, 13). The contribution of the H63D mutation to HH has been an area of controversy. Individuals with HH have been reported who demonstrate compound heterozygosity for H63D and other HFE mutations, particularly C282Y (14). Nonetheless, normal iron parameters are observed in approximately half of individuals compound heterozygous for H63D and C282Y (15). Homozygosity for the H63D mutation is associated with increased serum transferrin saturations and ferritin levels in some populations (16-18). However, the majority of individuals homozygous for H63D have normal iron parameters. These observations raise the question as to whether the H63D mutation itself results in changes in iron parameters, or whether other associated risk factors are required. Parameters of iron homeostasis are strongly influenced by environmental (e.g., diet, alcohol consumption, inflammation) as well non-HFE genetic factors (19). Such confounding variables limit the ability to make definitive conclusions from human population studies when studying a low-penetrance genotype. For this reason, we generated a mouse model of the H63D orthologous mutation (H67D). To determine the consequences of compound heterozygosity, we also generated mice carrying the C282Y orthologous mutation (C294Y). We compared parameters of iron homeostasis in mice carrying one or two H67D alleles with wild mice, Hfe knock-out mice, and mice carrying one or two C294Y alleles.
Materials and Methods
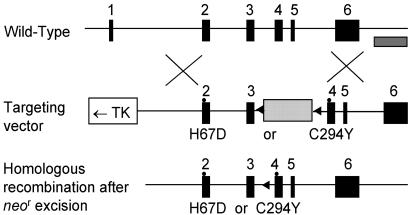
Site-Directed Mutagenesis and Targeting Vector Construction. The murine Hfe gene was isolated from a 129/SvJ mouse bacterial artificial chromosome (BAC) library as described (20). The 5′ 3.5-kb fragment and 3′ 2.8-kb fragment of the murine Hfe gene were subcloned into the pBS vector, containing exons 2 and 3 and exons 4 to 6, respectively. The mutation (underlined) was introduced into the appropriate fragment by using the mutagenic primers for H67D (199C to -G) in exon 2 (5′-TCTTTGTGTCCTACAATGATGAGAGTCGCCGTGCT-3′) and for C294Y (882G to -A) in exon 4 (5′-GACGAGACAAGGTTCACCTATCAAGTCGAGCACCCAGGCCTGC-3′). The H67D point mutation destroyed a BspHI restriction site whereas the C294Y mutation with an additional silent alteration (888G to -C) created a TaqI site. The presence of each point mutation and the exon 2-6 sequences in the targeting construct were confirmed by sequence analysis. The pPNT-lox2 vector containing a 3′ PGK neor cassette flanked by loxP sites and a 5′ thymidine kinase (TK) cassette was used as described (21, 22). The 5′ 3.5-kb fragment was introduced between the TK and neor genes of the pPNT-loxP2 vector. Next, the 3′ 2.8-kb fragment containing exons 4-6 of the Hfe gene was added downstream of the neor gene to create the complete targeting vector. This lox-neo-lox cassette can be eliminated by mating the heterozygotes with transgenic mice expressing the Cre recombinase enzyme (23). The final construct contained 3.5 kb and 2.8 kb of 5′ and 3′ homology of the Hfe gene, respectively, with each point mutation (Fig. 1).
Fig. 1.
Targeted mutagenesis of the Hfe gene. The structure of the endogenous gene, the targeting constructs, and the neo-excised allele are presented schematically on successive lines. Filled rectangles represent exons whereas an open rectangle and one dotted rectangle indicate thymidine kinase (TK) and the neor gene, respectively. The striped bar under the wild-type allele represents the probe used for Southern blots. The arrows show the loxP site.
Homologous Recombination in Embryonic Stem (ES) Cells and Generation of Germ-Line Chimeras. The targeting vector (25 μg) was linearized with NotI and introduced into the 129/Sv-derived ES cell line RW4 (Incyte Genomics Systems, St. Louis) (1 × 107 cells) by electroporation (230 V and 500 μF) in a Bio-Rad gene pulser. After 24 h, the cells were placed under selection with 200 μg/ml G418 (GIBCO/BRL) and 2 μM ganciclovir (Syntex Chemicals, Boulder, CO) for 6 days. ES cell colonies resistant to double selection were isolated, and isolated DNAs from ES cells were subjected to PCR and Southern blot analysis. The PCR method utilizes a forward primer in neor gene (LOXL (2)R: 5′-CTTCGTATAGCATACATTATACGAAGTTATGTCGA-3′) and a reverse primer in intron 6 outside the targeting sequences (HHPCRSc2R: 5′-TCATTTCATGCCACTCATATAGTCTGGTGGT-3′), which produces a 2.8-kb fragment in the mutant allele but no fragment in the wild-type allele. Genomic DNAs of the clones positive by PCR screening were digested with SacI, transferred, and hybridized with the 3′ probe (probe 900-bp), generating 6.0-kb fragments in the recombinants instead of 7.6-kb fragments in the normal allele (data not shown).
Two independent, targeted ES clones were injected into C57BL/6J blastocysts, and chimeric males were backcrossed for germ-line transmission to C57BL/6J females. The F1 mice were crossed with mice expressing Cre enzyme to remove the neor gene. The resultant neo-excised heterozygous mice were mated to produce homozygous mutant mice. This deletion leaves only the 34 bp of one loxP site in intron 3. Heterozygotes were either intercrossed for experimental use or backcrossed to C57BL/6 mice to put the mutation on a congenic background. Genotyping was performed by PCR analysis of DNA obtained by tail biopsies at 10 days. The resultant homozygous mice with H67D and C294Y point mutations were named as Hfetm(H67D)Stl (or Hfetm(H67D)Stl/tm(H67D)Stl) and Hfetm(C294Y)Stl (or Hfetm(C294Y)Stl/tm(C294Y)Stl), respectively, following the nomenclature recommended by The Jackson Laboratory (www.informatics.jax.org/mgihome/nomen/table.shtml).
We also bred the compound heterozygote of Hfetm(H67D)Stl/- and Hfetm(H67D)Stl/tm(C294Y)Stl mice by crossing Hfetm(H67D)Stl/+ with Hfe(+/-) or Hfetm(C294Y)Stl /+ mice. Mice were provided standard chow (Purina 5001) ad libitum that contains 0.02% iron. At 10 weeks of age, mice were fasted overnight and anesthetized before blood was collected by cardiac puncture and tissue samples were obtained. The studied populations consisted of 8 male and 7 female Hfetm(C294Y)Stl mice, 9 male and 10 female Hfetm(H67D)Stl mice, 8 male and 6 female Hfetm(H67D)Stl/tm(C294Y)Stl mice, 11 male and 13 female Hfe(-/-) knock-out mice, and 19 male and 12 female Hfetm(H67D)Stl/- mice. The studied control populations consisted of 8 male and 14 female Hfe(+/+) mice, 8 male and 5 female Hfe(+/-) mice, and 11 male and 9 female Hfetm(H67D)Stl/+ mice.
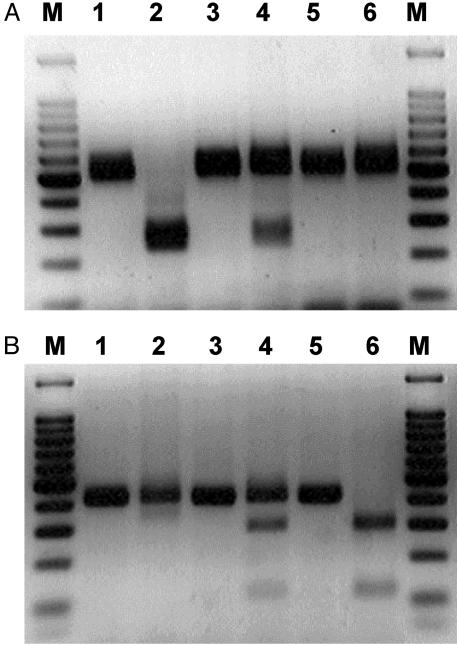
To amplify the murine Hfe gene, including the H67D mutation, a forward primer (mHHex2a: 5′-AGGACTCACTCTCTGGCAGCAGGAGGTAACCA-3′) and a reverse primer (mHHex2bR: 5′-TTTCTTTTACAAAGCTATATCCCCAGGGT-3′) were used, resulting in amplification of an ≈500-bp fragment. Digestion with BspHI revealed the 260- and 240-bp digested fragments for the wild-type allele and the uncleaved 500-bp PCR fragment for the mutant allele. To amplify the fragment of the murine Hfe gene containing the C294Y mutation, a forward primer (HH1: 5′-ATTCAGTTCTCTTCTCTGGAATAAGGCAG-3′) and a reverse primer (HH2R: 5′-AGCCCAGAGCAGACTACTCTCTCACTGCAAAGGTGAC-3′) were used. Digestion with TaqI showed the 128- and 289-bp fragments in the C294Y mutant allele and the uncleaved 417-bp PCR fragment in the wild-type allele (Fig. 2).
Fig. 2.
Detection of H67D and C294Y point mutations in the murine Hfe gene by genomic PCR amplification and subsequent BspHI and TaqI restriction site digestion. The H67D (CAT to GAT) destroys a BspHI site and C294Y (TGT to TAT) produces a TaqI new restriction site. Restriction enzyme analysis for each mutation introduced was performed by using homozygous, heterozygous, and normal control mice. (A) H67D mutation. Lane 1, undigested amplified PCR product from a normal control (500 bp); lane 2, DNA digested with BspHI from a normal control (240 and 260 bp); lane 3, undigested amplified PCR product from a heterozygote (500 bp); lane 4, DNA digested with TaqI from a heterozygote (240 bp, 260 bp, and 500 bp); lane 5, undigested amplified PCR product from a H67D homozygote (500 bp); lane 6, DNA digested with BspHI from a homozygote (500 bp). (B) C294Y mutation. Lane 1, undigested amplified PCR product from a normal control (417 bp); lane 2, DNA digested with TaqI from a normal control (417 bp); lane 3, undigested amplified PCR product from a heterozygote (417 bp); lane 4, DNA digested with TaqI from a heterozygote (128 bp, 289 bp, and 417 bp); lane 5, undigested amplified PCR product from a C294Y homozygote (417 bp); lane 6, DNA digested with TaqI from a homozygote (128 bp and 289 bp). The 100-bp markers were run at both ends of each gel.
Measurement of Iron Parameters. To determine the effect of each mutant allele on iron homeostasis, we analyzed F2-F4 mice of each genotype at 10 weeks of age that had been maintained on a standard chow diet (0.02% wt/wt iron) from weaning at age 3 weeks. Serum iron and total iron binding capacity (TIBC) were measured by using the protocol of Fielding (24). For the results shown in Fig. 3, 200 μl of serum from each animal was used for analysis of iron and TIBC using a kit from Sigma, and the assays were performed by Genox, by using a Cobas Fara II chemical analyzer. Transferrin saturation was calculated as (serum iron/TIBC) × 100%. Non-heme iron concentration in liver tissue was measured by the bathophenanthroline method as described (25), and the values were expressed as μg of iron per g of dry tissue.
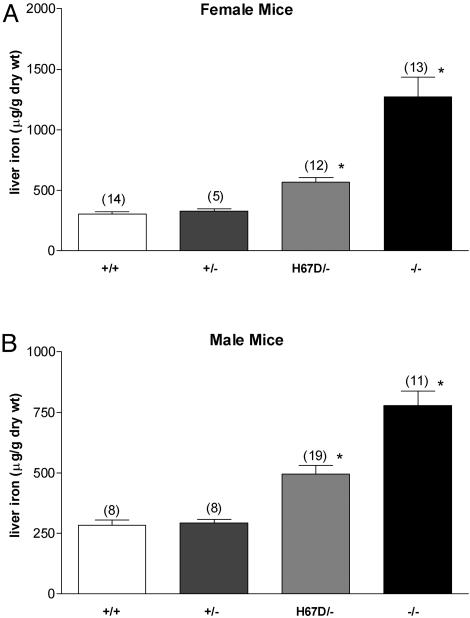
Fig. 3.
Effect of wild-type and H67D Hfe alleles on hepatic iron content. Non-heme iron concentrations were determined in liver specimens from female (A) and male (B) wild-type mice (+/+), mice carrying a single wild-type Hfe allele (+/-), mice carrying a single H67D Hfe allele (H67D/-), and Hfe knock-out mice (-/-). Numbers in parentheses represent number of mice analyzed. Bars represent mean ± SEM. *, P < 0.05 compared with wild-type mice of same sex by ANOVA followed by Dunnett's posttest analyses.
Histology of Liver Iron Deposition. Liver tissue samples were fixed in neutral-buffered 10% formalin for 18 h and subjected to routine histological processing. The sections were stained with Perls' Prussian blue followed by exposure to diaminobenzidine and counterstained with nuclear fast red. Iron distribution was determined by light microscopy.
Statistical Analysis. Because the consequences of functional loss of Hfe are greater in female than in male mice (26), each gender was analyzed separately. Mean values for transferrin saturations, and liver non-heme iron concentrations, were each compared across mice with different Hfe genotypes (but same gender) by one-way ANOVA, followed by Dunnet's posttest analyses. Liver iron concentrations were reciprocally transformed before ANOVA to account for unequal variability among the analyzed groups. P < 0.05 was considered statistically significant.
Results
Generation of Hfetm(H67D)Stl and Hfetm(C294Y)Stl Knock-In Mice. To introduce each point mutation in the Hfe gene in mouse ES cells, we designed a targeting vector with a total of 6.3 kb of homologous genomic sequence. The mutation was cotransferred with neor flanked by two loxP sites through homologous recombination in ES cells. After selection with G418 and ganciclovir, doubly resistant clones were screened for homologous recombination by PCR and Southern blotting hybridized with a 3′ external probe. The two targeting vectors gave a targeting efficiency of 10-20% of doubly resistant ES clones analyzed for homologous recombination. Of 96 clones screened by PCR, 10 for H67D and 20 for C294Y were homologous recombinant clones, which showed the 2.8-kb PCR fragment diagnostic of homologous recombination in one allele. The presence of the introduced point mutation in the targeted clones was analyzed by PCR amplification followed by restriction enzyme digestion. Three of 10 (30%) clones contained the H67D mutation (BspHI digestion); 10 of 20 (50%) clones contained the C294Y mutation (TaqI digestion).
Targeted ES cells containing one mutant allele were injected into C57BL/6 blastocysts, and chimeric males were obtained, followed by germ-line transmission of the mutant allele (F1). Heterozygous F1 offspring were independently intercrossed with C57BL/6 mice to generate F2 homozygous mice that were 129/Sv×C57BL/6 hybrids. The neor gene was then excised by using Cre-loxP recombination by crossing with Cre transgenic mice. This strategy generated mice heterozygous for the mutation, with an expected Mendelian segregation (wild-type and mutant 1:1) at birth. Two independent mouse lines for each mutation, derived from two separate ES cell clones, had identical, grossly normal phenotypes. All mice heterozygous and homozygous for the Hfe knock-in mutation appeared healthy, grew, reproduced normally, and produced homozygous offspring in the number expected. The colonies of Hfetm(H67D)Stl and Hfetm(C294Y)Stl mice were maintained by brother-sister matings and genotyped by PCR analysis of genomic DNA (Fig. 2). To confirm that Hfetm(H67D)Stl, Hfetm(C294Y)Stl, and Hfetm(H67D)Stl/tm(C294Y)Stl mice express the Hfe gene product, we performed Northern blot analyses on total RNA isolated from liver from mice of each genotype. The murine Hfe transcript of 1.9 kb, present in multiple tissues from +/+ mice, was present in normal amounts in all mutants (data not shown). Transcripts from Hfetm(H67D)Stl, Hfetm(C294Y)Stl, and Hfetm(H67D)Stl/tm(C294Y)Stl mice were amplified by RT-PCR and sequenced. To amplify mouse Hfe cDNA, including the H67D and C294Y mutation, a forward primer (HH17: 5′-GAAGACCGGTGGACCCAGCTGAGG-3′) and a reverse primer (HH13R: 5′-TCACTCACAGTCTGTTAAGACATA-3′) were used, resulting in amplification of a 1,080-bp fragment covering the whole coding region. Sequencing showed no alterations except the introduced nucleotide alterations on each construct (data not shown).
Consequences of a Single Wild-Type or H67D Hfe Allele on Hepatic Iron Levels. To characterize the consequences of the H67D allele on iron homeostasis, we generated mice containing a single functional Hfe allele, either wild-type or H67D. This result was accomplished by breeding Hfetm(H67D)Stl mice with Hfe knock-out mice. Mice were analyzed at 10 weeks of age, maintained on a standard diet. Because the consequences of functional loss of Hfe are greater in female than in male mice (26), each gender was analyzed separately. As previously reported, the Hfe knockout mice have significantly higher hepatic iron levels than wild-type mice. Mice containing a single wild-type allele had liver iron concentrations similar to mice with two wild-type alleles (Fig. 3). However, mice containing a single H67D allele had intermediate liver iron concentrations, higher than wild-type mice, but lower than mice with no functional Hfe allele.
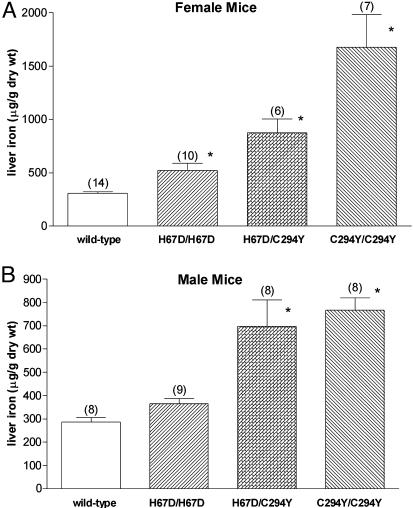
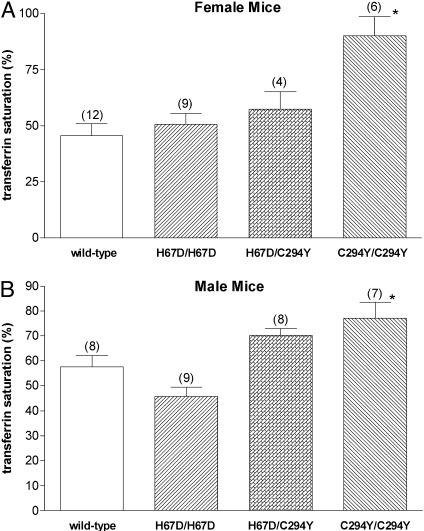
Iron Loading in Hfe H67D Homozygous and H67D/C294Y Compound Heterozygous Mice. We compared the function of the wild-type, H67D, and C292Y Hfe proteins by generating mice homozygous for the H67D allele, homozygous for the C294Y allele, or compound heterozygous for each allele. Mice heterozygous for the H67D or the C294Y allele had hepatic iron levels similar to that of wild-type mice (not shown). Female mice homozygous for either the H67D allele or the C294Y allele had significantly higher hepatic iron levels than did wild-type mice (Fig. 4A). The same trend was true for male mice homozygous for H67D, but the difference did not reach statistical significance. However, in both male and female mice, the severity of hepatic iron loading in H67D homozygotes was less than that observed in mice homozygous for C294Y or compound heterozygous for H67D and C294Y. The effects of the H67D and C294Y mutations on serum transferrin saturation were less marked than those observed on hepatic iron levels (Fig. 5). Thus, only the C294Y homozygous mice demonstrated statistically significant differences in transferrin saturation compared with wild-type mice (Fig. 5).
Fig. 4.
Hepatic iron content in mice carrying H67D and C294Y Hfe alleles. Non-heme iron concentrations were determined in liver specimens from female (A) and male (B) wild-type mice, mice homozygous for the H67D Hfe allele (H67D/H67D), mice compound heterozygous for the H67D and C294Y alleles (H67D/C294Y), and mice homozygous for the C294Y allele (C294/C294). Numbers in parentheses represent number of mice analyzed. Bars represent mean ± SEM. *, P < 0.05 compared with wild-type mice of same sex by ANOVA, followed by Dunnett's posttest analyses.
Fig. 5.
Transferrin saturations in mice carrying H67D and C294Y Hfe alleles. Serum transferrin saturations were measured in female (A) and male (B) wild-type mice, mice homozygous for the H67D Hfe allele (H67D/H67D), mice compound heterozygous for the H67D and C294Y alleles (H67D/C294Y), and mice homozygous for the C294Y allele (C294/C294). Numbers in parentheses represent number of mice analyzed. Bars represent mean ± SEM. *, P < 0.05 compared with wild-type mice of same sex by ANOVA, followed by Dunnett's posttest analyses.
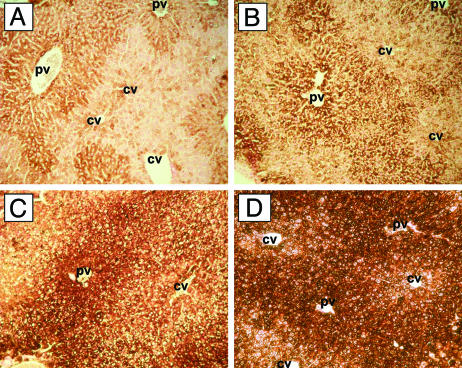
Histopathology of Hepatic Iron in Hfe Mutant Mice. Distribution of iron in liver sections from the Hfe mutant mice was examined by Perls' Prussian blue staining. Standard staining methods revealed little iron in either wild-type or H67D homozygous mice (not shown). However, when signal enhancement was performed by using diaminobenzidine, hepatocellular iron was visualized even in wild-type mice (Fig. 6A). Mice containing two Hfe mutant alleles demonstrated increased hepatocellular iron, with a predominantly periportal distribution (Fig. 6). The relative intensity of hepatocellular iron staining was as follows: wild-type < H67D/H67D << H67D/C294Y < C294Y/C294Y, in agreement with biochemical determination of liver iron content (Fig. 4A).
Fig. 6.
Enhanced Perls' Prussian blue staining of liver sections. Liver sections were stained with Perls' Prussian blue staining followed by diaminobenzidine enhancement, counterstained with nuclear fast red, and analyzed by light microscopy. By this technique, iron is represented by brown staining. Shown are wild-type (A), H67D/H67D (B), H67D/C294Y (C), and C294Y/C294Y (D). PV, portal vein; CV, central vein.
Discussion
Although the HFE mutation most commonly associated with a clinical diagnosis of HH is C282Y, the most common HFE mutation in the general population is H63D. Whether the H63D mutation can lead to consequential alterations in parameters of iron homeostasis has been controversial. Carella et al. (27) suggested three lines of evidence to support the argument that H63D is a polymorphic change rather than a pathologic mutation: (i) H63D attains a similar frequency in patients and controls, (ii) homozygotes are rare among patients, and (iii) compound heterozygotes have also been found in the normal population. In contrast, Beutler argued that the H63D is a disease-related mutation having very low clinical penetrance (28). Subsequent population studies have supported the argument that H63D has a mild effect on parameters of iron homeostasis (16-18). Scarce data are available on the effect of the H63D mutation on hepatic iron concentration. Nonetheless, individuals carrying the H63D mutation (usually in combination with another HFE mutation) seem to be at increased risk for hemochromatosis. The specific contribution of H63D to iron overload without other risk factors is difficult to assess from human population studies.
Biochemical studies on the HFEH63D protein have likewise been inconclusive. Although some studies examining the effects of expressed HFE in cell culture have found differences between HFEH63D and wild-type HFE (6, 29, 30), others have not (31, 32). A putative structural abnormality in HFEH63D was suggested by crystallography studies, demonstrating that H63 normally forms a salt bridge with a residue in the HFE α2 loop, which is involved in the interaction of HFE with TfR1 (33). This bridge would be predicted to be lost in HFEH63D. Nonetheless, both the wild-type and HFEH63D proteins, when expressed in cell culture, are capable of forming stable complexes with TfR1 (2). Furthermore, wild-type HFE and HFEH63D (but not HFEC282Y) are both able to interact with β2m and traffic normally to the cell surface (3). The histidine residue at this position is conserved in all species (human, mouse, rat, chimpanzee, and rhinoceros) in which HFE cDNA sequences have been submitted to GenBank to date.
Because the human population studies and ex vivo biochemical studies have not yielded consistent results regarding the consequences of the H63D mutation, we generated a mouse model carrying the analogous mutation H67D. These studies allowed us to determine the effect of the H67D allele on iron homeostasis independent of environmental risk factors that potentially confound human population studies. Our results demonstrate that the H67D allele in the mouse has measurable consequences on hepatic iron levels. The consequences of the H67D allele on iron homeostasis were significantly less than those of the C294Y allele, suggesting that the H63D mutation leads to only partial loss of Hfe function. Thus, these observations support those human population studies that suggested that homozygosity or compound heterozygosity for the H63D mutation can contribute to increased iron loading. In the mouse model, measurements of hepatic iron concentration make this conclusion even more convincing. This mouse model may also prove useful in determining the consequences of the H63D mutation on HFE-TfR interaction and on transferrin-mediated iron uptake when the mutant protein is expressed at physiologic levels in naturally expressing cell types.
Acknowledgments
We acknowledge Elizabeth Snella and Tammi Holmes (Transgenic Mouse Facility of the Saint Louis University/Cardinal Glennon Children's Hospital Pediatric Research Institute) and Rosemary O'Neill. This work was supported by National Institutes of Health Grants RO1 DK-53405 (to W.S.S.), RO1 HL-66225 (to R.E.F.), and RO1 DK-41816 (to B.R.B).
Abbreviations: HH, hereditary hemochromatosis; β2M, β2-microglobulin; TfR, transferrin receptor; ES cell, embryonic stem cell.
References
- 1.Feder, J. N., Gnirke, A., Thomas, W., Tsuchihashi, Z., Ruddy, D. A., Basava, A., Dormishian, F., Domingo, R., Jr., Ellis, M. C., Fullan, A., et al. (1996) Nat. Genet. 13, 399-408. [DOI] [PubMed] [Google Scholar]
- 2.Feder, J. N., Tsuchihashi, Z., Irrinki, A., Lee, V. K., Mapa, F. A., Morikang, E., Prass, C. E., Starnes, S. M., Wolff, R. K., Parkkila, S., et al. (1997) J. Biol. Chem. 272, 14025-14028. [DOI] [PubMed] [Google Scholar]
- 3.Waheed, A., Parkkila, S., Zhou, X. Y., Tomatsu, S., Tsuchihashi, Z., Feder, J. N., Schatzman, R. C., Britton, R. S., Bacon, B. R. & Sly, W. S. (1997) Proc. Natl. Acad. Sci. USA 94, 12384-12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkkila, S., Waheed, A., Britton, R. S., Feder, J. N., Tsuchihashi, Z., Schatzman, R. C., Bacon, B. R. & Sly, W. S. (1997) Proc. Natl. Acad. Sci. USA 94, 2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waheed, A., Parkkila, S., Saarnio, J., Fleming, R. E., Zhou, X. Y., Tomatsu, S., Britton, R. S., Bacon, B. R. & Sly, W. S. (1999) Proc. Natl. Acad. Sci. USA 96, 1579-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feder, J. N., Penny, D. M., Irrinki, A., Lee, V. K., Lebron, J. A., Watson, N., Tsuchihashi, Z., Sigal, E., Bjorkman, P. J. & Schatzman, R. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross, C. N., Irrinki, A., Feder, J. N. & Enns, C. A. (1998) J. Biol. Chem. 273, 22068-22074. [DOI] [PubMed] [Google Scholar]
- 8.Waheed, A., Grubb, J. H., Zhou, X. Y., Tomatsu, S., Fleming, R. E., Costaldi, M. E., Britton, R. S., Bacon, B. R. & Sly, W. S. (2002) Proc. Natl. Acad. Sci. USA 99, 3117-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burt, M. J., George, P. M., Upton, J. D., Collett, J. A., Frampton, C. M., Chapman, T. M., Walmsley, T. A. & Chapman, B. A. (1998) Gut 43, 830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg, K. K., Cogswell, M. E., Chang, J. C., Caudill, S. P., McQuillan, G. M., Bowman, B. A., Grummer-Strawn, L. M., Sampson, E. J., Khoury, M. J. & Gallagher, M. L. (2001) J. Am. Med. Assoc. 285, 2216-2222. [DOI] [PubMed] [Google Scholar]
- 11.de Juan, D., Reta, A., Castiella, A., Pozueta, J., Prada, A. & Cuadrado, E. (2001) Eur. J. Hum. Genet. 9, 961-964. [DOI] [PubMed] [Google Scholar]
- 12.Merryweather-Clarke, A. T., Pointon, J. J., Shearman, J. D. & Robson, K. J. (1997) J. Med. Genet. 34, 275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochette, J., Pointon, J. J., Fisher, C. A., Perera, G., Arambepola, M., Arichchi, D. S., De Silva, S., Vandwalle, J. L., Monti, J. P., Old, J. M., et al. (1999) Am. J. Hum. Genet. 64, 1056-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassanelli, S., Pignatti, E., Montosi, G., Garuti, C., Mariano, M., Campioli, D., Carbonieri, A., Baldini, E. & Pietrangelo, A. (2001) J. Hepatol. 34, 523-528. [DOI] [PubMed] [Google Scholar]
- 15.Koziol, J. A., Ho, N. J., Felitti, V. J. & Beutler, E. (2001) Clin. Chem. 47, 1804-1810. [PubMed] [Google Scholar]
- 16.Gochee, P. A., Powell, L. W., Cullen, D. J., Du, S. D., Rossi, E. & Olynyk, J. K. (2002) Gastroenterology 122, 646-651. [DOI] [PubMed] [Google Scholar]
- 17.Cogswell, M. E., Gallagher, M. L., Steinberg, K. K., Caudill PhD, S. P., Looker, A. C., Bowman, B. A., Gunter, E. W., Franks, A. L., Satten, G. A., Khoury, M. J., et al. (2003) Genet. Med. 5, 304-310. [DOI] [PubMed] [Google Scholar]
- 18.Njajou, O. T., Houwing-Duistermaat, J. J., Osborne, R. H., Vaessen, N., Vergeer, J., Heeringa, J., Pols, H. A., Hofman, A. & van Duijn, C. M. (2003) Eur. J. Hum. Genet. 11, 225-231. [DOI] [PubMed] [Google Scholar]
- 19.Fleming, R. E., Holden, C. C., Tomatsu, S., Waheed, A., Brunt, E. M., Britton, R. S., Bacon, B. R., Roopenian, D. C. & Sly, W. S. (2001) Proc. Natl. Acad. Sci. USA 98, 2707-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, X. Y., Tomatsu, S., Fleming, R. E., Parkkila, S., Waheed, A., Jiang, J., Fei, Y., Brunt, E. M., Ruddy, D. A., Prass, C. E., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 2492-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomatsu, S., Orii, K. O., Vogler, C., Grubb, J. H., Snella, E. M., Gutierrez, M. A., Dieter, T., Sukegawa, K., Orii, T., Kondo, N., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14982-14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomatsu, S., Orii, K. O., Vogler, C., Grubb, J. H., Snella, E. M., Gutierrez, M., Dieter, T., Holden, C. C., Sukegawa, K., Orii, T., et al. (2003) Hum. Mol. Genet. 12, 961-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marth, J. D. (1996) J. Clin. Invest. 97, 1999-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fielding, J. (1980) Methods Hematol. 1, 15-43. [Google Scholar]
- 25.Torrance, J. D. & Bothwell, T. H. Methods Hematol. 1, 90-115.
- 26.Sproule, T. J., Jazwinska, E. C., Britton, R. S., Bacon, B. R., Fleming, R. E., Sly, W. S. & Roopenian, D. C. (2001) Proc. Natl. Acad. Sci. USA 98, 5170-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carella, M., D'Ambrosio, L., Totaro, A., Grifa, A., Valentino, M. A., Piperno, A., Girelli, D., Roetto, A., Franco, B., Gasparini, P., et al. (1997) Am. J. Hum. Genet. 60, 828-832. [PMC free article] [PubMed] [Google Scholar]
- 28.Beutler, E. (1997) Am. J. Hum. Genet. 61, 762-764. [PMC free article] [PubMed] [Google Scholar]
- 29.Feeney, G. P. & Worwood, M. (2001) Biochim. Biophys. Acta 1538, 242-251. [DOI] [PubMed] [Google Scholar]
- 30.Drakesmith, H., Sweetland, E., Schimanski, L., Edwards, J., Cowley, D., Ashraf, M., Bastin, J. & Townsend, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 15602-15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy, C. N., Carlson, E. J., Anderson, E. L., Basava, A., Starnes, S. M., Feder, J. N. & Enns, C. A. (2000) FEBS Lett. 484, 271-274. [DOI] [PubMed] [Google Scholar]
- 32.Wang, J., Chen, G. & Pantopoulos, K. (2003) Biochem. J. 370, 891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebron, J. A., Bennett, M. J., Vaughn, D. E., Chirino, A. J., Snow, P. M., Mintier, G. A., Feder, J. N. & Bjorkman, P. J. (1998) Cell 93, 111-123. [DOI] [PubMed] [Google Scholar]