Abstract
We used electron tomography to directly visualize trilobed presumptive envelope (env) glycoprotein structures on the surface of negatively stained HIV type 1 (HIV-1) and simian immunodeficiency virus (SIV) virions. Wild–type HIV-1 and SIV virions had an average of 8–10 trimers per virion, consistent with predictions based on biochemical evidence. Mutant SIVs, biochemically demonstrated to contain high levels of the viral env proteins, averaged 70–79 trimers per virion in tomograms. These correlations strongly indicate that the visualized trimers represent env spikes. The env trimers were without obvious geometric distribution pattern or preferred rotational orientation. Combined with biochemical analysis of gag/env ratios in virions, these trimer counts allow calculation of the number of gag molecules per virion, yielding an average value of ≈1,400. Virion and env dimensions were also determined. Image-averaging analysis of SIV env trimers revealed a distinct chirality and strong concordance with recent molecular models. The results directly demonstrate the presence of env trimers on the surface of AIDS virus virions, albeit at numbers much lower than generally appreciated, and have important implications for understanding virion formation, virus interactions with host cells, and virus neutralization.
Despite substantial progress in characterizing the structures of the envelope (env) glycoproteins, surface env glycoprotein of Mr 120 (gp120) (SU) and transmembrane env gp41 or gp32 (TM) of HIV type 1 (HIV-1) and simian immunodeficiency virus (SIV) at the protein level (1–9), the actual structure of the native env on virions has remained elusive. Crystallographic studies of the HIV and SIV TM glycoproteins support a trimeric structure, as do structure–function analogies to other viruses with fusogenic env (1, 2, 10). The anti-HIV activity, including in vivo activity (11), of various fusion inhibitors whose mechanism of action is based on interference with the formation of the six-helix bundle form of TM is also consistent with a trimeric structure for the env on virions (reviewed in ref. 12). In biophysical studies of env purified from HIV and SIV virions, both sedimentation–equilibrium profiles and scanning transmission electron microscopy (EM) observations were also consistent with a trimeric organization (13, 14). However, although early EM studies provided tantalizing hints and led to much largely theoretical speculation (15–17), state-of-the-art EM techniques have not been applied to the direct elucidation of structural characteristics of env on actual HIV-1 and SIV particles.
To directly characterize the nature of env on the surface of HIV and SIV virions, we used negative-stain transmission EM analysis coupled with electron tomography and image processing. The performance and interpretation of these studies were facilitated by three recent developments. First, we have shown that treatment of retroviruses with the mild oxidizing agent 2,2′-dithiodipyridine (aldrithiol-2, AT-2) inactivates retroviral infectivity through preferential covalent modification of internal viral proteins while preserving the structural and functional properties of the viral env (18, 19). Use of AT-2-inactivated sucrose gradient-purified preparations of SIV and HIV-1 virions allowed safe specimen processing for EM studies while preserving authentic env structure. Second, we have recently established methods for quantitative analysis of viral proteins in purified HIV and SIV preparations, including quantitation of virion gag and SU and TM content, allowing calculation of average gag/env ratios on particles. Third, by using these methods, we have characterized numerous HIV and SIV preparations, in the process identifying SIV isolates that vary in their gag/env ratios, and by implication, in their average per-virion env content, by 10-fold. Taking advantage of these approaches, we directly visualized, characterized, and enumerated trimeric structures on the surfaces of virions of a panel of HIV-1 and SIV isolates, known to differ in their env content, while performing parallel quantitative biochemical analyses of the different viruses (20).
Materials and Methods
Viruses. All viruses were provided by the AIDS Vaccine Program, SAIC-Frederick, National Cancer Institute, Frederick, MD. Virus production and purification procedures were as described (20). All viruses were treated with AT-2 to eliminate infectivity while preserving env structure and function (18), followed by sucrose gradient purification to remove all residual AT-2 and to purify the inactivated virions.
Biochemical Analysis. Purified virus preparations were lysed with 8 M guanidine·HCl, and viral proteins were resolved by microbore HPLC methods. The identity of viral- and host-cell-derived proteins was confirmed by immunoblot and/or amino acid sequencing, and quantification was based on amino acid analysis of HPLC-purified proteins and calibrated immunoblots, as described (20).
EM of Whole Viruses. All work with viruses (before or after AT-2 inactivation) was conducted by using BSL-2+ facilities and procedures. Aliquots (20 μl) of AT-2-treated viruses (≈1.5–2 mg/ml total protein) were washed twice in 100 μl of PBS and fixed for 30 min in 20 μl of 2.5% glutaraldehyde/PBS at 4°C. Viruses were pelleted by using an Airfuge centrifuge (Beckman Coulter) at 100,000 × g for 10 min after each wash and fixation step. Viruses were resuspended in 20 μl of saline, and 3 μl was placed on carbon-covered grids for 30 sec. Excess virus and buffer were removed by capillary action, and 6 μl of 1% uranyl formate stain was immediately added and incubated for 60 sec (21). Excess stain was then removed, and the samples were allowed to air dry. The sample grids were screened at ×20,000, and electron micrographs were recorded at a nominal magnification of ×100,000 at 100 kV on a JEOL JEM 1200EX electron microscope.
Electron Tomography Data Collection and 3D Reconstruction of Viruses. Well prepared grids were subjected to electron tomographic analysis on a Philips (Eindhoven, The Netherlands) CM300 FEG electron microscope equipped with a goniometer, a Gatan Model 670 Ultrahigh tilt analytical holder (Gatan, Pleasanton, CA) and a Tietz Tem-Cam F224 slow scan charge-coupled device camera (2,048 × 2,048 pixels, Tietz Video, Gauting, Germany). Electron tomography techniques were used for the 3D data collection and reconstruction of viruses (22). By using a computerized automated tracking system (EM-MENU 3.0, TVIPS, Tietz) and a low-dose technique, we recorded each tomographic series over a tilt range of ± 70° at 4° intervals at an instrumental magnification of ×45,000. Reconstruction of 3D volumes from tilt series was performed by the weighted back-projection method by using a tomography software package (24, 25).§ The 3D image was obtained and imported into software package IMOD for interpretation (26).
Image Analysis and Modeling. Sequential images corresponding to multiple sections of the selected virions were viewed for characterization and quantification of viral surface env. Where indicated, multiple sections were summed to reduce noise and reinforce env image signal. env spikes protruding laterally from the equatorial sections of the virions (i.e., side views of env; see Fig. 2B) were used for env height measurements, and only those spikes with obvious trilateral symmetry (i.e., top and bottom views) were included for diameter measurements and image averaging. Because the cross-sectional diameter through the layers (i.e., distal, medial, proximal) of an env spike would be expected to differ, several sections through the 3D reconstruction of each env spike were measured, and the section with the largest diameter was selected for inclusion in the final analysis of the basic profile. To assess rotational orientation, we superimposed an asymmetric three-armed stick trimer over each well delineated env trimer, avoiding those near the edges. The angle formed between the 12:00 position (0°) on an artificially imposed (vertical) viral axis and the arm closest to it in the 12:00 to 4:00 position (0–120°) were then determined for comparison.
Fig. 2.
Representative 3D tomograms from virions with high and low env content. (A) Representative tomogram sections from a SIVmac239 virion with truncated TM glycoprotein and high env content, produced from Sup-T1-CCR5 cells. Trilobed propeller-like presumptive trimer structures are readily apparent (n ≈ 73). (B) Representative tomogram sections from an HIV-1 MN (wild-type) virion from Sup-T1-CCR5 cells (n ≈ 17 trimers). For each tomogram series, the top and bottom sections are in Upper Left and Lower Right, respectively. A representative env spike on the bottom face (black arrow) and projecting from an equatorial section (white arrow) of each virus type is indicated. A cluster of env spikes is located to the right of the bracket (Upper Left) of HIV-1 MN (B). (Bar = 100 nm.)
The virion size, trimer size, and center-to-center distance of adjacent trimers were measured by using the image analysis software, IMAGEPRO (www.mediacy.com). For trimer image averaging and image analysis, images from multiple sequential sections were sorted into top hemisphere or bottom hemisphere sets, brought into alignment with each other, and the two sets were independently averaged by using the SPIDER software package (27). Windowed env trimer images were first normalized by scaling to adjust the image pixel values to a mean of 1. Env trimers were then centered and masked for an alignment through classification (multiple reference alignment) (27). For this alignment procedure, the particles were first assigned to one of six classes by using k means clustering. The images in each class were then aligned by rotating, translating, and averaging, and the six class averages were taken as the multireferences for the realignment of all of the original windowed env trimers. The newly aligned particles were then reclassified, and class averages were calculated as the new references. All of the trimers were then realigned based on the new multiple references. The classification and multireference alignment processes were repeated until no further improvement was evident. The gp120 trimer model (6) was displayed by using the software programs O (28) and VIEWERLITE (www.accelrys.com), adjusted to scale, and superimposed on the averaged env images.
Results and Discussion
EM analysis was performed on HIV-1 and SIV virions known from biochemical analysis to vary in their env content. Consistent with previous observations (20), biochemical analysis of mutant SIV isolates with truncated transmembrane env (gp32™) showed increased levels of virion env content, whereas HIV samples and those SIV specimens having wild-type full-length TM env (gp41™) had lower levels of env (Fig. 1 and Table 1).
Fig. 1.
HPLC analysis of the SIV and HIV-1 viruses. Details for virus preparations are shown in Table 1. The viruses include two preparations of the same HIV-1 isolate (HIV-1MN) with a full-length TM and low env content, produced from MHC-II-positive (A) and MHC-II-negative (B) cells, one SIV (SIVmac239) with low env content (C), and two SIV preparations with truncated TM glycoproteins and high env content (SIVmac239/tail/CEMX174, D); and SIVmac239/SupT1, E). All show a 1:1 molar ratio of SU/TM. Peaks for viral gag proteins [p1, p6, p7/p8, matrix (MA), capsid (CA)] are labeled. Peaks for gp120SU and gp41/32™ are indicated in red and dark blue, respectively. Peaks containing cellular proteins of interest are indicated as follows: MHC-I, light blue; β2-microglobulin, turquoise; actin (for virus produced from MHC-II cells), lavender; actin and MHC-II (for virus produced from MHC-II+ cells), yellow and green.
Table 1. Characteristics of virions analyzed.
| Virus | Cell line* | gag/env | TM form | Estimated env trimers per virion† | EM counted env trimers per virion ± SD (range)‡ | Calculated gag per virion§ | Virus diameter¶, nm | Trimer diameter, nm |
|---|---|---|---|---|---|---|---|---|
| HIV-1 MN | CEM×174 | 42:1 | gp41 | 10–19 | 8 ± 2 (3–12) | 1,008 | 135.0 ± 17.4 | 11.7 ± 1.3 |
| HIV-1 MN | Sup-T1-CCR5 | 56:1 | gp41 | 7–14 | 10 ± 3 (5–17) | 1,512 | 116.9 ± 8.5 | 12.6 ± 0.9 |
| SIVmac239 | CEM×174 | 56:1 | gp41 | 7–14 | 9 ± 5 (2–17) | 1,344 | 128.0 ± 7.1 | 12.6 ± 1.4 |
| SIVmac239-tail∥ | CEM×174 | 6:1 | gp32 | 67–133 | 79 ± 12 (61–96) | 1,422 | 134.4 ± 23.0 | 12.3 ± 1.2 |
| SIVmac239** | Sup-T1-CCR5 | 8:1 | gp32 | 50–100 | 70 ± 9 (52–84) | 1,680 | 121.1 ± 22.6 | 11.6 ± 0.8 |
| Overall mean ± SD | 1,393 ± 249 | 126.5 ± 16.4 | 12.4 ± 1.3 |
Cell lines in which viruses were propagated. Virus from CEM×174 cells contained MHC-II, and virus from SupT1-CCR5 cells contained no detectable MHC-II molecules
Trimer estimation based on HPLC, amino acid analysis, and immunoblot analysis, assuming 1,200–2,400 gag molecules per virion (20)
Trimer estimation based on direct counting from 3D EM tomograms, mean value ± SD, for 10–15 independent virions counted for each different virus sample
Gag molecule per virion values calculated based on EM determined per virion trimer counts and biochemically determined gag:env molar ratios for each virus preparation
From membrane surface to membrane surface (excluding env spikes)
SIVmac239-tail is a variant of SIVmac239 with a truncated TM env glycoprotein (gp32) and increased virion env content
Propagation of SIVmac239 in Sup-T1-CCR5 cells resulted in truncation of the transmembrane glycoprotein and increased levels of virion env glycoprotein content
To determine the contribution of MHC class II (MHC-II) molecules incorporated during virus assembly and budding (29) to the number of projections on the virion surface observed by EM, the viruses studied were propagated in cells that either did (CEMx174) (30) or did not (Sup-T1-CCR5) (31) express MHC-II molecules, and comparative EM analyses were performed on samples of the same virus isolate produced from the MHC-II-positive and -negative cells.
For EM examination, observers were blinded to the identity of virus samples and expectations of virion env content based on biochemical analyses. Initially, we examined an SIV isolate (SIVmac239-tail) for which biochemical evidence indicated a high level of env content. By EM analysis, we found multiple molecular masses, many displaying trilobed symmetric propeller-like features, covering the surface of the virions (see Fig. 4, which is published as supporting information on the PNAS web site). Because each of these images represents a projection through the entire virion, the structures on the tops and bottoms of the virions were superimposed, interfering with detailed analysis. We thus examined subsequent preparations by using multiple EMs of each virion taken at various tilt angles, which we subjected to tomographic analysis to yield 3D images.
By viewing individual sections through the reconstructed 3D image of each virion, we could independently analyze env spikes residing on the top, middle, and bottom of the particle. Fig. 2A shows representative sequential sections through a tomogram of a virion of SIVmac239 produced from Sup-T1 cells, a virus preparation characterized by a truncated TM (gp32™), and high virion env content based on biochemical analyses (Fig. 1, Table 1). Because each tomogram is constructed from multiple EM images of the same virion, fine details such as the shape of the putative env trimers are more readily apparent compared to 2D projection images. Individual presumptive env spikes can be clearly identified and display an obvious symmetric three-part propeller-like structure. These direct EM observations provide compelling support for predictions from various indirect analyses that HIV-1 and SIV env are likely to be present as trimers on the surface of virions.
Given quantitative biochemical data indicating that virions of different SIV isolates can vary in their env content by as much as 10-fold (20), we were also interested to directly assess by EM tomography the numbers of trimeric structures on the surface of virions of different virus isolates. Correlation between EM-determined trimer numbers and biochemically determined virion env content would support the idea that the trimeric propeller-like structures do indeed represent env trimers. Considerably fewer trimers were observed by EM tomography on wild-type HIV-1 and SIV compared to SIVs with truncated TM (Table 1, Fig. 2B; Fig. 5, which is published as supporting information on the PNAS web site). SIVs with high env content showed an average of 70–79 (range, 52–96) trimers per virion by EM analysis. HIV-1 virions and SIV virions with full-length TM showed an average of 8–10 trimers per virion (range 2–17) by EM. There was an excellent correlation between the numbers of trimers observed by EM and levels of relative virion env content determined by quantitative biochemical analysis. The presence of MHC-II molecules in virions did not influence the number of trimers observed for a given virus isolate (Table 1). Although Table 1 shows EM and quantitative biochemical results for preparations of HIV-1MN, an X4 HIV-1 produced from two different T cell lines, our previous biochemical analyses showed comparable levels of relative virion env content for several other X4 and R5 HIV-1 isolates produced from T cell lines (20).
Direct EM enumeration of trimers per virion, in combination with determination of the ratio of gag/env proteins by biochemical methods (20) for the different virus preparations, allowed calculation of the number of gag molecules per virion, yielding an average value of 1,393 ± 249 (SD) (Table 1). The number of env trimers per virion, determined by counting env trimers on EM images, showed good agreement with previous estimates, based on biochemical determination of gag/env ratios and assuming 1,200–2,400 gag molecules per virion, for all viruses evaluated (20) (Table 1).
EM analyses also allowed determination of virion size. No distinct or consistent difference in diameter was noted between the virions of the different SIV and HIV isolates analyzed, regardless of the host cells used for their production. The overall size distribution (average 126.5 nm ± 16.4, excluding env spikes; Table 1; Fig. 6 A and B, which is published as supporting information on the PNAS web site) is consistent with prior estimates (32) but may slightly overestimate actual virion size due to sample processing related factors (32).
Exploiting the opportunity to clearly visualize env trimers on the surface of virions with defined properties, we also evaluated trimer center-to-center distances, surface distribution patterns, and relative orientations, factors that may reflect the underlying structural geometry of the virions. To maximize the ability to detect patterns of env distribution and orientation, we summed multiple sections representing a given hemisphere of each selected virion so that all hemispheric env trimers would be displayed in one section. Env axial dots were applied to the EMs to aid in distance measurements and pattern recognition (see Fig. 7, which is published as supporting information on the PNAS web site). For SIV high env mutants, the center-to-center distance measurements show a Gaussian distribution centering ≈16 nm and ranging from 10 to 24 nm (Fig. 6 C and D). The env trimers appeared to be fairly evenly distributed and regularly spaced, although a distinct geometric pattern was not obvious (see Fig. 7); patterns of distribution of env trimers on the virion surface might be more apparent on immature particles, before cleavage of the gag polyprotein precursor (33). No obvious pattern of rotational orientation was observed (see Fig. 7). For wild-type SIV and HIV-1, the trimers appeared randomly dispersed, with most isolated and a few in clusters (Figs. 2B and 5).
The ability to visualize env trimers in their native state afforded the opportunity to analyze some of their basic structural features, including the diameter of the SU trimer array, its basic profile, and its mass density distribution. The height of the env spikes was determined to be 8.6 ± 1.0 nm (n = 55), comparable to that reported Gelderblom et al. (15) (9 nm), but greater than observed by Grief et al. (32) (6 nm). The thickness of the presumptive SU portion is 6.0 ± 0.8 nm, which compares well with the molecular models of Kwong et al. (6) (4–6 nm for trimer core and 5–7 nm for glycosylated trimer). The height of the exposed TM ectodomain stalk was 2.6 nm, and the diameter was 5.6 ± 0.7 nm. The diameter measurements for TM are somewhat greater than the dimension reported for trimeric HIV gp41 crystal and NMR structures (≈3.5 nm) (10, 34, 35). The diameter of env SU trimers in our EMs (12.4 ± 1.3 nm, Fig. 6 E and F, Table 1) is close to the size predicted based on previous EM estimates of 10 nm (17, 32) to 15 nm (15, 36) and the molecular modeling estimates from x-ray crystallography data of the gp120 protein core structure (11 nm) and glycosylated protein (15 nm) (4, 37). These values are all somewhat less than those reported for scanning transmission EM measurements of purified HIV and SIV env trimers (mean of 15.4–22.7 nm in published figures (figure 3 in ref. 13, figure 2C in ref. 14).
Fig. 3.
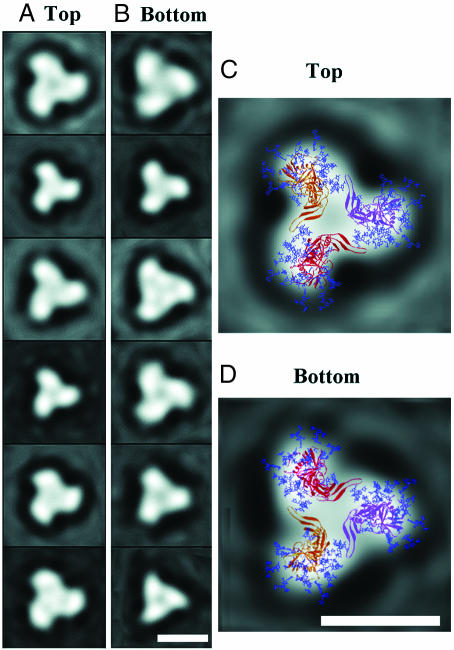
Image-averaged and model-fitted SIV env trimers. Individual trimer images from the top (A) and bottom (B) halves of SIV virions with truncated TM glycoproteins and high env content (SIVmac239/Sup-T1-CCR5 and SIVmac239-tail/CEMx174) were classified, rotated, aligned, and averaged. Trimers were sorted into six categories based on their similarity, aligned to each other by rotation and translation in each category, and then averaged. From top to bottom, the images are from the average of 23, 23, 17, 15, 13, and 8 (A) and 25, 16, 15, 9, 8, and 5 (B) individual trimers, respectively. The glycosylated env trimer molecular model of Kwong et al. (6) was superimposed on the trimer profiles for the corresponding top (as viewed from the outside of the virion) (C) and bottom (as viewed from the inside of the virion) (D) averaged images. The averaged images selected for comparison were both the largest in profile and represented the sets with greatest number of images (i.e., the top image from A and B). (Bars = 10 nm.)
Trimer tomograms were also subjected to computational alignment and image averaging in an attempt to discern additional detail. Unlike the diameter and orientation measurements, multiple individual sections from each env spike were included in this analysis to sample throughout the z axis. Each image was also self-aligned in the 0°, 120°, and 240° rotational orientation before sorting into classes. The env spikes from the tops and bottoms of each virion were analyzed independently, because these two classes would display opposite chirality if present. The results demonstrate some heterogeneity when sorted and averaged but reveal three consistent features (Fig. 3 A and B). First, most of the images show pronounced protrusions of mass from the threefold axes of symmetry, consistent with the previously noted propeller-like morphology observed for the individual unprocessed trimer images. Second, the central region appears less biochemically dense (i.e., displaces less stain) than the laterally projecting arms. The degree of contact between the gp120 SU subunits in situ is unknown, although biochemical evidence and modeling suggest at least some interaction (6). A final observation is that there appears to be a degree of chirality to most of the image averages in each hemispheric set. Significantly, the top views (i.e., as viewed from the outside of the virus) of SIV env show a counterclockwise inflection of the distal ends of the SU subunits (Fig. 3A), and the bottom views (i.e., as viewed from the viral membrane) show a clockwise inflection (Fig. 3B). Thus, the chiral nature of these data is internally consistent and not likely to derive from a sample preparation or image-processing artifact. To compare our averaged images to a recent molecular model of the HIV core structure with added glycosylation (6), we superimposed the two, demonstrating remarkably good agreement in mass distribution, including top/bottom chirality, between the predicted and directly visualized structures (Fig. 3 C and D).
Direct EM enumeration of per virion env trimer content, along with biochemical determination of the molar ratio of gag/env in different viruses, allows estimation of the number of gag molecules per virion, yielding an average value of ≈1,400 molecules per particle for the viruses studied. These values for per virion env and gag content are broadly consistent with at least some earlier estimates of per particle gag and env trimer content for HIV-1 based on indirect assessments (38, 39), although the level of virion env trimer content has remained controversial (15, 17, 20, 38, 40, 41). It should be noted that these earlier estimates of env trimer number were based on the assumption of incorporation of larger amounts of env into virions, followed by shedding of gp120, leaving a residual virion gp120 content corresponding to ≈10 trimers per virion. However, we recently demonstrated that env incorporation, not shedding of gp120, is the primary determinant of virion env content, because such virions contain approximately equimolar amounts of gp120SU and gp41™, whereas appreciable shedding of gp120SU would result in a molar excess of membrane-embedded gp41™ relative to the gp120SU remaining virion-associated after any shedding. In aggregate, the available data indicate that most HIV-1 and SIV virions with full length TM glycoproteins are formed with only ≈10 trimers. Both the relevant mechanisms involved in morphogenesis of virions with limited env content and the implications for interactions of such virions with target cells and the defense mechanisms of the infected host will be important areas for future study.
Considerable heterogeneity was observed between individual virions and trimers for both HIV-1 and SIV, with no apparent preferred rotational orientation or pattern of distribution of trimers across the virion surface. The truncated TM env of the mutant SIV viruses studied may be less constrained for lateral diffusion in the viral envelope membrane compared with full-length TM molecules, based on interactions of the cytoplasmic tail of the latter with the viral matrix protein (42). Nevertheless, the env on these virions were rather uniformly dispersed as although constrained by interaction with the matrix. It may be that the unavoidable deformations resulting from binding to the carbon film and subsequent drying during EM preparation have obscured more subtle patterns. The observation that wild-type HIV-1 and SIV isolates with full-length TM displayed low numbers of env trimers distributed across the surface of the virions suggests that very limited numbers of env interactions with cellular receptors may be sufficient to mediate infection. Although wide spacing of limited numbers of trimers seems difficult to reconcile with models that postulate a requirement for cooperative binding of multiple trimers to mediate infection (7), the low particle infectivity ratio of retroviruses, including HIV-1 (23), leaves open the possibility that a small fraction of virions, potentially with much higher env content per particle or those bearing stochastically or structurally fostered microclusters of env, might be responsible for most or all of the infectivity in virus preparations. Depending on their frequency, particles with high env content, if present, might not have been sampled in the analyses reported here. Some wild-type virions did appear to have several env trimers in close proximity.
In summary, we have used detailed EM tomography analysis to study the env on the surface of individual virions in HIV-1 and SIV preparations having a range of biochemically determined env content. We directly visualized and enumerated trilobed propeller-like structures, supporting the idea that env does indeed exist in trimeric form on the surface of virions. HIV-1 and SIV isolates with full-length TM glycoproteins had an average of only 8–10 env trimers per virion, whereas SIV isolates with truncated TM glycoproteins had an average of 70–79 env trimers per virion. The striking correlation between trimers per virion enumerated by EM and the relative env content of virions based on quantitative biochemical methods, across a panel of defined viruses varying by nearly 10-fold in env content, strongly supports the idea that the trimeric structures visualized by EM are indeed env trimers.
The results provide insights into the native structure of env on the surface of HIV-1 and SIV virions and have important implications for the development of vaccines and antiviral therapeutics targeted to the viral env. To date, advanced 3D EM analysis methods have been most productively applied to non-enveloped viruses, whose consistent underlying geometry lends itself to statistical image averaging approaches. However, the current studies demonstrate the feasibility of applying such methods to the study of more variable enveloped viruses to allow detailed structural analysis of env trimers on the surface of AIDS virus virions. Extension of these and related approaches, such as cryo-EM, to the analysis of additional virus isolates produced from primary cells and analysis of virions complexed with neutralizing antibodies and other env-directed antiviral reagents should allow even more precise definition of structural parameters relevant to both the understanding of virion morphogenesis and the optimization of treatment and vaccine approaches.
Supplementary Material
Acknowledgments
We thank Peter Kwong (Vaccine Research Center, National Institute of Allergy and Infectious Diseases) for providing the env trimer model and Kimberly Riddle and Jon Ekman of the Imaging Resources Facility at Florida State University for excellent technical support. This work was supported by National Institutes of Health Grants R21 AI 44291 and R01 AI 055461 (to P.Z. and K.H.R.), GM 30598 (to K.A.T.), and in part by federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-124000.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EM, electron microscopy; env, envelope; SU, surface env gp120; TM, transmembrane env gp41 or gp32; HIV-1, HIV type 1; SIV, simian immunodeficiency virus; AT-2, aldrithiol-2; MHC-II, MHC class II.
Footnotes
Winkler, H., Tang, J., Taylor, D. W. & Taylor, K. A. (1998) Biophys. J. 74, A45.
References
- 1.Blacklow, S. C., Lu, M. & Kim, P. S. (1995) Biochemistry 34, 14955-14962. [DOI] [PubMed] [Google Scholar]
- 2.Lu, M., Blacklow, S. C. & Kim, P. S. (1995) Nat. Struct. Biol. 2, 1075-1082. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt, R., Kwong, P. D., Desjardins, E., Sweet, R. W., Robinson, J., Hendrickson, W. A. & Sodroski, J. G. (1998) Nature 393, 705-711. [DOI] [PubMed] [Google Scholar]
- 4.Kwong, P. D., Wyatt, R., Robinson, J., Sweets, R. W., Sodroski, J. & Hendrickson, W. A. (1998) Nature 393, 648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyatt, R. & Sodroski, J. G. (1998) Science 280, 1884-1888. [DOI] [PubMed] [Google Scholar]
- 6.Kwong, P. D., Wyatt, R., Sattentau, Q. J., Sodroski, J. & Hendrickson, W. A. (2000) J. Virol. 74, 1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhmann, S. E., Platt, E. J., Kozak, S. L. & Kabat, D. (2000) J. Virol. 74, 7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poignard, P., Saphire, E. O., Parren, P. & Burton, D. (2001) Annu. Rev. Immunol. 19, 253-274. [DOI] [PubMed] [Google Scholar]
- 9.Sanders, R. W., Vesanen, M., Schuelke, N., Master, A., Schiffner, L., Kalyanaraman, R., Paluch, M., Berkhout, B., Olson, W. C., Lu, M., et al. (2002) J. Virol. 76, 8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 11.Kilby, J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302-1307. [DOI] [PubMed] [Google Scholar]
- 12.Chan, D. C. & Kim, P. S. (1998) Cell 93, 681-684. [DOI] [PubMed] [Google Scholar]
- 13.Center, R., Leapman, R., Lebowitz, J., Arthur, L., Earl, P. & Moss, B. (2002) J. Virol. 76, 7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Center, R. J., Schuck, P., Leapman, R. D., Arthur, L. O., Earl, P. L., Moss, B. & Lebowitz, J. (2001) Proc. Natl. Acad. Sci. USA 98, 14877-14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelderblom, H. R., Hausmann, E. H. S., Ozel, M., Pauli, G. & Koch, M. A. (1987) Virology 156, 171-176. [DOI] [PubMed] [Google Scholar]
- 16.Ozel, M., Pauli, G. & Gelderblom, H. R. (1988) Arch. Virol. 100, 255-266. [DOI] [PubMed] [Google Scholar]
- 17.Gelderblom, H. R., Ozel, M. & Pauli, G. (1989) Arch. Virol. 106, 1-13. [DOI] [PubMed] [Google Scholar]
- 18.Rossio, J. L., Esser, M. T., Suryanarayana, K., Schneider, D. K., Bess, J. W., Jr., Vasquez, G. M., Wiltrout, T. A., Chertova, E., Grimes, M. K., Sattentau, Q., et al. (1998) Virology. 72, 7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthur, L. O., Bess, J. W., Jr., Chertova, E. N., Rossio, J. L., Esser, M. T., Benveniste, R. E., Henderson, L. E. & Lifson, J. D. (1998) AIDS Res. Hum. Retroviruses 14, 8311-8319. [PubMed] [Google Scholar]
- 20.Chertova, E., Bess, J. W., Jr., Crise, B. J., Sowder, R. C., II, Schaden, T. M., Hilburn, J. M., Hoxie, J. A., Benveniste, R. E., Lifson, J. D., Henderson, L. E., et al. (2002) J. Virol. 76, 5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roux, K. H. (1989) Methods Enzymol. 178, 130-144. [DOI] [PubMed] [Google Scholar]
- 22.Radermacher, M. (1992) in Electron Tomography, ed. Frank, J. (Plenum, New York), pp. 91-115.
- 23.Piatak, M., Saag, M. S., Yang, L. C., Clark, S. J., Kappes, J. C., Luk, K. C., Hahn, B. H., Shaw, G. M. & Lifson, J. D. (1993) Science 259, 1749-1754. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, K. A., Schmitz, H., Reedy, M. C., Goldman, Y. E., Franzini-Armstrong, C., Sasaki, H., Tregear, R. T., Poole, K., Lucaveche, C., Edwards, R. J., et al. (1999) Cell 99, 421-431. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, K. A., Tang, J., Cheng, Y. & Winkler, H. (1997) J. Struct. Biol. 120, 372-386. [DOI] [PubMed] [Google Scholar]
- 26.Kremer, J., Mastronarde, D. & McIntosh, J. (1996) J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- 27.Frank, J. (1996) in Three-Dimensional Electron Microscopy of Macromolecular Assemblies (Academic, New York), pp. 1-341.
- 28.Jones, T., Zou, J.-Y., Cowan, S. & Kjeldgarrd, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 29.Arthur, L. O., Bess, J. W., Jr., Sowder, R. C., Jr., Benveniste, R. E., Mann, D. L., Chermann, J. C. & Henderson, L. E. (1992) Science 258, 1935-1938. [DOI] [PubMed] [Google Scholar]
- 30.Ting, J. P., Carrington, M. N., Salter, R. D., DeMars, R. & Cresswell, P. (1985) Immunogenetics 22, 571-583. [DOI] [PubMed] [Google Scholar]
- 31.Means, R. E., Matthews, T., Hoxie, J. A., Malim, M. H., Kodama, T. & Desrosiers, R. C. (2001) J. Virol. 75, 3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grief, C., Hockley, D. J., Fromholc, C. E. & Kitchin, P. A. (1989) J. Gen. Virol. 70, 2215-2219. [DOI] [PubMed] [Google Scholar]
- 33.Schatzl, H., Gelderblom, H. R., Nitschko, H. & von der Helm, K. (1991) Arch. Virol. 120, 71-81. [DOI] [PubMed] [Google Scholar]
- 34.Caffrey, M., Cai, M., Kaufman, J., Stahl, S. J., Wingfield, P. T., Covell, D. G., Gronenborn, A. & Clore, G. M. (1998) EMBO J. 17, 4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caffrey, M. (2001) Biochhim. Biophys. Acta 1536, 116-122. [DOI] [PubMed] [Google Scholar]
- 36.Forster, M. J., Mulloy, B. & Nermut, M. V. (2000) J. Mol. Biol. 298, 841-857. [DOI] [PubMed] [Google Scholar]
- 37.Kwong, P. D., Wyatt, R., Majeed, S., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (2000) Structure (London) 8, 1329-1339. [DOI] [PubMed] [Google Scholar]
- 38.Layne, S. P., Merges, M. J., Dembo, M., Spouge, J. L., Conley, S. R., Moore, J. P., Raina, J. L., Renz, H., Gelderglom, H. R. & Nara, P. L. (1992) Virology 189, 695-714. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien, W. A., Mao, S. H., Cao, Y. & Moore, J. P. (1994) J. Virol. 68, 5264-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider, J., Kaaden, O., Copeland, T. D., Oroszlan, S. & Hunsmann, G. (1986) J. Gen. Virol. 67, 2533-2538. [DOI] [PubMed] [Google Scholar]
- 41.Fu, Y.-K., Hart, T. K., Jonak, Z. L. & Bugelski, P. J. (1993) J. Virol. 67, 3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freed, E. O. & Martin, M. A. (1996) J. Virol. 70, 341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.