Abstract
Leprosy, which has afflicted human populations for millenia, results from infection with Mycobacterium leprae, an unculturable pathogen with an exceptionally long generation time. Considerable insight into the biology and drug resistance of the leprosy bacillus has been obtained from genomics. M. leprae has undergone reductive evolution and pseudogenes now occupy half of its genome. Comparative genomics of four different strains revealed remarkable conservation of the genome (99.995% identity) yet uncovered 215 polymorphic sites, mainly single nucleotide polymorphisms, and a handful of new pseudogenes. Mapping these polymorphisms in a large panel of strains defined 16 single nucleotide polymorphism-subtypes that showed strong geographical associations and helped retrace the evolution of M. leprae.
Keywords: drug-resistance, leprosy, molecular epidemiology, Mycobacterium leprae, phylogeography, pseudogene
Mycobacterium leprae is the etiologic agent of leprosy, a chronic but curable human disease affecting the skin, peripheral nerves, eyes and mucosa of the upper respiratory tract [1]. The unique predilection of the leprosy bacillus for the PNS causes numbness, nerve damage and gradually the disfigurement of limbs and tissue decay. These outcomes of leprosy have been the root-cause of the fear and stigma associated with this disease for centuries. Leprosy manifests as a spectrum of disease with two poles known as lepromatous and tuberculoid leprosy on the basis of their histopathology by Ridley and Jopling [2]. More recently, the WHO introduced an operationally simpler classification comprising multibacillary and paucibacillary leprosy. Lepromatous leprosy patients have numerous skin lesions, with abundant bacilli, whereas lesions are rare in tuberculoid leprosy and the bacilli are often undetectable. In addition to differences in the bacillary load, these two forms exhibit reciprocal immune responses as multibacillary patients mount a strong humoral response against M. leprae but little cell-mediated immunity, whereas these immune responses are reversed in paucibacillary cases [1]. This dichotomy implies that cell-mediated immunity is critical for restricting bacterial growth.
Global leprosy control programs have been impressively successful, as more than 14 million people have been cured of the disease in last 20 years through multidrug therapy (MDT) implemented by the WHO and others. Although this correlates to more than a 90% reduction in the prevalence rate, the chain of transmission has not been broken as approximately 250,000 new cases of leprosy are still recorded annually. This translates into a new case being documented approximately every 2 min, thus highlighting the need for continued commitment towards control of the disease and to leprosy research. The countries worst affected by leprosy, in terms of case load, are India, Brazil, Indonesia, Bangladesh, Democratic Republic of the Congo, Ethiopia, Nepal and Nigeria [3].
The oldest written records describing the disease are from the ancient Indian texts in the Sushruta Samhita around 600 BC, which were certainly preceded (perhaps by centuries) by orally transmitted accounts. These records are impressively descriptive and accurate about the characteristic features, diagnosis and traditional treatment, for instance with Chaulmoogra oil, of leprosy [4–6]. There are also descriptions of the disease in ancient texts from Egypt [7]. The oldest leprosy skeletal remains found, from Balathal in India, are 4000 years old [8]. However, confirmation of the ‘diagnosis’ by molecular methods, such as identification of M. leprae DNA, is not yet available for this specimen. Such information would certainly be very helpful for understanding the spread of disease.
Armauer Hansen discovered the leprosy bacillus in 1873, in Bergen, Norway. With this landmark discovery, M. leprae became the first bacterial pathogen to be associated with any human disease. In honor of his discovery and in an attempt to eliminate the age-old stigma attached to the disease, leprosy is often referred to as Hansen’s Disease or Hanseniasis. A rod-shaped acid-fast bacterium, M. leprae is an obligate intracellular pathogen with a tropism for macrophages and, characteristically, for the Schwann cells of peripheral nerves. M. leprae prefers the cooler regions of the body and, for diagnostic purposes, slit skin smears are often taken from the earlobe of potential cases.
Despite over 100 years of research we still lack a clear understanding of the pathogenesis and physiology of this pathogen. Even basic questions regarding its mechanism of transmission, the role of environmental reservoirs (e.g., soil, water, animals), or differential outcomes of infection in different individuals are yet to be resolved completely. Major obstacles to progress in leprosy research are our inability to culture the bacterium in vitro and the very long incubation periods required, owing to its extremely slow doubling rate (2 weeks).
Attempts to exploit the cool body temperature of armadillos (30–35°C) led in 1973 to the discovery that this animal is a suitable host for propagation of M. leprae [9]. The armadillo was used initially to produce large numbers of bacilli for leprosy vaccine development, with surplus bacterial cells being used for research purposes. Armadillo-derived M. leprae made it possible to extract sufficient DNA to carry out whole genome sequencing of the TN strain of M. leprae, originally isolated from a patient in Tamil Nadu, India [10].
This article focuses on some of the important advances in leprosy research in the postgenomic era, outlines progress made in molecular epidemiology and drug-susceptibility testing (DST), and describes the phylogeography of M. leprae.
Genome biology
The genome sequence of the TN strain of M. leprae contains 3,268,210 bp and has an average G+C content of 57.8% [10]. The leprosy bacillus thus has the smallest and most A+T-rich genome of any known mycobacterium. Bioinformatic analysis uncovered 1614 genes coding for proteins and a further 50 that encode stable RNAs. These comprise a mere 49.5% of the genome with the remainder occupied by pseudogenes, inactive reading frames with functional counterparts in other mycobacteria, or regulatory sequences. Initially 1116 pseudo-genes were found [10] but this figure increased to 1293 when other genome sequences became available for comparison [11].
Analysis of the genome sequence has improved our understanding of the physiology, pathogenesis and genetics of M. leprae and is contributing towards the development of better diagnostics and molecular epidemiological tools for monitoring transmission dynamics and drug-resistance trends [12–15]. For instance, 165 of the seemingly functional genes have no ortholog in Mycobacterium tuberculosis. The corresponding proteins offer potential as specific immunodiagnostic reagents but since this has been reviewed elsewhere [16,17], the topic will not be covered here.
Physiology & biochemistry
While genomics did not provide a precise explanation for the slow growth of M. leprae, a number of clues arose. All of the functional categories of M. leprae are considerably smaller than their counterparts in the tubercle bacillus, and other mycobacteria, and some have almost disappeared completely. Although all the main anabolic pathways are relatively intact there has been extensive downsizing of the genetic repertoire required for lipolysis, the main catabolic route used by intracellular mycobacteria to derive their energy from degradation of host-derived lipids and fatty acids. Likewise, the pathways for central and energy metabolism are also predicted to be impaired with the result that the use of common carbon sources, such as acetate and galactose, is lost to M. leprae, and no ATP can be generated from oxidation of NADH. Furthermore, the anaerobic and microaerophilic electron transfer systems, together with the biosynthetic and transport systems required to produce the cognate prosthetic groups, have all been lost. This implies that M. leprae is restricted to growth on a very few carbon sources and that catabolism is severely limited [10,15].
Reductive evolution
A large number of pseudogenes, accumulation of insertion sequence (IS) elements and lowered G+C content are all hallmarks of reductive evolution, and may reflect passage through an evolutionary bottleneck [18,19] There are more than 26 defunct IS elements in the genome together with four families of dispersed repeats: RLEP (37 copies); REPLEP (15 copies); LEPREP (eight copies); and LEPRPT (five copies) [20]. Downsizing of the M. leprae genome was mainly due to recombination events between these repetitive sequences, which occupy nearly 2% of the TN genome. The presence of RLEP at the 3´-end of genes and often within pseudogenes suggests that these sequences are the remnants of the transposons that have now lost their ability to undergo transposition, as early RLEP-based restriction fragment length polymorphism studies detected no diversity [21], as was later confirmed by comparative genomics of M. leprae strains. Furthermore, the chromosomal rearrangements (inversions, translocations and gene deletions) mediated by recombinational events between these repetitive elements explain the loss of synteny (discontinuities in gene order, compared with that in the M. tuberculosis genome) in the regions they flank [20].
Comparative genomics of M. leprae strains
Genome decay and the presence of such a large number of pseudogenes suggested that much genetic diversity should exist among M. leprae strains, however, comparative genomics revealed variation to be exceptionally rare. Whole genome sequencing of strain Br4923, from a Brazilian patient, showed the genome to comprise 3,268,071 bp. This 141 bp decrease with respect to the TN genome is mostly attributable to the consistently higher repeat copy numbers at variable number tandem repeat (VNTR) loci in the TN strain [22]. In total, these two genomes differ at 185 loci, which include 146 single nucleotide polymorphisms (SNPs), 31 VNTR regions, and eight insertion-deletion (InDel) events.
Whole genome re-sequencing of two additional strains Thai53 (Thailand) and NHDP63 (USA) was later performed using the Illumina Solexa technology, and their sequences compared with the reference strains, TN and Br4923. This comparison revealed only 215 polymorphic sites (including SNPs and InDels) with over 99.995% genome sequence identity among these geographically distant strains. The polymorphisms observed are summarized in Table 1 and their use for strain typing presented below. In summary, the gene content is essentially identical between all four genomes in terms of sequence, number and order of genes, with no evidence found of inversion, translocation, duplication or transposition [22]. Possible recombination events between dispersed repeats were detected, however, as almost 25% of the total SNPs occur in these repeats, occupying only 1.16% of the genome. The over-representation of SNPs in elements such as RLEP may indicate that recombination events between different copies of the repetitive elements result in the dispersal of a particular SNP.
Table 1.
Polymorphisms observed in four Mycobacterium leprae genomes.
| Strain | SNP subtype |
SNPs and InDels specific to a strain or a group of strains |
Sequencing method used |
Specific pseudogenes |
|---|---|---|---|---|
| TN (India) | 1A | 21 | ABI | |
| Thai-53 (Thailand) | 1A | 23 | Illumina | ML0472 ML2472c ML2687c |
| NHDP63 (USA) | 3I | 65 | Illumina | ML2678 ML2472c |
| Br4923 (Brazil) | 4P | 65 | ABI | ML0825c |
SNPs in more than one strain: 50; total number of polymorphic loci: 215.
InDel: Insertion-deletion; SNP: Single nucleotide polymorphism.
Pseudogenes
M. leprae has undergone reductive evolution, whereby many genes and associated functions were lost through shrinkage of the genome and gene decay, possibly due to a drastic change in lifestyle from free-living, like most mycobacteria, to host-associated [11]. Reductive evolution has also been reported for other obligate intracellular pathogens such as Mycoplasma, Chlamydia, Rickettsia spp. and for Mycobacterium ulcerans, a close relative of the leprosy bacillus present in aquatic ecosystems, which causes Buruli ulcer in humans [23]. However, with 50% of the genome seemingly devoid of function, M. leprae has the largest proportion of pseudogenes in comparison to other pathogenic and nonpathogenic bacteria and archaea [24].
While the bulk of the pseudogenes are conserved, recent work has shown that some are restricted to certain strains (Table 1) [22]. Orthologs of these genes in M. tuberculosis are all nonessential, thus their loss should not impose a fitness defect on the corresponding strain. However, the presence of an intact gene in some strains, but not in others, suggests that, even if most pseudogenes arose in a single event (see below), this process still occurs. This is exemplified by the presence of intact ML0472 and ML2687c in the TN strain but not in Thai-53, two very closely related Asian strains with the same SNP subtype (1A). The mechanism by which pseudogenes arise is not yet known but different explanations, invoking altered or deficient responses to environmental or host-mediated stresses that lead to DNA damage, have been proposed. These include the loss of sigma-factors [25] or two-component systems [26]. Perhaps a more plausible mechanism is the lack of the DnaQ-mediated proofreading activity of DNA polymerase III, due to pseudogenization, as this probably contributes to mutations accumulating in genes [24].
Transcription of pseudogenes
Although the functional roles and significance of pseudogenes and other noncoding regions are unknown, some of them are highly expressed and their expression levels change following macrophage infection [27] and vary greatly among different leprosy patients [28]. While studying the transcriptome of M. leprae, Williams et al. found that as many as 36% of the total transcripts belong to pseudogenes and demonstrated that 43% of all pseudogenes were transcriptionally active [29]. Independently, using tiling arrays, it was recently demonstrated that only half of the transcripts are derived from coding genes, while the remaining half are contributed by pseudogenes and noncoding regions; M. leprae-specific RLEP sequences are also among these expressed noncoding regions [30]. An important feature of M. leprae pseudogenes is the presence of a large number (ranging from 1 to 40) of in-frame stop codons. Most (75%) transcribed pseudogenes lack conventional start codons, while 67% of expressed pseudogenes contain more than five stop codons [28].
The metabolic cost of transcribing these apparently nonfunctional sequences could be detrimental for cell growth and physiology or, on the other hand, it may be of biological importance to M. leprae. The rate of pseudogene deletion in M. leprae appears to be slower than in other bacteria [31] and this suggests that these sequences could provide back-up functions or be activated by events such as gene conversion in unusual circumstances. Pseudogenes might thus represent a reservoir for genetic diversity. It has also been speculated that some of these transcribed noncoding regions and pseudogenes play roles in the regulation of infection, intracellular parasitism and replication in M. leprae [28] but evidence for this is lacking. In higher eukaryotes, the significance of expressed pseudogenes as noncoding RNAs or riboregulators, and their importance in transcriptional and post-transcriptional gene regulation, are well documented in cancer and the CNS [32–36], and also in the cell growth and organogenesis [37]. With the improvement in our awareness of various regulatory roles of non-coding regions and pseudogenes, the biological significance of so called ‘junk’ DNA is being increasingly appreciated. Further studies in this direction can add to our understanding of the biology of M. leprae.
When did pseudogenes accumulate in M. leprae?
The half life of pseudogenes in some eukaryotes may be hundreds of million years [38], but they are generally lost faster from bacterial genomes [39]. However, because of this tendency of pseudo-genes to be eliminated from the genome due to deletion events, it is intriguing that M. leprae has accumulated and retained so many of them. Gomez-Valero et al. calculated that 89% of the nucleotide content of pseudogenes is still present in M. leprae genomes [11]. From genome comparisons, these and other investigators have attempted to reconstruct the evolution of M. leprae and to predict whether pseudogene formation was a gradual or stochastic event [25,39]. With the help of a novel approach, based on the number of nonsynonymous substitutions per site in pseudogenes, it was estimated that M. leprae and M. tuberculosis diverged 66 million years ago, with a single pseudogenization event occurring in the leprosy bacillus in the last 10–20 million years [11]. Given this vast timeframe it is truly puzzling that so little diversity is observed within both genes and pseudogenes of M. leprae, indicating that its emergence as a human pathogen may have been a recent event, such as passing through an evolutionary bottleneck or introduction from another host.
Mycobacterium lepromatosis: a sibling with pseudogenes
Recently, Mycobacterium lepromatosis, a new unculturable mycobacterial species associated with diffuse lepromatous leprosy, also known as Lucio’s leprosy or Latapi’s leprosy, was described by Han et al. [40,41]. Lucio’s leprosy is a very rare, unique form of leprosy apparently restricted to Mexico and the Caribbean [42]. Although Koch’s postulates have not yet been fulfilled for this pathogen, M. lepromatosis has been proposed to be responsible for Lucio’s phenomenon [40]. It is characterized clinically by diffuse, non-nodular cutaneous infiltration and pathologically by mycobacterial invasion into the endothelium. A PCR-based test to detect M. lepromatosis in biopsies has been described [40,41]. Limited DNA sequence information is available from M. lepromatosis, which has a G+C content of 58.6%, close to that of M. leprae, and also harbors pseudogenes [41]. The levels of sequence identity between the two bacteria for the 16S rRNA and 14 protein-encoding genes were 98.0% and 93.1%, respectively, but only 79.1% for the five pseudogenes examined. The greater divergence seen for the latter suggests that, unlike the functional genes, the pseudogenes are less constrained by selective pressure. Based on a phylogenetic comparison of five protein-coding genes, M. leprae and M. lepromatosis form a tight cluster with long terminal branches, and it has been hypothesized that they diverged after the unique pseudogenization event occurred in their last common ancestor.
The discovery of Lucio’s leprosy bacillus may have important implications for the disease spectrum and patient management. It has been hypothesized that some cases of leprosy may well be due to M. lepromatosis [40]. However, it is important that further epidemiological and clinical evidence be gathered and independent replication of this finding is also desirable. Whole genome sequencing of this new species and its much-awaited comparison with M. leprae is also a priority, to obtain better insight into the evolution of both pathogens and their pseudogenes.
Drug resistance
Multidrug therapy for multibacillary leprosy involves three antibiotics: monthly rifampicin, and dapsone and clofazimine on a daily basis for 12 months. However, paucibacillary cases are treated with rifampicin and dapsone only with the duration of treatment limited to 6 months. The emergence of drug resistance poses a serious threat to any infectious disease control program that mainly relies upon a secondary intervention, such as chemotherapy. While MDT has been very effective, there are a few reports of M. leprae strains resistant to one or more anti-leprosy drugs [43–46]. Although drug resistance among new cases is extremely rare, it has occurred in some relapsed cases, occasionally with multidrug-resistant strains [47,48]. In the absence of suitable phenotypic tests, the actual magnitude of drug resistance in leprosy remains largely undetermined. Proper surveillance of drug resistance becomes even more important considering that we have very limited alternative drugs to MDT. Acknowledging the growing concern on this issue, the WHO issued guidelines for the global surveillance for drug resistance in leprosy [49] and implemented a sentinel surveillance network in eight countries: Brazil, China, Colombia, India, Myanmar, Pakistan, Philippines and Vietnam.
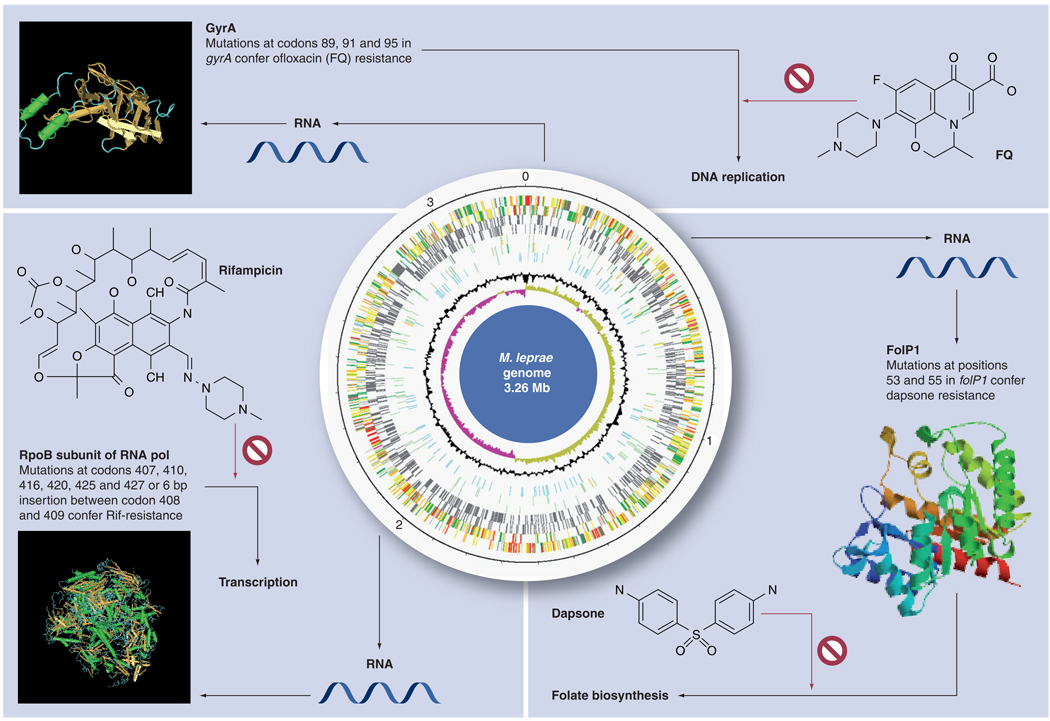
Since M. leprae cannot be cultured in vitro, DST relied initially on the laborious and lengthy (6–12 months) mouse foot pad technique [50,51] and later on a much faster (ς weeks) radio–respirometric method [52]. However, the molecular methods for detecting drug resistance-associated mutations have proved their immense value in faster and efficient detection of drug resistance, and their reliability has also been confirmed in mouse foot pad experiments. The availability of the M. leprae genome sequence and improved understanding of the genetic basis of drug resistance in mycobacteria led to the development of molecular methods for DST. These can be used to monitor resistance to the MDT components, rifampicin and dapsone, and fluoroquinolones (FQ) new leprosy drugs, but not to clofazimine as the resistance mechanism remains unknown. A scheme, showing the genomic location of mutations in the targets associated with resistance to various anti-leprosy drugs and their mechanism of action, is depicted in Figure 1. A brief description now follows.
Figure 1. Drug resistance mutations in the target gene loci in Mycobacterium leprae genome.
The genome map and the positions of three genes known to be associated with drug resistance, their RNA and protein products together with the antibiotics responsible for inhibiting the corresponding enzymatic functions are shown.
Drug-resistance mechanisms
Rifampicin resistance is mainly due to mutations in a small stretch of the rpoB gene, encoding the β-subunit of RNA polymerase, known as the rifampicin resistance determining region (RRDR) that includes codons 407–427. Rifampicin blocks transcription. The most commonly found mutation in the RRDR is Ser425Leu [45,47,53–56]. Out of many thousands of rifampicin-susceptible isolates, none harbored mutations at the RRDR locus in rpoB.
Sequencing of the M. leprae genome revealed two potential dihydropteroate synthase genes, folP1 and folP2 [10], involved in folate biosynthesis. Dapsone targets dihydropteroate synthase by acting as a competitive inhibitor of the substrate p-aminobenzoic acid (PABA), thus inhibiting folic acid biosynthesis. Mutations in codon 53 and 55 in folP1 have been associated with high or intermediate levels of dapsone resistance in over 90% of the isolates in different studies [47,48,56–61]. However, mutations in folP2 were not found to be associated with dapsone resistance [62]. Fluoroquinolones, such as ofloxacin, bind to the A subunit of DNA gyrase (GyrA) and inhibit DNA replication. Ofloxacin is a component of a new regimen for paucibacillary leprosy known as ROM (rifampicin, ofloxacin and minocycline) that is used occasionally. Ofloxacin-resistant isolates harbor the mutation Ala91Val [43,44,46]. Based on knowledge of M. tuberculosis gyrA mutations, it is likely that mutations at codon 89, 92, and 95 may also confer quinolone resistance [53].
Molecular drug-susceptibility testing
The currently preferred methods of performing DST employ molecular biology, as these are simpler, considerably quicker and less expensive than phenotypic techniques (for a recent review see [53]). Many laboratories now perform direct DNA sequencing of PCR fragments derived from the RDR of the target genes in order to detect mutations associated with resistance. Others have developed alternative methods, such as real time PCR or microarrays, based on oligonucleotide probes corresponding to each mutation in rpoB, folP1, and gyrA. Results from the latter exhibited high concordance with those from sequencing when tested in the Philippines and Myanmar [63]. For resource-limited settings, a simple line-probe reverse hybridization assay has been developed by Hain Lifescience GmbH, like that employed with great success for molecular DST in tuberculosis [64], and this is now undergoing evaluation.
Transmission & strain typing
The route of transmission of M. leprae remains obscure but early identification of infectious leprosy cases is very important in order to implement MDT effectively, to prevent disabilities and to limit contagion. Rapid, sensitive and specific diagnostic tests targeting the multicopy repeat RLEP have been successfully used for detection of low amounts of M. leprae DNA in paucibacillary biopsies and archaeological specimens [65]. The molecular typing of strains can help to understand better the transmission source(s) and dynamics. Different molecular epidemiological tools are being developed for tracing the possible sources of infection and spread of disease, for differentiating cases of relapse from re-infection, and for unravelling possible links between human and non-human reservoirs (Table 2). Comparative evaluation of the performance of these tools is in progress.
Table 2.
Different molecular typing systems for Mycobacterium leprae strains.
| Typing system | Principle and applications | Merits | Limitations | Ref. |
|---|---|---|---|---|
| Variable number of tandem repeats | ||||
| TTC repeats | Ten to 37 repeats of trinucleotides (TTC); also known as (GAA) 20 | Very high allelic number variation at a single locus Easy and cost effective |
Poor geographical associability Inherent instability at some loci due to slipped stranded mispairing Low resolution Hypervariability making them unreliable for some loci Not all the strains can be clustered Stuttering phenomenon at some loci has been observed when the stability was studied |
[72,73,103–107] |
| rpoT (sigA) | Based upon biased geographical distribution of three and four copies of hexanucleotide GACATC repeat | Geographical relationship, but very limited discriminations Easy and cost effective |
[108–111] | |
| Study of many loci or multiple-locus VNTR analysis | 16 polymorphic STRs are studied together Genotyping based on various polymorphic loci |
12 (of 13) STRs were found to be stable over the 5 years of passage in armadillo tissues | [74,75,112] | |
| 15 VNTRs (including six mini- and nine microsatellites) multi-locus coverage for increased discriminatory capacity | Multiplex-PCR approach and hence saves the amount of reagents and the DNA template FLA-based detection Easy and cost effective Automation of work-flow possible |
[77,78,80,113,114] | ||
| SNP and single base insertion/deletion | ||||
| SNP typing | Based on informative SNPs/InDel, four principle types are further differentiated into 16 subtypes Phylogeographical analysis of M. leprae strains around the globe, correlation with known routes of ancient human migrations |
Stable, strong geographical association In nonendemic countries, strains in immigrant population can be related to their origin and source of autochthonous transmission can be traced |
Too little diversity for monitoring short-range transmission Differentiation of closely related strains difficult |
[22,66,72,115] |
FLA: Fragment length analysis; InDel: Insertion-deletion; SNP: Single nucleotide polymorphism; STR: Short tandem repeat; VNTR: Variable number of tandem repeats.
It was known from RFLP studies that strains of M. leprae exhibited very little genetic diversity. Now, genome-derived knowledge of polymorphic sites has opened new avenues to pursue the molecular epidemiology of leprosy. At first site, the very low SNP frequency (one SNP approximately every 28 kb as against one every 3 kb in M. tuberculosis [66]) represents a challenge for strain typing but genome stability has also proved advantageous. By contrast, exploiting the VNTRs present in the genome provides a better dynamic range [20] but may be error-prone due to the homoplasies frequently associated with these minisatellites [67]. Many such polymorphic loci have been identified in the M. leprae genome but in order to be exploited successfully as molecular tools they must behave in a reproducible, stable and discriminatory manner [68,69].
SNP for phylogeography & long-range mapping
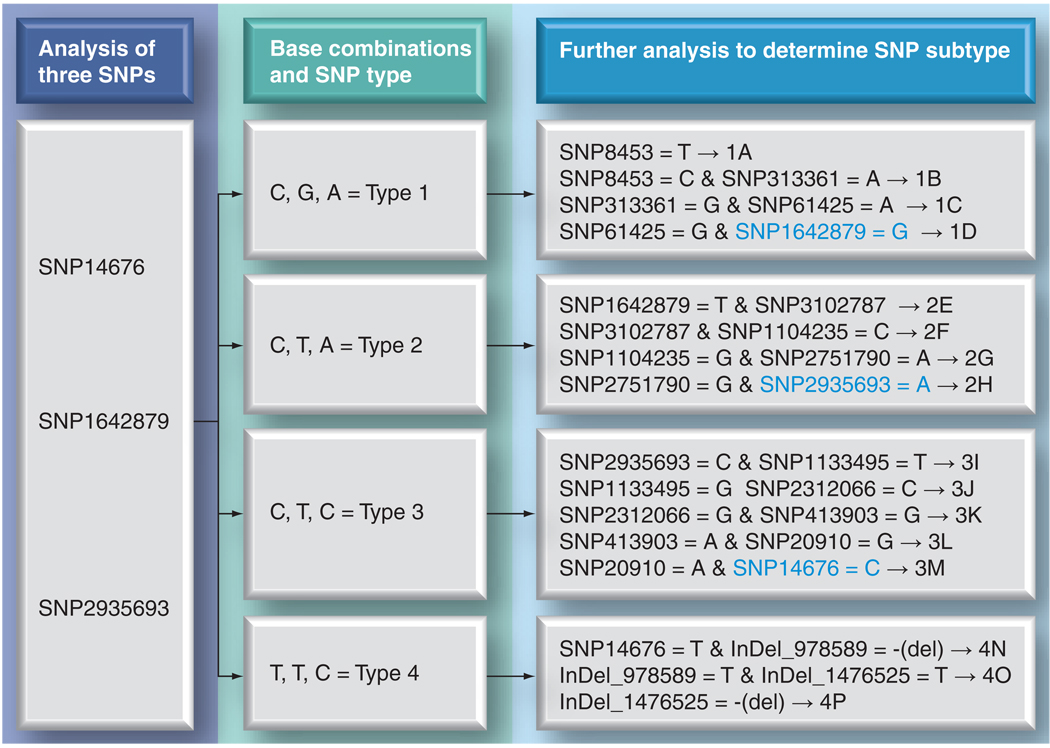
Single nucleotide polymorphisms can be restricted to a particular strain and are, therefore, relatively uninformative, or shared by two or more related strains in which case they may serve as more informative markers. The first SNP typing system was based upon three informative SNPs and used to screen approximately 400 different strains from 28 different countries worldwide. This approach revealed only four combinations (SNP types 1, 2, 3, and 4) but with strong geographical associations [66]. Subsequently, a further 84 informative markers (78 SNPs and six InDels in homopolymeric tracts) were found, thereby enabling the classification of M. leprae strains into 16 different SNP genotypes (1A–4P), again displaying a phylogeographical relationship [22]. The details of this SNP subtyping scheme are shown in Figure 2 and will be discussed further below. In a country with very few endemic cases of leprosy, this system can help trace the possible sources of infection through immigration or foreign residence history of the patients. While the SNP-based typing system is very stable and reliable for classifying strains over broader geographical regions, short-range transmission studies are difficult with this typing system due to the limited discrimination among the strains.
Figure 2. Single nucleotide polymorphism-typing scheme for Mycobacterium leprae.
Using the first three SNPs shown on the left (SNP14676, 1642879, and 2935693), a strain is first typed into one of the four SNP types (1–4) and then subtyped using three or four markers shown at right/SNP type to give 16 subtypes (A–P). SNPs shown in blue are the same as those used for typing into type 1, 2, 3 & 4 (left side of the figure).
InDel: Insertion-deletion; SNP: Single nucleotide polymorphism.
VNTR-based genotyping for tracing short-range transmission
The inherent variability of VNTRs, which are also named short tandem repeats (STRs) or microsatellites (repeat length 2–5 bp) and minisatellites (repeat length 6–50 bp) has been the focus of a lot of molecular epidemiological studies. In comparison with the low rate of SNPs, VNTRs exhibit a much higher mutation frequency because of slipped strand mispairing during DNA replication [70,71], leading to clonal instability [72]. While this dynamism provides much broader discriminatory power, the lower stability and reproducibility of some of these loci needs to be considered; loci must, therefore, be carefully selected in order to extract epidemiologically meaningful information/conclusions. Owing to this inherent instability, specimens from different sites on the same patient or from different passages in animals can exhibit variations, rendering such loci unattractive targets for genotyping [72–74]. This limitation has been addressed by testing various such loci for their stability over a period of 5 years in animal passage [75]. In other work, the intrapatient stability of VNTRs has been tested [75–77]. In a study from China and the Philippines, M. leprae was genotyped in biopsy specimens and slit skin smears before the start of treatment and at 2-monthly intervals during treatment. These studies uncovered intrapatient variability in the polymorphic and stutter-prone hypervariable microsatellites (mainly di- or tri-nucleotide repeats), whereas other loci remained stable, thus allowing the authors to establish transmission links between the study isolates from multicase families or the same town [76,78].
Thus, apart from a few microsatellite loci, which gave either irreproducible results or were not stable, for example AT(15), TA(18) and 18–8, others offer potential for molecular epidemiology of M. leprae. Use of reliable VNTRs has been encouraged as a standardized methodology [79] and some of the limitations can be overcome by surveying multiple sites. More recently, using a neighbor clustering method, VNTR patterns have been shown to cluster in groups in accordance with their geographic origin [80]. This two-stage method first assigns individual isolates to a cluster, comprised of a generalized population sharing similar genotypes, then sub-clusters individuals by using a nearest neighbor network. However, it is clear that this approach is far from optimal as on the one hand it generates an unrealistically high number of genotypes (417 from 465 strains) while on the other, not all isolates (101 out of 465) could be assigned to the clusters and it is not possible to establish a phylogenetic relationship based upon the VNTR patterns [80]. This phylogenetic component can be easily imparted by merging SNP subtyping with VNTR data.
Recommended system for M. leprae genotyping
Considering the limitations of the individual methods, the combined use of SNP- and multilocus VNTR-typing appears to be the best option. As shown in Figure 2, initial typing of a strain into one of four SNP types requires analysis of only three SNPs, and further sub-typing into one of the 16 subtypes needs interrogation of four more SNPs, at most. Once a strain is subtyped with these six or seven SNPs, information is generated for all 84 polymorphic sites [22] as these share linkage with the tested set of SNPs. To further resolve SNP subtypes, multi-locus VNTR analysis should be performed by multiplexed-fragment length analysis or, better still, by DNA sequencing. Thus, a phylogeographic component imparted by SNP subtypes together with dynamic branching provided by VNTRs, could be ideal for the molecular typing of M. leprae strains. The combined SNP–VNTR data can be analyzed and presented using phylogenetic tools, such as minimum spanning trees, maximum likelihood analysis or other suitable methods.
The merits and limitations of some of the most widely used genotyping methods are compared and contrasted in Table 2. Various applications of these methods based on SNP and VNTR have also been reviewed recently by Matsuoka [81].
Phylogeographical inferences from M. leprae genotyping
The histories of microbes and humans have been entwined since the very beginning and infectious diseases have contributed significantly to shaping the human genome. As humans, we are always curious to know where this evolutionary journey began and when it started. Various datasets, based on haploid markers from mitochondrial DNA and the Y chromosome, along with archaeological, anthropological and linguistic evidence, support East Africa as being the cradle of mankind. After initially spreading within Africa, during the first half of the last 100,000 years, humans migrated to the rest of the world through Asia [82]. According to this “standard model for evolution of modern humans”, the humans from East Africa first started moving to the Near East approximately 60,000 years ago before spreading to Southern Asia and Australasia, Europe and Central Asia, until finally reaching the Americas via the Bering Strait approximately 14–20,000 years ago.
Many commensal and parasitic microorganisms accompanied humans in the course of this journey. By tracing their genetic diversity and evolutionary divergence, microbial phylogeographical studies have helped improve our understanding of ancient human migrations [83,84]. Genotyping studies of viral agents, for example, hepatitis B [82], hepatitis G [85], JC polyomavirus [86,87], the soil fungus Coccidioides immitis [88] and, above all, the Gram-negative bacterium Helicobacter pylori [89–91], have provided convincing and fascinating insights into man’s eternal quest to inhabit new niches. The latter pathogen, which displays exceptionally high genetic diversity, has proved especially informative for retracing the routes of human migrations.
In contrast to, but nonetheless like, H. pylori, the more stable genome of M. leprae has its own advantages for human and mycobacterial phylogeography. Leprosy has visited every civilization and left its distinctive mutilations that are visible even today as lesions in the skeletal remains from those civilizations. Such remains can be used for paleomicrobiological studies as M. leprae, and its DNA can be detected in bone fragments, particularly from the faciomaxillary region [22,92–99]. Because of the remarkable genomic conservation, even extinct strains from skeletal remains of leprosy victims (as old as 1500 years) could be genotyped along with extant strains, thus allowing us to draw solid inferences about the prevalent genotypes in areas where the disease has long disappeared, such as Europe. Such paleomicrobiological studies of ancient M. leprae DNA, present in skeletal remains not only bridged gaps in our understanding of the dissemination routes of leprosy but also established the relationship of extinct with extant strains, present today in other parts of the world [22,96].
Phylogeographical studies of M. leprae have shed light on human migration and on the evolution of the disease itself. According to the phylogenetic scheme derived from the SNP data, it is likely that ancestral M. leprae in East Africa or the Near East had a genotype that most probably resembled the present SNP type 2 strains. With successive human migrations, it spread eastwards to Asia giving rise to SNP type 1, while a west-ward spread to the Middle East and Europe gave rise to SNP type 3. Colonialism, emigration and the slave trade introduced M. leprae of SNP type 3 to the Americas, whereas SNP type 4 arose in West Africa but was then transported to the Americas by infected slaves [66].
These four SNP types could be further classified into 16 SNP subtypes [22] that exhibit a strong geographical association. The phylogenetic inferences drawn from this typing scheme about the spread of leprosy are largely in agreement with the known routes of ancient human migrations [82]. The trade route between Europe and Asia, known as the silk road, may have contributed to the spread of leprosy from Europe and the Near East to the Far East and to China. The reverse direction is thought to be responsible for the entry of the Black Death, caused by Yersinia pestis, into Europe in the 14th Century [100]. Previously, leprosy was believed to have been introduced to China from the Indian subcontinent but this interpretation is not borne out by genotyping studies of M. leprae [22]. Nor is the earlier proposition that Alexander the Great’s soldiers brought leprosy back from India to Europe [4–6]. Likewise, as stated above, it appears that colonialism, rather than the Asian migration via the Bering Strait, introduced leprosy into the New World.
Retracing evolution of M. leprae
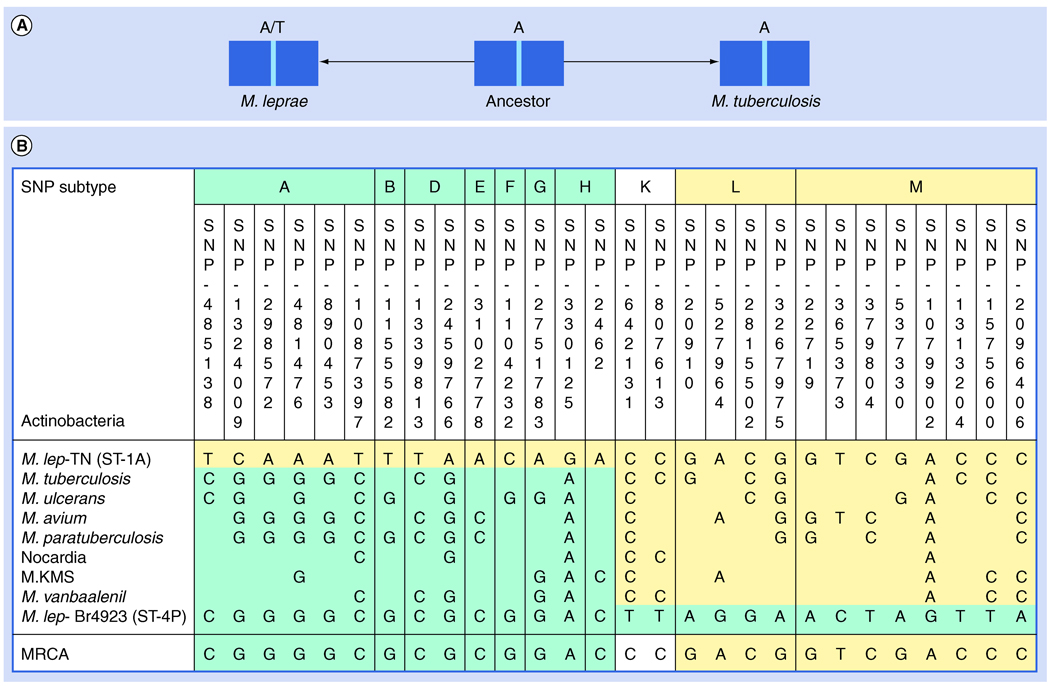
Single nucleotide polymorphism typing M. leprae strains could also be used to deduce the evolutionary descendence of these strains from each other. This has been done by the outgroup analysis, which is based upon determination of the corresponding ancestral base in other actinobacteria (Figure 3A) [22]. Out of the 15 groups of markers for SNP subtyping, this could be achieved for ten groups. Together, these comprise 28 positions (Figure 3B). Based upon this analysis, the most recent common ancestor of M. leprae should have been closest to the SNP subtypes 2H or 3K. Availability of the whole genome sequence of M. lepromatosis can shed more light on the evolution of M. leprae.
Figure 3. Outgroup analysis to deduce ancestral bases for Mycobacterium leprae single nucleotide polymorphism positions.
(A) A polymorphic site is shown and comparisons performed between different Mycobacterium leprae strains and other actinobacteria which serve as an outgroup. The strain showing identity to the base occupying this site in the outgroup is considered to be closer to the MRCA. (B) The results of outgroup analysis at 28 sites defined by SNP and the bases occupying those sites in the TN (subtype 1A) or Br4923 (subtype 4P) strains and the outgroups. Note that not all genes are present in outgroup members. At the bottom the deduced ancestral base is shown (MRCA).
MRCA: Most recent common ancestor; SNP: Single nucleotide polymorphism.
Phylogenetic analysis of the related pathogen M. tuberculosis has revealed six main lineages [101] that also show geographic association and it is interesting to note that a particular lineage can transmit more efficiently among members of certain ethnic groups. Some of these associations may reflect ancient human migrations and evolution but host adaptation also seems likely [102]. It is of interest to determine whether such ‘ethnic’ adaptation occurs among strains of M. leprae.
Future perspective
Genomics has provided considerable insight into the genes, pseudogenes and genetics of M. leprae, and its human host, whereas comparative genomics has generated a robust phylogeny of strains of the leprosy bacillus of different geographical origin. In order to challenge and refine this phylogenetic model it is necessary to sequence more M. leprae genomes particularly from the as-yet unrepresented SNP type 2 and also from strains at, or near, the intersection between type 2H and 3I-3K, which are purported to be closest to the most recent common ancestor (Figure 3). The availability of a genome sequence for M. lepromatosis may also be informative in terms of further defining the core gene set of a pathogenic mycobacterium and in retracing the proposed common history with M. leprae. Comparison of the genomes of these two leprosy bacilli may indeed clarify whether pseudogenes arose as a single stochastic event or if this was a gradual process.
Working with M. leprae remains immensely challenging and one of the reasons why there are still gaps in our phylogenomics repertoire stems from the fact that even though the next-generation sequencing technologies provide truly colossal output they still require significant quantities of DNA (i.e., several micrograms) as input material for library construction. A powerful technique that would allow DNA amplification from the nanogram amounts present in slit skin biopsies of patients, in an efficient, unbiased and error-free manner, would revolutionize such analysis by providing increased sensitivity, reducing the time requirement and eliminating the need to culture bacilli in animals. In our hands, the amplification technologies currently available do not meet these requirements but may be suitable for more focused approaches [97].
In the meanwhile one could envision using high-throughput bead-based SNP typing technologies or array-based resequencing approaches for multiplexing and genotyping but the restrictions outlined above would still apply. Ideally, it would be advantageous to dispose of a tool that could simultaneously determine genotypes and perform DST as this would be of great epidemiological value to leprosy. Such tools are already finding widespread application for genotyping in human genetics and we anticipate that as better clinically defined cohorts are established with paucibacillary patients or, for instance, individuals prone to erythema nodosum leprosum, much deeper understanding of the genetic ‘risk factors’ for leprosy will be obtained. The ability to predict reliably which patients may present with complications, following reconstitution of the immune responses that often follows MDT, and are hence at risk of painful reversal reactions, is highly desirable. Such knowledge would benefit patients considerably by enabling better follow-up and timely modification of therapy.
Executive summary.
Genome biology
-
■
The leprosy bacillus has undergone massive genome decay and downsizing.
-
■
The core gene set has been defined and many pseudogenes identified.
-
■
Repetitive DNA played a role in reducing genome size and loss of synteny.
-
■
Almost no variation was seen on sequencing the genome of four strains.
-
■
Very few new pseudogenes were found.
Pseudogenes
-
■
Mycobacterium lepromatosis appears to cause diffuse lepromatous leprosy (Lucio’s or Latipa leprosy), a rare disease related to classical leprosy.
-
■
Limited sequence information reveals the closest relative is Mycobacterium leprae.
-
■
M. lepromatosis appears to have many pseudogenes.
-
■
Pseudogenes arose in the last common ancestor of both bacilli before their divergence.
-
■
Many pseudogenes are transcribed in M. leprae.
Drug resistance
-
■
Despite treatment of 14 million patients with multidrug therapy drug resistance is very rare.
-
■
Resistance is due to missense mutations in genes encoding drug targets.
-
■
By providing information about drug target genes genomics has generated molecular tools for detecting drug resistance.
-
■
These tools provide results in less than 48 hours rather than the 1–2 weeks (radio–respirometry) or 6–12 months (mouse footpad model) required previously.
Transmission & strain typing
-
■
Genomics is underpinning the development of tools for molecular epidemiology.
-
■
Single nucleotide polymorphism (SNP) typing defined 16 different subtypes of M. leprae.
-
■
Subtypes show geographical associations.
-
■
Variable number of tandem repeats (VNTR) typing can be useful but is fallible owing to homoplasies and intrapatient variation.
-
■
Surveying multiple VNTR loci simultaneously can help overcome these limitations.
-
■
Combining SNP and VNTR typing is a powerful way forward.
Phylogeographical inferences from M. leprae genotyping
-
■
M. leprae strains are highly related and appear to have originated in East Africa or the Near East.
-
■
An association exists between the SNP type and the region of origin.
-
■
Routes of dissemination of M. leprae can be traced.
-
■
Sequencing of M. leprae genomes from East Africa (SNP type 2) is required.
Acknowledgments
We thank Philippe Busso, Jeffrey Chen, Alberto Paniz and Swapna Uplekar for helpful discussions.
This work received the financial support of the Foundation Raoul Follereau, and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant RO1-AI47197-01A1).
Footnotes
Financial & competing interest disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1. Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363(9416):1209–1219. doi: 10.1016/S0140-6736(04)15952-7. ▪ Comprehensive review summarizing all the major aspects of the biology of the leprosy bacillus and clinical features of the disease.
- 2.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 1966;34(3):255–273. [PubMed] [Google Scholar]
- 3.World Health Organization. Global leprosy situation, 2010. Wkly Epidemiol. Rec. 2009;85(35):337–348. [PubMed]
- 4.Lowe J. Comments on the history of leprosy. Indian Medical Gazette. 1942;77:680–685. reprinted in Lepr. Rev. 1918, 1954–1964 (1947). [PMC free article] [PubMed] [Google Scholar]
- 5.Dharmendra . Ministry of Health, India (2nd Edition) 1967. History of spread and decline of leprosy. [Google Scholar]
- 6.Browne SG. The history of leprosy. In: Hastings RC, editor. Leprosy. Churchill Livingstone, UK: 1985. pp. 1–14. [Google Scholar]
- 7.Hulse EV. Leprosy and ancient Egypt. Lancet. 1972;2(7785):1024–1025. doi: 10.1016/s0140-6736(72)92433-6. [DOI] [PubMed] [Google Scholar]
- 8.Robbins G, Tripathy VM, Misra VN, et al. Ancient skeletal evidence for leprosy in India (2000 B.C.) PLoS ONE. 2009;4(5):e5669. doi: 10.1371/journal.pone.0005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchheimer WF, Sanchez RM. Leprosy-susceptibility testing of armadillos. 2. Late cell and bacterial responses at inoculation site of living leprosy bacilli in ‘resistant’ armadillos. Microbios. 1973;8(31):241–246. [PubMed] [Google Scholar]
- 10. Cole ST, Eiglmeier K, Parkhill J, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409(6823):1007–1011. doi: 10.1038/35059006. ▪▪ Genome sequencing and annotation of first strain of Mycobacterium leprae and its comparison with Mycobacterium tuberculosis to reveal massive gene decay and reductive evolution.
- 11. Gomez-Valero L, Rocha EP, Latorre A, Silva FJ. Reconstructing the ancestor of Mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. 2007;17(8):1178–1185. doi: 10.1101/gr.6360207. ▪ Annotation of additional 177 pseudogenes by comparative analysis with other mycobacterial species and timing of pseudogene formation.
- 12.Curtiss R, 3rd, Blower S, Cooper K, Russell D, Silverstein S, Young L. Leprosy research in the post-genome era. Lepr. Rev. 2001;72(1):8–22. doi: 10.5935/0305-7518.20010004. [DOI] [PubMed] [Google Scholar]
- 13.Eiglmeier K, Parkhill J, Honore N, et al. The decaying genome of Mycobacterium leprae. Lepr. Rev. 2001;72(4):387–398. [PubMed] [Google Scholar]
- 14.Vissa VD, Brennan PJ. The genome of Mycobacterium leprae: a minimal mycobacterial gene set. Genome Biol. 2001;2(8) doi: 10.1186/gb-2001-2-8-reviews1023. REVIEWS1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler PR. The microbial physiologist’s guide to the leprosy genome. Lepr. Rev. 2001;72(4):399–407. doi: 10.5935/0305-7518.20010048. [DOI] [PubMed] [Google Scholar]
- 16.Araoz R, Honore N, Banu S, et al. Towards an immunodiagnostic test for leprosy. Microbes Infect. 2006;8(8):2270–2276. doi: 10.1016/j.micinf.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Geluk A, Spencer JS, Bobosha K, et al. From genome-based in silico predictions to ex vivo verification of leprosy diagnosis. Clin. Vaccine Immunol. 2009;16(3):352–359. doi: 10.1128/CVI.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkhill J, Wren BW, Thomson NR, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413(6855):523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill J, Sebaihia M, Preston A, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35(1):32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 20.Cole ST, Supply P, Honore N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 2001;72(4):449–461. [PubMed] [Google Scholar]
- 21.Williams DL, Gillis TP, Portaels F. Geographically distinct isolates of Mycobacterium leprae exhibit no genotypic diversity by restriction fragment-length polymorphism analysis. Mol. Microbiol. 1990;4(10):1653–1659. doi: 10.1111/j.1365-2958.1990.tb00542.x. [DOI] [PubMed] [Google Scholar]
- 22. Monot M, Honore N, Garnier T, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 2009;41(12):1282–1289. doi: 10.1038/ng.477. ▪ Three new genome sequences reveal limited diversity, few new pseudogenes and deepen phylogeographical understanding.
- 23.Stinear TP, Seemann T, Pidot S, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17(2):192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Harrison PM, Kunin V, Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5(9):R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu MM. Did the loss of sigma factors initiate pseudogene accumulation in Mycobacterium leprae? Trends Microbiol. 2003;11(2):59–61. doi: 10.1016/s0966-842x(02)00031-8. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi JS, Saini DK. Did the loss of two-component systems initiate pseudogene accumulation in Mycobacterium leprae? Microbiology. 2004;150(Pt 1):4–7. doi: 10.1099/mic.0.26863-0. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki K, Nakata N, Bang PD, Ishii N, Makino M. High-level expression of pseudogenes in Mycobacterium leprae. FEMS Microbiol. Lett. 2006;259(2):208–214. doi: 10.1111/j.1574-6968.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Akama T, Bang PD, et al. Detection of RNA expression from pseudogenes and non-coding genomic regions of Mycobacterium leprae. Microb. Pathog. 2009;47(3):183–187. doi: 10.1016/j.micpath.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Williams DL, Slayden RA, Amin A, et al. Implications of high level pseudogene transcription in Mycobacterium leprae. BMC Genomics. 2009;10:397. doi: 10.1186/1471-2164-10-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akama T, Suzuki K, Tanigawa K, et al. Whole-genome tiling array analysis of Mycobacterium leprae RNA reveals high expression of pseudogenes and noncoding regions. J. Bacteriol. 2009;191(10):3321–3327. doi: 10.1128/JB.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mira A, Pushker R, Rodriguez-Valera F. The Neolithic revolution of bacterial genomes. Trends Microbiol. 2006;14(5):200–206. doi: 10.1016/j.tim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2(12):919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 33.Erdmann VA, Barciszewska MZ, Hochberg A, de Groot N, Barciszewski J. Regulatory RNAs. Cell Mol. Life Sci. 2001;58(7):960–977. doi: 10.1007/PL00000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korneev SA, Park JH, O’Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19(18):7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu W, Zhou D, Glusman G, et al. KLK31P is a novel androgen regulated and transcribed pseudogene of kallikreins that is expressed at lower levels in prostate cancer cells than in normal prostate cells. Prostate. 2006;66(9):936–944. doi: 10.1002/pros.20382. [DOI] [PubMed] [Google Scholar]
- 36.Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem. Biophys. Res. Commun. 2005;337(4):1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 37.Kandouz M, Bier A, Carystinos GD, Alaoui-Jamali MA, Batist G. Connexin43 pseudogene is expressed in tumor cells and inhibits growth. Oncogene. 2004;23(27):4763–4770. doi: 10.1038/sj.onc.1207506. [DOI] [PubMed] [Google Scholar]
- 38.Graur D, Shuali Y, Li WH. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J. Mol. Evol. 1989;28(4):279–285. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Valero L, Latorre A, Silva FJ. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 2004;21(11):2172–2181. doi: 10.1093/molbev/msh232. [DOI] [PubMed] [Google Scholar]
- 40. Han XY, Seo YH, Sizer KC, et al. A new Mycobacterium species causing diffuse lepromatous leprosy. Am. J. Clin. Pathol. 2008;130(6):856–864. doi: 10.1309/AJCPP72FJZZRRVMM. ▪▪ Description of a new mycobacterial species as the likely causative agent for Lucio’s leprosy
- 41.Han XY, Sizer KC, Thompson EJ, et al. Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis. J. Bacteriol. 2009;191(19):6067–6074. doi: 10.1128/JB.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin. Microbiol. Rev. 2006;19(2):338–381. doi: 10.1128/CMR.19.2.338-381.2006. ▪ Exhaustive review detailing various aspects of leprosy and leprosy research.
- 43.Cambau E, Perani E, Guillemin I, Jamet P, Ji B. Multidrug-resistance to dapsone, rifampicin, and ofloxacin in Mycobacterium leprae. Lancet. 1997;349(9045):103–104. doi: 10.1016/S0140-6736(05)60888-4. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka M, Kashiwabara Y, Namisato M. A Mycobacterium leprae isolate resistant to dapsone, rifampin, ofloxacin and sparfloxacin. Int. J. Lepr. Other Mycobact. Dis. 2000;68(4):452–455. [PubMed] [Google Scholar]
- 45.Cambau E, Bonnafous P, Perani E, Sougakoff W, Ji B, Jarlier V. Molecular detection of rifampin and ofloxacin resistance for patients who experience relapse of multibacillary leprosy. Clin. Infect. Dis. 2002;34(1):39–45. doi: 10.1086/324623. [DOI] [PubMed] [Google Scholar]
- 46.Matsuoka M, Kashiwabara Y, Liangfen Z, Goto M, Kitajima S. A second case of multidrug-resistant Mycobacterium leprae isolated from a Japanese patient with relapsed lepromatous leprosy. Int. J. Lepr. Other Mycobact. Dis. 2003;71(3):240–243. doi: 10.1489/1544-581x(2003)71<240:ascomm>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 47.Maeda S, Matsuoka M, Nakata N, et al. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob. Agents Chemother. 2001;45(12):3635–3639. doi: 10.1128/AAC.45.12.3635-3639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka M, Budiawan T, Aye KS, et al. The frequency of drug resistance mutations in Mycobacterium leprae isolates in untreated and relapsed leprosy patients from Myanmar, Indonesia and the Philippines. Lepr. Rev. 2007;78(4):343–352. [PubMed] [Google Scholar]
- 49.World Health Organization. Guidelines for global surveillance of drug resistance in leprosy. SEA-GLP. 2009:1–32. [Google Scholar]
- 50.Shepard CC, Congdon CC. Increased growth of Mycobacterium leprae in thymectomized-irradiated mice after foot pad inoculation. Int. J. Lepr. Other Mycobact. Dis. 1968;36(2):224–227. [PubMed] [Google Scholar]
- 51.Pettit JHS, Rees RJW. Sulphone resistance in leprosy: an experimental and clinical study. Lancet. 1964;284(7361):673–674. doi: 10.1016/s0140-6736(64)92482-1. [DOI] [PubMed] [Google Scholar]
- 52.Franzblau SG. Drug susceptibility testing of Mycobacterium leprae in the BACTEC 460 system. Antimicrob. Agents Chemother. 1989;33(12):2115–2117. doi: 10.1128/aac.33.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuoka M. Drug resistance in leprosy. Jpn. J. Infect. Dis. 2010;63(1):1–7. [PubMed] [Google Scholar]
- 54.Honore N, Cole ST. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob. Agents Chemother. 1993;37(3):414–418. doi: 10.1128/aac.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honore N, Roche PW, Grosset JH, Cole ST. A method for rapid detection of rifampicin-resistant isolates of Mycobacterium leprae. Lepr. Rev. 2001;72(4):441–448. doi: 10.5935/0305-7518.20010052. [DOI] [PubMed] [Google Scholar]
- 56.Williams DL, Gillis TP. Molecular detection of drug resistance in Mycobacterium leprae. Lepr. Rev. 2004;75(2):118–130. [PubMed] [Google Scholar]
- 57.Williams DL, Spring L, Harris E, Roche P, Gillis TP. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob. Agents Chemother. 2000;44(6):1530–1537. doi: 10.1128/aac.44.6.1530-1537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams DL, Pittman TL, Gillis TP, Matsuoka M, Kashiwabara Y. Simultaneous detection of Mycobacterium leprae and its susceptibility to dapsone using DNA heteroduplex analysis. J. Clin. Microbiol. 2001;39(6):2083–2088. doi: 10.1128/JCM.39.6.2083-2088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gillis TP, Williams DL. Dapsone resistance in Mycobacterium leprae. Lepr. Rev. 2000;71 Suppl.:S91–S95. [PubMed] [Google Scholar]
- 60.Lee SB, Kim SK, Kang TJ, et al. The prevalence of folP1 mutations associated with clinical resistance to dapsone, in Mycobacterium leprae isolates from South Korea. Ann. Trop. Med. Parasitol. 2001;95(4):429–432. doi: 10.1080/000349801300188447. [DOI] [PubMed] [Google Scholar]
- 61.Cambau E, Carthagena L, Chauffour A, Ji B, Jarlier V. Dihydropteroate synthase mutations in the folP1 gene predict dapsone resistance in relapsed cases of leprosy. Clin. Infect. Dis. 2006;42(2):238–241. doi: 10.1086/498506. [DOI] [PubMed] [Google Scholar]
- 62.Gillis TP, William DL. Dapsone resistance does not appear to be associated with a mutation in the dihydropteroate synthase-2 of Mycobacterium leprae. Indian J. Lepr. 1999;71:11–18. [PubMed] [Google Scholar]
- 63.Matsuoka M, Aye KS, Kyaw K, et al. A novel method for simple detection of mutations conferring drug resistance in Mycobacterium leprae, based on a DNA microarray, and its applicability in developing countries. J. Med. Microbiol. 2008;57(Pt 10):1213–1219. doi: 10.1099/jmm.0.2008/002600-0. [DOI] [PubMed] [Google Scholar]
- 64.Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur. Respir. J. 2008;32(5):1165–1174. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
- 65.Donoghue HD, Holton J, Spigelman M. PCR primers that can detect low levels of Mycobacterium leprae DNA. J. Med. Microbiol. 2001;50(2):177–182. doi: 10.1099/0022-1317-50-2-177. [DOI] [PubMed] [Google Scholar]
- 66. Monot M, Honore N, Garnier T, et al. On the origin of leprosy. Science. 2005;308(5724):1040–1042. doi: 10.1126/science/1109759. ▪ Single nucleotide polymorphism-typing scheme of M. leprae and its association with human migration patterns
- 67.Filliol I, Motiwala AS, Cavatore M, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 2006;188(2):759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunter PR. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 1990;28(9):1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lovett ST. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004;52(5):1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 71.Reyes JF, Tanaka MM. Mutation rates of spoligotypes and variable number tandem repeat loci in Mycobacterium tuberculosis. Infect. Genet. Evol. 2010;10(7):1046–1051. doi: 10.1016/j.meegid.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 72.Monot M, Honore N, Baliere C, et al. Are variable-number tandem repeats appropriate for genotyping Mycobacterium leprae? J. Clin. Microbiol. 2008;46(7):2291–2297. doi: 10.1128/JCM.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Truman R, Fontes AB, de Miranda AB, Suffys P, Gillis T. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 2004;42(6):2558–2565. doi: 10.1128/JCM.42.6.2558-2565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young SK, Ponnighaus JM, Jain S, et al. Use of short tandem repeat sequences to study Mycobacterium leprae in leprosy patients in Malawi and India. PLoS Negl. Trop. Dis. 2008;2(4):e214. doi: 10.1371/journal.pntd.0000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gillis T, Vissa V, Matsuoka M, et al. Characterisation of short tandem repeats for genotyping Mycobacterium leprae. Lepr. Rev. 2009;80(3):250–260. [PubMed] [Google Scholar]
- 76.Xing Y, Liu J, Sakamuri RM, et al. VNTR typing studies of Mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr. Rev. 2009;80(3):261–271. [PubMed] [Google Scholar]
- 77.Shinde V, Newton H, Sakamuri RM, et al. VNTR typing of Mycobacterium leprae in South Indian leprosy patients. Lepr. Rev. 2009;80(3):290–301. [PubMed] [Google Scholar]
- 78.Sakamuri RM, Harrison J, Gelber R, et al. A continuation: study and characterisation of Mycobacterium leprae short tandem repeat genotypes and transmission of leprosy in Cebu, Philippines. Lepr. Rev. 2009;80(3):272–279. [PubMed] [Google Scholar]
- 79.Hall BG. Molecular epidemiology of Mycobacterium leprae: a solid beginning. Lepr. Rev. 2009;80(3):246–249. [PubMed] [Google Scholar]
- 80.Hall BG, Salipante SJ. Molecular epidemiology of Mycobacterium leprae as determined by structure-neighbor clustering. J. Clin. Microbiol. 2010;48(6):1997–2008. doi: 10.1128/JCM.00149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsuoka M. Recent advances in the molecular epidemiology of leprosy. Nihon Hansenbyo Gakkai Zasshi. 2009;78(1):67–73. doi: 10.5025/hansen.78.67. [DOI] [PubMed] [Google Scholar]
- 82.Cavalli-Sforza LL, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nat. Genet. 2003;33 Suppl.:266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 83.Pavesi A. Microbes coevolving with human host and ancient human migrations. J. Anthropol. Sci. 2005;83:9–28. [Google Scholar]
- 84.Rinaldi A. Tiny travel companions. As microorganisms have accompanied mankind’s journeys around the globe, they could help scientists to unravel our past. EMBO Rep. 2007;8(2):121–125. doi: 10.1038/sj.embor.7400908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pavesi A. Origin and evolution of GBV-C/hepatitis G virus and relationships with ancient human migrations. J. Mol. Evol. 2001;53(2):104–113. doi: 10.1007/s002390010198. [DOI] [PubMed] [Google Scholar]
- 86.Sugimoto C, Kitamura T, Guo J, et al. Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proc. Natl Acad. Sci. USA. 1997;94(17):9191–9196. doi: 10.1073/pnas.94.17.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pavesi A. Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times. J. Gen. Virol. 2005;86(Pt 5):1315–1326. doi: 10.1099/vir.0.80650-0. [DOI] [PubMed] [Google Scholar]
- 88.Fisher MC, Koenig GL, White TJ, et al. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc. Natl Acad. Sci. USA. 2001;98(8):4558–4562. doi: 10.1073/pnas.071406098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Falush D, Wirth T, Linz B, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 90.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284(5418):1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 91.Linz B, Balloux F, Moodley Y, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rafi A, Spigelman M, Stanford J, Lemma E, Donoghue H, Zias J. Mycobacterium leprae DNA from ancient bone detected by PCR. Lancet. 1994;343(8909):1360–1361. [PubMed] [Google Scholar]
- 93.Montiel R, Garcia C, Canadas MP, Isidro A, Guijo JM, Malgosa A. DNA sequences of Mycobacterium leprae recovered from ancient bones. FEMS Microbiol. Lett. 2003;226(2):413–414. doi: 10.1016/S0378-1097(03)00617-7. [DOI] [PubMed] [Google Scholar]
- 94.Matheson CD, Vernon KK, Lahti A, et al. Molecular exploration of the first-century Tomb of the Shroud in Akeldama, Jerusalem. PLoS ONE. 2009;4(12):e8319. doi: 10.1371/journal.pone.0008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watson CL, Lockwood DNJ. Single nucleotide polymorphism analysis of European archaeological Mycobacterium leprae DNA. PLoS ONE. 2009;4(10):e7547. doi: 10.1371/journal.pone.0007547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor GM, Blau S, Mays S, et al. Mycobacterium leprae genotype amplified from an archaeological case of lepromatous leprosy in Central Asia. J. Archaeol. Sci. 2009;36(10):2408–2414. [Google Scholar]
- 97.Suzuki K, Takigawa W, Tanigawa K, et al. Detection of Mycobacterium leprae DNA from archaeological skeletal remains in Japan using whole genome amplification and polymerase chain reaction. PLoS ONE. 2010;5(8) doi: 10.1371/journal.pone.0012422. e12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donoghue HD, Marcsik A, Matheson C, et al. Co-infection of Mycobacterium tuberculosis and Mycobacterium leprae in human archaeological samples: a possible explanation for the historical decline of leprosy. Proc. Biol. Sci. 2005;272(1561):389–394. doi: 10.1098/rspb.2004.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haas CJ, Zink A, Pálfi G, Szeimies U, Nerlich AG. Detection of leprosy in ancient human skeletal remains by molecular identification of Mycobacterium leprae. Am. J. Clin. Pathol. 2000;114(3):428–436. doi: 10.1093/ajcp/114.3.428. [DOI] [PubMed] [Google Scholar]
- 100.Achtman M, Morelli G, Zhu P, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA. 2004;101(51):17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7(5):328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 102.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 103.Shin Y-C, Lee H, Lee H, Walsh GP, Kim J-D, Cho S-N. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J. Clin. Microbiol. 2000;38(12):4535–4538. doi: 10.1128/jcm.38.12.4535-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Groathouse NA, Rivoire B, Kim H, et al. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 2004;42(4):1666–1672. doi: 10.1128/JCM.42.4.1666-1672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Young SK, Taylor GM, Jain S, et al. Microsatellite mapping of Mycobacterium leprae populations in infected humans. J. Clin. Microbiol. 2004;42(11):4931–4936. doi: 10.1128/JCM.42.11.4931-4936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weng X, Wang Z, Liu v, et al. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J. Clin. Microbiol. 2007;45(6):1728–1734. doi: 10.1128/JCM.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Srisungngam S, Rudeeaneksin J, Wattanpokayakit S, et al. Typing of Thai clinical isolates of Mycobacterium leprae and analysis of leprosy transmission by polymorphism of tandem repeats. Southeast Asian J. Trop. Med. Public Health. 2007;38(4):714–720. [PubMed] [Google Scholar]
- 108.Matsuoka M, Maeda S, Kai M, et al. Mycobacterium leprae typing by genomic diversity and global distribution of genotypes. Int. J. Lepr. Other Mycobact. Dis. 2000;68(2):121–128. [PubMed] [Google Scholar]
- 109.Matsuoka M, Zhang L, Morris MF, Legua P, Wiens C. Polymorphism in the rpoT gene in Mycobacterium leprae isolates obtained from Latin American countries and its possible correlation with the spread of leprosy. FEMS Microbiol. Lett. 2005;243(2):311–315. doi: 10.1016/j.femsle.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 110.Lavania M, Katoch K, Singh H, et al. Predominance of three copies of tandem repeats in rpoT gene of Mycobacterium leprae from Northern India. Infect. Genet. Evol. 2007;7(5):627–631. doi: 10.1016/j.meegid.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 111.Lavania M, Lal R, Joseph G, et al. Genotypic analysis of Mycobacterium leprae strains from different regions of India on the basis of rpoT. Indian J. Lepr. 2009;81(3):119–124. [PubMed] [Google Scholar]
- 112.Zhang L, Budiawan T, Matsuoka M. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 2005;43(10):5221–5229. doi: 10.1128/JCM.43.10.5221-5229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kimura M, Sakamuri RM, Groathouse NA, et al. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J. Clin. Microbiol. 2009;47(6):1757–1766. doi: 10.1128/JCM.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balagon MF, Cellona RV, Cruz E, et al. Long-term relapse risk of multibacillary leprosy after completion of 2 years of multiple drug therapy (WHO-MDT) in Cebu, Philippines. Am. J. Trop. Med. Hyg. 2009;81(5):895–899. doi: 10.4269/ajtmh.2009.09-0189. [DOI] [PubMed] [Google Scholar]
- 115.Suzuki K, Udono T, Fujisawa M, Tanigawa K, Idani Gi, Ishii N. Infection during infancy and long incubation period of leprosy suggested in the case of a chimpanzee used for medical research. J. Clin. Microbiol. 2010;48(9):3432–3434. doi: 10.1128/JCM.00017-10. [DOI] [PMC free article] [PubMed] [Google Scholar]