Abstract
Epigenetic regulation represents a fundamental mechanism to maintain cell-type specific gene expression during development and serves as an essential mediator to interface the extrinsic environment and the intrinsic genetic program. Adult neurogenesis occurs in discrete regions of the adult mammalian brain and is known to be tightly regulated by various physiological, pathological and pharmacological stimuli. Emerging evidence suggests that various epigenetic mechanisms play important roles in fine-tuning and coordinating gene expression during adult neurogenesis. Here we review recent progress in our understanding of various epigenetic mechanisms, including DNA methylation, histone modifications and non-coding RNAs, as well as cross-talk among these mechanisms, in regulating different aspects of adult mammalian neurogenesis.
Keywords: adult neural stem cells, DNA methylation, histone modifications, non-coding RNAs
Introduction
Epigenetics can be loosely defined as heritable changes in the function of genetic elements without changes in the actual genetic or underlying DNA sequence (Bird, 2007). There are three predominant mechanisms, including DNA methylation, histone modifications and non-coding RNAs. There are now numerous studies demonstrating significant roles of epigenetic regulation in biological systems, ranging from modulation of normal embryonic development to plasticity in the adult nervous system. For example, with the same set of genomic DNA, epigenetic regulation allows one zygote to differentiate into hundreds of distinct functional cell types in the body. Aberrant epigenetic regulation has also been implicated in cancer, congenital diseases, neurodegenerative diseases and neuropsychiatric disorders (Jones and Baylin, 2002; Feng and Fan, 2009; Urdinguio et al., 2009).
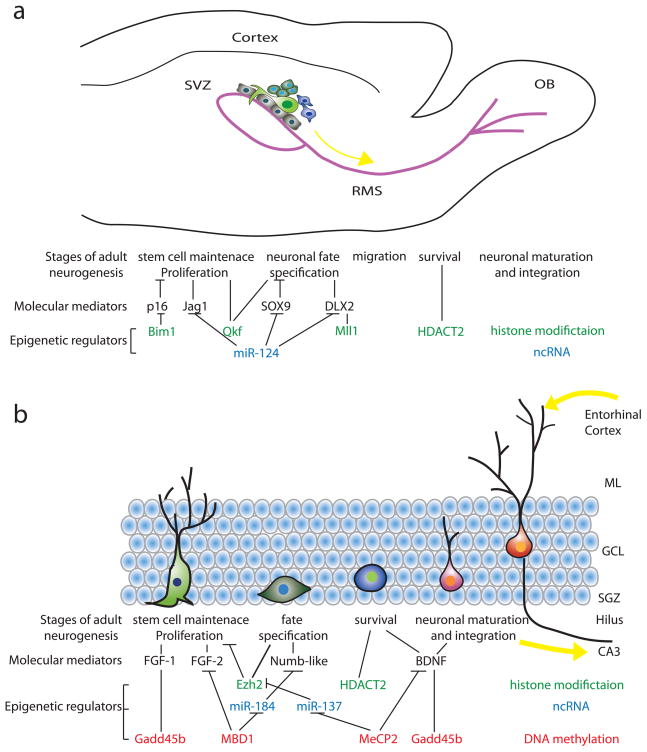
Adult neurogenesis occurs throughout life in two discrete regions of adult mammalian brain (Ming and Song, 2005; Lledo et al., 2006; Ma et al., 2009a). In the subventricular zone (SVZ) of the lateral ventricles, neural progenitors proliferate and give rise to neuroblasts, which then migrate through the rostral migratory stream (RMS) and differentiate into granule neurons and periglomerular neurons in the olfactory bulb (Figure 1a)(Alvarez-Buylla and Lim, 2004). In the subgranular zone (SGZ) of the dentate gyrus in the hippocampus, neural progenitors produce immature new neurons that migrate locally into the granule cell layer to become dentate granule cells (Figure 1b)(Zhao et al., 2008). Adult neurogenesis appears to recapitulate the complete process of neuronal development, ranging from neural progenitor activation and fate determination, to differentiation, migration, axonal and dendritic development of newborn neurons, to synapse formation and functional integration into the existing neural circuitry (Duan et al., 2008). Adult neurogenesis is tightly regulated by their local environment, or “neurogenic niche” (Doetsch, 2003; Alvarez-Buylla and Lim, 2004). This niche is composed of the extracellular matrix and various cell types, including astroglia, ependymal cells, endothelial cells, immature progeny of adult neural stem cells, and mature neurons within the local circuitry (Ma et al., 2005; Jordan et al., 2007). The niche is also a target of many physiological, pathological and pharmacological stimuli that regulate adult neurogenesis. Until recently, the analysis of adult neural stem cells and neurogenesis has largely focused on cellular signaling studies. We have just begun to understand the critical roles of nuclear epigenetic regulation in linking external environmental influence of the niche with the internal transcriptional and post-transcriptional control of gene expression in neural progenitors and their progeny in the adult brain. In this review, we highlight three major epigenetic mechanisms, including DNA methylation, histone modifications and non-coding RNAs, in both cell intrinsic and extrinsic regulation of different aspects of adult neurogenesis in the SVZ/olfactory bulb and SGZ/dentate gyrus. Interested readers are encouraged to read additional reviews on related topics (Ma et al., 2009b; Covic et al., 2010; Hsieh and Eisch, 2010; Ma et al., 2010).
Figure 1.
Epigenetic regulation of neurogenesis in the adult mammalian brain. Shown are schematic diagrams of the neurogenesis process in the adult SVZ/olfactory bulb system (a) and the SGZ/hippocampal system (b), respectively. A summary of current knowledge on major epigenetic regulations in regulating gene expression and different aspects of adult neurogenesis is also presented. SVZ: subventricular zone; OB: olfactory bulb; RMS: rostral migratory stream; RGL: radial glia-like cell; TA cell: transient amplifying cell; ncRNA: non-coding RNA; ML: molecular layer; GCL: granule cell layer; SGZ: subgranular zone.
DNA methylation
DNA methylation was first described in the late 1940s (Hotchkiss, 1948) and early 1950s (Wyatt, 1951) and is probably the most intensively studied type of epigenetic modification. DNA methylation in the mammalian genomes predominantly occurs at the cytosine residue of CpG dinucleotides to generate 5-methylcytosine on the pyrimidine ring (Jaenisch and Bird, 2003; Suzuki and Bird, 2008; Zhu, 2009). Limited non CpG methylation has been found in embryonic stem cells (Ramsahoye et al., 2000; Lister et al., 2009), but has not yet been characterized in detail in other cell types, such as neural progenitors and their progeny. A family of DNA methyltransferases (Dnmts) is responsible for the catalysis of DNA methylation (Goll and Bestor, 2005; Reik, 2007). Dnmt3a and Dnmt3b act as de novo methyltransferases to transfer methyl groups from S-adenosyl-L-methionine (SAM) to unmethylated target DNA, whereas Dnmt1 recognizes hemi-methylated DNA after DNA replication and maintains the DNA methylation state in daughter cells. DNA methylation has been traditionally viewed as a very stable epigenetic mark compared to other epigenetic modifications, yet the loss of DNA methylation, or demethylation, has been observed in many biological processes (Wu and Zhang, 2010). The loss of the methyl group from 5-methylcytosine DNA can occur through a passive process during DNA replication in the absence of functional Dnmt1 activity. The mechanism of active demethylation, a process of enzymatic removal of the methyl group from genomic DNA without DNA replication, is under intensive debate (Ooi and Bestor, 2008; Wu and Zhang, 2010). Multiple mechanisms for active DNA demethylation have been proposed for the mammalian system (Ma et al., 2009c; Wu and Zhang, 2010), including DNA excision repair-based mechanisms involving the Growth arrest and DNA damage-inducible protein 45 (Gadd45) family of proteins (Morgan et al., 2004; Rai et al., 2008; Gehring et al., 2009; Ma et al., 2009e) and Radial SAM mechanisms involving the elongator complex (Okada et al., 2010).
The methylation status of DNA, depending on its location and density, has been shown to play critical roles in gene regulation, including tissue-specific gene expression, X chromosome inactivation, gene imprinting and cell reprogramming (Jaenisch and Bird, 2003; Edwards and Ferguson-Smith, 2007; Reik, 2007; Suzuki and Bird, 2008). Deficiencies in Dnmts result in embryonic lethality or other profound developmental defects (Goll and Bestor, 2005; Reik, 2007). Recent studies have implicated a critical role of DNA methylation in regulating neurogenesis (Meissner et al., 2008; Mohn et al., 2008). During the embryonic development, Dnmt1 is critical for the timing of the switch from neurogenesis to gliogenesis; conditional deletion of Dnmt1 in embryonic cortical neural progenitors results in DNA hypomethylation and precocious astrogliogenesis (Fan et al., 2005). Furthermore, proliferating neural progenitors with Dnmt1 deletion exhibit DNA hypomethylation and are rapidly eliminated within three weeks of postnatal life (Fan et al., 2001). Thus, the proper DNA methylation status is critical for both maintenance and fate choice of neural progenitors during early development. Whether Dnmts play similar roles in regulating adult neurogenesis remains to be examined.
Methyl-CpG-binding proteins are major mediators of DNA methylation in regulating gene expression. Methyl-CpG-binding domain protein 1 (Mbd1) null mice exhibit defects in neurogenesis in the dentate gyrus of the adult hippocampus (Zhao et al., 2003). Mbd1 directly binds to the promoter region of fibroblast growth factor 2 (Fgf2), a major mitogen for adult neural progenitors in vitro and in vivo (Ma et al., 2009d), resulted in a tight regulation of Fgf2 expression in adult neural progenitors (Figure 1b)(Li et al., 2008a). A recent study has revealed an even more complex picture wherein Mbd1 suppresses several microRNAs (miRNAs) and interaction between two epigenetic mechanisms helps control the balance between proliferation and differentiation of adult neural progenitors of the hippocampus (Figure 1b; See below)(Liu et al., 2010).
Methyl-CpG-binding protein 2 (Mecp2), a gene mutated in Rett Syndrome, is also a major regulator of gene expression in the nervous system. In the developing brain, MeCP2 functions both cell-autonomously and non-cell-autonomously to regulate maturation and dendritic arborization of cortical pyramidal neurons (Kishi and Macklis, 2010). In the adult dentate gyrus of Mecp2 knockout mice, newborn neurons exhibit pronounced deficits in their neuronal maturation and spine formation (Smrt et al., 2007), suggesting a conserved function of Mecp2 during different developmental stages. One of the best known targets of Mecp2 is brain-derived neurotrophic factor (Bdnf), which has been implicated in regulating several aspects of adult hippocampal neurogenesis, including proliferation of neural progenitors (Li et al., 2008b) and development of newborn neurons (Schmidt and Duman, 2007). MeCP2 binds to the Bdnf promoter and suppresses its expression. After neuronal activation and membrane depolarization, two potential mechanisms may facilitate the release of MeCP2 from the Bdnf promoter to facilitate its transcription. First, in cultured primary neurons, neuronal activity induces DNA demethylation at specific CpG sites in the promoter IV of Bdnf, resulting in a dissociation of the MeCP2-histone deacetylase-mSin3A complex from the promoter and increased Bdnf transcription (Martinowich et al., 2003). Secondly, neuronal activity-induced calcium influx triggers phosphorylation of MeCP2 at serine 421 to facilitate its release from the promoter and subsequent transcription of Bdnf (Chen et al., 2003; Zhou et al., 2006). While MeCP2 was previously believed to regulate neuronal maturation, but not fate choice of neural progenitors (Kishi and Macklis, 2004), a recent study has suggested a novel role for MeCP2 in regulating proliferation and differentiation of adult hippocampal neural progenitors both in vitro and in vivo through modulating the expression of one miRNA, miR-137 (Figure 1b; See below)(Szulwach et al., 2010). Similar to the action of Mbd1, MeCP2 represents another example that interaction between two epigenetic mechanisms regulates the balance between proliferation and differentiation of neural progenitors in the adult hippocampus.
Adult neurogenesis is known to be regulated by many stimuli, including neuronal activity and antidepressant treatments. Transient neuronal activation, such as seizures, leads to sustained up-regulation of adult hippocampal neurogenesis over a period of weeks (Parent et al., 1997). Using electroconvulsive stimulation as a paradigm for synchronous neuronal activation of dentate granule cells, Gadd45b was identified as an immediate early gene that requires depolarization-induced calcium influx and CaM kinase activity for its induction (Ma et al., 2009e). A family of Gadd45 proteins has recently been implicated in promoting active DNA demethylation in the vertebrate system, likely through its interaction with the DNA excision-repair based DNA demethylation mechanism (Ma et al., 2009c). Interestingly, Gadd45b knockout mice exhibit specific defects in neuronal activity-induced proliferation of neural progenitors and dendritic growth of newborn dentate granule cells in the adult hippocampus, by either electroconvulsive stimulation or physical exercise (Ma et al., 2009e). Mechanistically, Gadd45b knockout mice exhibit defects in neuronal activity-induced CpG demethylation in the brain-specific promoter B of the Fgf1 gene and promoter IX of the Bdnf gene, as well as subsequent sustained expression of these specific gene isoforms (Ma et al., 2009e). In vitro, FGF-1 exhibits similar potency as FGF-2 in regulating the self-renewal of adult hippocampal neural progenitors. Interestingly, Gadd45b appears to be largely induced in mature neurons, but not in proliferating neuronal progenitors in vivo, suggesting a niche-based regulatory mechanism instead of an intrinsic mechanism. Thus, Gadd45b-dependent DNA demethylation may serve as a key mechanism to translate transient environment signaling to epigenetic changes in neurogenic niche cells for sustained regulation of neural progenitor and their neuronal development over the long-term.
Histone modifications
The building block of chromatin, nucleosome core particle, each consists of approximately 147 base pairs of DNA wrapped around two copies of four distinct histone proteins: H2A, H2B, H3 and H4 (Luger et al., 1999). The N-terminal tails of histones are subject to at least six distinct post-translational modifications, including acetylation, methylation, ubiquitination, phosphorylation, ribosylation, and SUMOylation. These histone tail amino acid-specific modifications, named “histone codes”, regulate the genomic accessibility and provide a platform for binding of other factors to control the activation or repression of associated genes.
Acetylation of histones occurs at lysine residues and is catalyzed by histone acetyltransferases (HATs). Histone acetylation is a reversible process and deacetylation is catalyzed by histone deacetylases (HDACs). Importantly, HDACs can act on many lysine-acetylated proteins, in addition to their prototype substrate, histones. Thus, any manipulations of HDAC activity might affect acetylation of a wide variety of intracellular targets. Recent studies have implicated HATs and HDACs in regulating adult neurogenesis. Knockout mice for Querkopf (Qkf, Myst4, Morf), a MYST family transcriptional co-activator with HAT activity (Champagne et al., 1999), exhibit reduced proliferation of neural progenitors in the SVZ, a decreased number of both migrating neuroblasts in the RMS and new interneurons in the olfactory bulb in middle-aged mice (9 months old), but not in young adult mice (3 months old)(Merson et al., 2006). Thus, Qkf-dependent epigenetic mechanisms may help to maintain the neurogenic capacity of neural progenitors in the adult brain (Figure 1a). In support of this view, neurospheres derived from Qkf knockout mice exhibit a decreased capacity for neuronal differentiation, whereas overexpression of Qkf increases neuronal production from both wild-type and Qkf mutant neurospheres in vitro (Merson et al., 2006). In a rodent model of human temporal lobe epilepsy, kainic acid-induced seizures lead to increased proliferation of neural progenitors, defective migration, and aberrant axonal and dendritic development of newborn dentate granule cells in the adult hippocampus (Jessberger et al., 2005; Jessberger et al., 2007b; Jessberger et al., 2007a). Interestingly, treatment with the HDAC inhibitor valproic acid blocks seizure-induced proliferation of neural progenitors in the adult dentate gyrus and protects animals from seizure-induced cognitive impairment in a hippocampus-dependent learning task (Jessberger et al., 2007b). In a separate study, treatment with sodium butyrate, another HDAC inhibitor, right after cerebral ischemia promotes proliferation of neural progenitors in both SVZ and SGZ (Kim et al., 2009). Given the non specificity of these drugs, studies using conditional knockout mice of different HATs and HDACs will provide more insight into the direct role of histone acetylation in regulating specific aspects of adult neurogenesis and the potential molecular mechanisms. Indeed, a recent study with inducible deletion of HDAC2 in neural progenitors using GLAST-CreERT2 mice shows abnormal maturation of newborn neurons in both the SGZ/hippocampus and the SVZ/olfactory bulb system, leading to increased cell death in both neurogenic regions (Jawerka et al., 2010). Further characterization suggests that HDAC2 plays an cell autonomous role during the immature neuronal stage only in adult neurogenesis, but not embryonic neurogenesis (Jawerka et al., 2010). One potential mechanism underlying HDAC2-dependent regulation is the silencing of Sox2 expression after neuronal differentiation of neural progenitors (Jawerka et al., 2010).
Histone methylation represents another important epigenetic mechanism for gene expression. The polycomb and trithorax group (PcG and TrxG) proteins are antagonistic chromatin complexes: members of the PcG complex catalyze trimethylation of lysine 27 of histone 3 (H3K27me3) that leads to transient transcriptional repression through local heterochromatin formation, whereas the TrxG complex is recruited by RNA polymerase II and catalyzes H3K4 trimethylation (H3K4me3) of promoter proximal nucleosomes to activate their target loci (Ringrose and Paro, 2007; Ng and Gurdon, 2008). The PcG and TrxG complexes have also been implicated in regulating specific aspects of adult neurogenesis. For example, in knockout mice for Bmi-1, a member of the PcG complex, neural progenitors, but not transient amplifying cells, are depleted in the SVZ (Molofsky et al., 2003). The effect of Bmi1 appears to be mediated by the cell cycle inhibitor p16 protein, since Bmi1 and p16 double knockouts have largely restored numbers of adult neural progenitors. Furthermore, Bmi1 over-expression in vitro significantly expands the number of adult SVZ neural progenitors and maintains their developmental potential to generate neuronal lineages (Fasano et al., 2007; Fasano et al., 2009). In another example, Mll1 (mixed-lineage leukaemia 1), a TrxG member that encodes an H3K4 methyltransferase, is specifically required for neuronal differentiation, but not glial differentiation, from SVZ neural progenitors (Lim et al., 2009). Dlx2 was identified as a direct target of Mll1 and is crucial for SVZ neurogenesis.
Our understanding of the roles of histone modifications in adult neurogenesis is just beginning to be developed. Rapid progress in the field has led to the identification of many histone methylatransferases and demethylases (Klose and Zhang, 2007). With an increasing number of animal models available to determine roles of specific histone modifications in different stages of adult neurogenesis, we expect to uncover an expanded role of such epigenetic mechanisms in regulating adult neurogenesis as well as underlying molecular mechanisms.
Non coding RNAs
Recent large-scale genome sequencing projects have revealed that only a very small percentage of the mammalian genome encode mRNAs that translate into proteins. Non-coding RNAs, which are transcribed from non-protein coding regions, have emerged as an important class of epigenetic regulators that interact with chromatin modifiers and transcription factors to regulate gene expression (Hobert, 2008; Morris, 2009; Iorio et al., 2010). There are both long and small non-coding RNAs, which include small nucleolar RNAs (snoRNAs), miRNAs, small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs). This list continues to grow with the recent identification of promoter-associated small RNAs (PASRs), transcription initiation RNAs (tiRNAs), microRNA-offset RNAs (moRNAs), MSY2-associated RNAs (MSY-RNAs), telomere small RNAs (tel-sRNAs) and centrosome-associated RNAs (crasiRNAs)(Taft et al., 2010). The non-coding RNAs regulate gene expression largely by modulating chromatin modifications, DNA transcription, RNA modifications, splicing, mRNA translation, and RNA stability. Recent evidence has revealed multiple members of the small RNA family that regulate adult neurogenesis.
One major class of non-coding RNAs is miRNAs, which inhibit gene expression through post-transcriptional mechanisms (Makeyev and Maniatis, 2008).A brain-specific miRNA, miR-124, while not detectable in neural progenitors, is up-regulated during the transition from transient amplifying cells to proliferating neuroblasts in the adult SVZ and further up-regulated in immature neurons in the RMS and olfactory bulb (Cheng et al., 2009). Gain- and loss-of-function analysis suggests that miR-124 functions in transient amplifying cells to promote their differentiation into neuroblasts, and specifically regulates the timing of lineage progression, instead of fate specification per se (Figure 1a). Mechanistically, miR-124 has three direct targets in the SVZ lineage, including Dlx2, Jag1 and Sox9 (Cheng et al., 2009). Dlx2 is a transcription factor involved in interneuron formation (Doetsch et al., 2002), whereas Jag1 is a Notch ligand that is important for self-renewal of neural progenitors in the postnatal SVZ (Nyfeler et al., 2005). On the other hand, Sox9 promotes the generation of GFAP+ cells while suppressing neuronal production. Importantly, miR-124 appears to eliminate Sox9 and Jag1 expression while only reducing Dlx2 expression in SVZ neuroblasts. Thus, a single miR-124 can fine-tune both the amount and timing of neurogenesis from transient amplifying cells to interneurons during postnatal SVZ neurogenesis (Figure 1a).
Two miRNAs have so far been implicated in regulating adult hippocampal neurogenesis. First, miR-184 is a direct target of Mbd1 in adult hippocampal neural progenitors (Liu et al., 2010). High levels of miR-184 promote proliferation and inhibit differentiation of adult hippocampal neural progenitors both in vitro and in vivo. Mechanistically, miR-184 acts through post-transcriptional repression of Numb-like, a known regulator of neuronal differentiation during development. Second, miR-137 has been identified as a direct target of Sox2 and MeCP2 in adult SGZ neural progenitors (Szulwach et al., 2010). Similar to miR-184, overexpression of miR-137 promotes the proliferation of adult hippocampal neural progenitors whereas a reduction of miR-137 enhances both neuronal and astrocyte differentiation from these progenitors in vitro. One major function of miR-137 appears to repress the translation of Ezh2, an H3K27 methyltransferase and PcG complex protein, leading to a global reduction of H3K27 methylation in adult SGZ neural progenitors. These interesting studies demonstrate a complex signaling network involving multiple epigenetic mechanisms to exquisitely regulate the fine balance between proliferation and differentiation of adult neural progenitors.
In addition to miRNAs, a novel small non-coding double stranded (ds) RNA containing the NRSE sequence, NRSE dsRNA, was suggested to be a key regulator of neuronal differentiation of adult hippocampal neural progenitors in vitro (Kuwabara et al., 2004). The NRSE sequence is a 21- to 23-base pair conserved DNA response element recognized by neuronal restricted silencing factor/RE-1 silencing transcription factor (NRSF/REST) (Schoenherr et al., 1996; Chen et al., 1998; Huang et al., 1999). NRSF/REST is a key transcriptional repressor for neuron-specific genes in non-neuronal cells and it mediates transcriptional repression through the association of the N-terminal repressor domain with the mSin3/histone deacetylase-1/2 (HDAC1/2) complex and through the association of C-terminal repressor domain with the CoREST complex (Huang et al., 1999; Naruse et al., 1999; Lunyak et al., 2002). One of the known REST/NRSF targets is miR-124, which regulates adult SVZ neurogenesis (Figure 1a)(Yoo et al., 2009; Juliandi et al., 2010). Surprisingly, NRSE dsRNA appears to convert NRSF/REST from a transcriptional repressor to an activatorin adult neural progenitors to promote neuronal differentiation (Kuwabara et al., 2004). In vitro analysis suggests that NRSE dsRNA is both sufficient and necessary for neuronal differentiation of adult neural progenitors. It remains to be determined whether this mechanism plays a critical role in regulating adult SGZ and SVZ neurogenesis in vivo.
Conclusions
Adult neurogenesis not only exemplifies the tremendous plasticity of the adult mammalian brain, but also provides a unique experimental model system to explore both intrinsic and extrinsic mechanisms regulating stem cell maintenance, activation and development. While the field of epigenetic analysis of adult neurogenesis is still in its nascent stage, significant progress has been made in the past few years and a number of basic principles have started to emerge. First, each stage of the adult neurogenesis process, ranging from maintenance and activation of neural progenitors, their fate specification, to maturation and development of neuronal progeny, is fine-tuned by multiple mechanisms involving various epigenetic regulators. In most cases, this regulation involves a complex interaction among different epigenetic mechanisms. Second, each epigenetic regulator appears to have multiple targets and affects multiple process of adult neurogenesis, especially at the transition stages. Third, epigenetic mechanisms regulate both intrinsic developmental stage-specific signaling and environmental niche signaling during adult neurogenesis. Fourth, certain epigenetic mechanisms regulating adult neurogenesis exhibit some unique features that are not present during embryonic and early postnatal neurogenesis.
Given the tremendous technical advances in the recent years, we expect to see an explosion of studies of epigenetics in the field of adult neurogenesis. First, we need to understand the profile of epigenetic status during different stages of adult neurogenesis. Such studies present a unique technical challenge since each diploid cell has only two such gene-specific modifications. A number of technologies are being developed to enrich specific cell types based on immunophenotyping (Rietze et al., 2001; Capela and Temple, 2002; Nagato et al., 2005; Corti et al., 2007; Pastrana et al., 2009) or transgenic reporter animals (Encinas and Enikolopov, 2008; Kanki et al., 2010). Emerging sequencing technologies also have made it possible for more sensitive, precise and genome-wide scale measurements of epigenetic DNA and histone modifications, and for profiling of mRNAs and various non-coding RNAs. Second, we need to gain knowledge on specific roles of different epigenetic regulators at distinct stages of adult neurogenesis in vivo. A number of Cre-ERT2-based driver mice now are available for inducible deletion or overexpression of many epigenetic regulators. Progress in the past decades has delineated the sequential steps during functional adult neurogenesis and such a detailed blueprint to adult neurogenesis will guide future analyses. These future endeavors will significantly enrich our knowledge regarding the basic mechanisms of adult neurogenesis and its regulation. Given the wealth of evidence implicating both epigenetics and adult neurogenesis in epileptogenesis, neuronal injury, degeneration and psychiatric disorders, futures studies may also lead to novel therapeutic targets and treatment strategies.
Acknowledgments
We thank D.K. Ma, K. Christian and H.S. Chu for comments and suggestions. Supported by the National Alliance for Research on Schizophrenia and Depression (NARSAD), National Institute of Health (HD069184, NS048271) and Miriam and Sheldon G. Adelson Medical Research Foundation to G.L.M., by National Institute of Health (NS047344, AG024984, MH087874) to H.J.S.
References
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y, Heng HH, Yang XJ. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Nardini M, Donadoni C, Locatelli F, Papadimitriou D, Salani S, Del Bo R, Ghezzi S, Strazzer S, Bresolin N, Comi GP. Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol. 2007;205:547–562. doi: 10.1016/j.expneurol.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Covic M, Karaca E, Lie DC. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity. 2010;105:122–134. doi: 10.1038/hdy.2010.27. [DOI] [PubMed] [Google Scholar]
- Doetsch F. A niche for adult neural stem cells. Curr Opin Genet Dev. 2003;13:543–550. doi: 10.1016/j.gde.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Duan X, Kang E, Liu CY, Ming GL, Song H. Development of neural stem cell in the adult brain. Curr Opin Neurobiol. 2008;18:108–115. doi: 10.1016/j.conb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Enikolopov G. Identifying and quantitating neural stem and progenitor cells in the adult brain. Methods Cell Biol. 2008;85:243–272. doi: 10.1016/S0091-679X(08)85011-X. [DOI] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, Sun YE. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, Trumpp A, Poon C, Wilson CB, Jaenisch R. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- Hsieh J, Eisch AJ. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol Dis. 2010;39:73–84. doi: 10.1016/j.nbd.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jawerka M, Colak D, Dimou L, Spiller C, Lagger S, Montgomery RL, Olson EN, Wurst W, Gottlicher M, Gotz M. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6:93–107. doi: 10.1017/S1740925X10000049. [DOI] [PubMed] [Google Scholar]
- Jessberger S, Romer B, Babu H, Kempermann G. Seizures induce proliferation and dispersion of doublecortin-positive hippocampal progenitor cells. Exp Neurol. 2005;196:342–351. doi: 10.1016/j.expneurol.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Jessberger S, Zhao C, Toni N, Clemenson GD, Jr, Li Y, Gage FH. Seizure-associated, aberrant neurogenesis in adult rats characterized with retrovirus-mediated cell labeling. J Neurosci. 2007a;27:9400–9407. doi: 10.1523/JNEUROSCI.2002-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S, Nakashima K, Clemenson GD, Jr, Mejia E, Mathews E, Ure K, Ogawa S, Sinton CM, Gage FH, Hsieh J. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J Neurosci. 2007b;27:5967–5975. doi: 10.1523/JNEUROSCI.0110-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jordan JD, Ma DK, Ming GL, Song H. Cellular niches for endogenous neural stem cells in the adult brain. CNS Neurol Disord Drug Targets. 2007;6:336–341. doi: 10.2174/187152707783220866. [DOI] [PubMed] [Google Scholar]
- Juliandi B, Abematsu M, Nakashima K. Chromatin remodeling in neural stem cell differentiation. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Kanki H, Shimabukuro MK, Miyawaki A, Okano H. “Color Timer” mice: visualization of neuronal differentiation with fluorescent proteins. Mol Brain. 2010;3:5. doi: 10.1186/1756-6606-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Leeds P, Chuang DM. The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. J Neurochem. 2009;110:1226–1240. doi: 10.1111/j.1471-4159.2009.06212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MeCP2 functions largely cell-autonomously, but also non-cell-autonomously, in neuronal maturation and dendritic arborization of cortical pyramidal neurons. Exp Neurol. 2010;222:51–58. doi: 10.1016/j.expneurol.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem. 2008a;283:27644–27652. doi: 10.1074/jbc.M804899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luikart BW, Birnbaum S, Chen J, Kwon CH, Kernie SG, Bassel-Duby R, Parada LF. TrkB regulates hippocampal neurogenesis and governs sensitivity to antidepressive treatment. Neuron. 2008b;59:399–412. doi: 10.1016/j.neuron.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Ma DK, Ming GL, Song H. Glial influences on neural stem cell development: cellular niches for adult neurogenesis. Curr Opin Neurobiol. 2005;15:514–520. doi: 10.1016/j.conb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ma DK, Bonaguidi MA, Ming GL, Song H. Adult neural stem cells in the mammalian central nervous system. Cell Res. 2009a;19:672–682. doi: 10.1038/cr.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Kim WR, Ming GL, Song H. Activity-dependent extrinsic regulation of adult olfactory bulb and hippocampal neurogenesis. Ann N Y Acad Sci. 2009b;1170:664–673. doi: 10.1111/j.1749-6632.2009.04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009c;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Ponnusamy K, Song MR, Ming GL, Song H. Molecular genetic analysis of FGFR1 signalling reveals distinct roles of MAPK and PLCgamma1 activation for self-renewal of adult neural stem cells. Mol Brain. 2009d;2:16. doi: 10.1186/1756-6606-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009e;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merson TD, Dixon MP, Collin C, Rietze RL, Bartlett PF, Thomas T, Voss AK. The transcriptional coactivator Querkopf controls adult neurogenesis. J Neurosci. 2006;26:11359–11370. doi: 10.1523/JNEUROSCI.2247-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Morris KV. Non-coding RNAs, epigenetic memory and the passage of information to progeny. RNA Biol. 2009;6:242–247. doi: 10.4161/rna.6.3.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagato M, Heike T, Kato T, Yamanaka Y, Yoshimoto M, Shimazaki T, Okano H, Nakahata T. Prospective characterization of neural stem cells by flow cytometry analysis using a combination of surface markers. J Neurosci Res. 2005;80:456–466. doi: 10.1002/jnr.20442. [DOI] [PubMed] [Google Scholar]
- Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci U S A. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, Taylor V. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. Embo J. 2005;24:3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Parent JM, Yu TW, Leibowitz RT, Geschwind DH, Sloviter RS, Lowenstein DH. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J Neurosci. 1997;17:3727–3738. doi: 10.1523/JNEUROSCI.17-10-03727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK, Bartlett PF. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 2001;412:736–739. doi: 10.1038/35089085. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Schmidt HD, Duman RS. The role of neurotrophic factors in adult hippocampal neurogenesis, antidepressant treatments and animal models of depressive-like behavior. Behav Pharmacol. 2007;18:391–418. doi: 10.1097/FBP.0b013e3282ee2aa8. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Eaves-Egenes J, Barkho BZ, Santistevan NJ, Zhao C, Aimone JB, Gage FH, Zhao X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27:77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt GR. Recognition and estimation of 5-methylcytosine in nucleic acids. Biochem J. 1951;48:581–584. doi: 10.1042/bj0480581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]