Abstract
Neurological symptoms are common manifestations of Lyme disease; however, the paucibacillary nature of the spirochete in this environment has precluded a molecular analysis of the spirochete in the CNS. We have now adapted differential expression analysis by using a custom-amplified library (DECAL) in conjunction with Borrelia burgdorferi whole-genome and subgenome arrays to examine in vivo gene expression by B. burgdorferi in a non-human primate (NHP) model of neuroborreliosis. The expression profile of B. burgdorferi was examined in the CNS and heart of steroid-treated and immunocompetent NHPs. Eighty-six chromosomal genes and 80 plasmid-encoded genes were expressed at similar levels in the CNS and heart tissue of both immunocompetent and steroid-treated NHPs. The expression of 66 chromosomal genes and 32 plasmid-encoded genes was increased in the CNS of both immunocompetent and steroid-treated NHPs. It is likely that the expression of these genes is governed by physiological factors specific to the CNS milieu. However, 83 chromosomal and 114 plasmid-encoded genes showed contrasting expression profiles in steroid-treated and immunocompetent NHPs. The effect of dexamethasone on the immune status of the host as well as on the host metabolic pathways could contribute to these differences in the B. burgdorferi transcriptome. Results obtained herein underscore the complex interplay of host factors on B. burgdorferi gene expression in vivo. The results provide a global snapshot of the spirochetal transcriptome in the CNS and should spur the design of experiments aimed at understanding the molecular basis of neuroborreliosis.
Lyme borreliosis, caused by Borrelia burgdorferi (1), can involve the skin, joints, heart, and nervous system (2). The manifestations of neuroborreliosis include facial palsy, aseptic meningitis, cranial neuritis, peripheral neuropathy, radiculopathy, and encephalitis (3, 4). The pathogenesis of neuroborreliosis is multifactorial and likely related to the persistence of a small number of spirochetes in the CNS and the host response to B. burgdorferi (5). Despite intensive clinical and basic research on neuroborreliosis, questions concerning its diagnosis, treatment, and pathogenesis remain unanswered.
The understanding of Lyme disease has been aided by several animal models, most notably the murine model (6–8). To date, however, the non-human primate (NHP) model has been the only system available for the study of nervous system involvement (4, 9, 10). Immunocompetent NHPs, as well as NHPs maintained on a low dose of dexamethasone, develop disseminated infection (4, 11, 12). B. burgdorferi-infected NHPs show both central and peripheral nervous system involvement, as evidenced by cerebrospinal fluid pleocytosis, mononuclear infiltration of cutaneous nerves, radiculopathy (10, 13), and mononeuropathy multiplex (14, 15).
B. burgdorferi preferentially expresses specific genes throughout its life cycle, both in the arthropod vector (Ixodes scapularis) and in the vertebrate host (16–21). It is generally considered that this process of differential gene expression facilitates spirochetal survival and disease pathogenesis (22, 23). B. burgdorferi gene expression is apparently orchestrated by environmental factors, including pH (24–26), temperature (27–30), and host immune responses (31). The analysis of tissue-specific expression of a limited number of genes has shown that B. burgdorferi also differentially express antigens in diverse tissues within the vertebrate host and the tick vector (16, 19–21, 32–37) invoking the additional role of physiological factors other than pH and temperature in modulating B. burgdorferi gene expression.
Although information gleaned from in vitro gene profiling of B. burgdorferi has been informative (25, 38), this exercise is limited by the inability to recapitulate the complex in vivo milieu of the diverse host tissues that B. burgdorferi infects. B. burgdorferi has been shown to colonize many tissues in the NHP (5, 11). The relatively few number of organisms in the CNS has further limited our ability to directly examine spirochete gene expression in the nervous system. To overcome this limitation, we have exploited differential expression analysis by using a custom-amplified library (DECAL) (36, 39). DECAL, a technique to selectively amplify specific prokaryotic transcriptomes, was first used for the global analysis of gene expression in Mycobacterium tuberculosis grown in vitro (39). The technique can be performed with as little as 10 ng of total bacterial RNA, can detect as low as 4-fold differences in gene expression, and can be used where contaminating host material is present (39). We have previously used this technique to characterize the gene expression profile of B. burgdorferi in engorging ticks in conjunction with a nylon membrane array consisting of clones from a B. burgdorferi genomic library (36). The uncharacterized nature of the clones spotted on this array, however, rendered the analysis slow and tedious. Recently, B. burgdorferi whole-genome microarrays have been used to examine the influence of pH and temperature on the transcriptome of spirochetes grown in vitro (25, 38) and of spirochetes grown in vivo within dialysis membrane chambers (DMCs) (40). Liang et al. (31) have used a subgenome microarray to assess the influence of host antibodies on gene expression. Therefore, DECAL was exploited in conjunction with whole-genome membrane arrays used in two previously published reports by Brooks et al. (38, 40) and subgenome glass slide arrays constructed by Liang et al. (41) to compare the transcriptome of B. burgdorferi in the central nervous tissue and heart in a NHP model of Lyme borreliosis. Hence, results obtained by Ojaimi et al. (38) and Liang et al. (31) serve as a baseline for comparison in this paper. Our results indicate that B. burgdorferi gene expression in vivo is predominantly modulated by physiological factors specific for the microenvironment of the particular tissue as well as by the metabolic and immune status of the host. This report provides the rationale for the design of hypothesis-driven experiments that will facilitate a molecular understanding of CNS–spirochete interactions and disease pathogenesis.
Materials and Methods
Array and Bb-CAL Construction. A whole-genome array containing 1,697 putative ORFs encoded by the B. burgdorferi B31 genome was printed onto a positively charged nylon membrane, as described (38). An array representing 137 genes encoding for putative lipoproteins in the B31 genome was printed onto glass slides as described (41). It has been determined that 68 genes downloaded on this array do not encode true lipoproteins. This glass slide array, representing a subset of B31 genes, is henceforth referred to as the B. burgdorferi subgenome array. A custom-amplified B. burgdorferi λ library was constructed as described (36) and schematically represented in Fig. 1A.
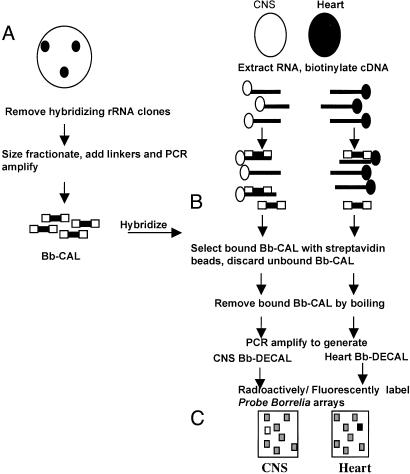
Fig. 1.
Schematic representation of DECAL. (A) Construction of Bb-CAL. A ZAP II B. burgdorferi genomic library was screened with radioactively labeled B. burgdorferi rRNA probes and hybridizing clones discarded. The nonribosomal clones were pooled and restriction digested with EcoRV and SmaI. The 200- to 2,000-bp fragments were gel-purified, ligated to PCR adapters, and PCR-amplified. (B) Positive selection and amplification. Total RNA isolated from heart and CNS of B. burgdorferi-infected NHPs was reverse transcribed in the presence of biotin-dATP. The biotinylated cDNA samples were hybridized to Bb-CAL under stringent conditions. The cDNA-Bb-CAL hybrids were bound to streptavidin-coated magnetic beads and then washed to remove unhybridized Bb-CAL. (C) The bound genomic Bb-CAL (representing the B. burgdorferi transcripts in the CNS/heart) was eluted by boiling and PCR-amplified. The PCR products were radioactively labeled and hybridized to a replicate genomic array.
Competent NHPs. Two 3- to 5-yr-old Chinese male rhesus monkeys (Macaca mulatta) were given 4 × 108 B. burgdorferi (strain JD1) per animal. The inoculum was administered i.p., s.c., and intradermally, as described (4). Infection was confirmed for both animals by culture of skin biopsy samples in BSK-H medium (Sigma) within the first 4 wk postinoculation. Animals were killed between 64 and 71 wk postinoculation. Medulla and heart specimens representing the CNS and peripheral tissues, respectively, were collected aseptically at necropsy, flash-frozen immediately after collection, and kept at –70°C until use.
Dexamethsone-Treated NHPs. Four male Rhesus macaques, M. mulatta, 3–4 yr of age were given dexamethasone (2 mg/kg) once daily i.m. 3 d before inoculation with 1 × 106 B. burgdorferi (N40) and maintained on this dose of dexamethasone for 28 days postinoculation. Subsequently, the monkeys were given 1 mg/kg of body weight of dexamethasone once daily for 1 wk followed by 0.5 mg/kg of body weight for 2 wk, after which the dexamethasone treatment was stopped. The animals were killed after 13 wk and necropsied as described (11). Infection was confirmed by culturing a sample of central nervous and peripheral tissue (12). Medulla representing the CNS and heart representing peripheral tissue were used for this study.
RNA Isolation. RNAs were isolated from the medulla and heart tissue of B. burgdorferi-infected NHPs (≈2–5 g of each tissue) by using the RNAWIZ RNA isolation reagent (Ambion, Austin, TX) according to the manufacturer's protocol. One microgram of total RNA was used to prepare biotinylated cDNA by using biotinylated random hexamer primers, biotin-dATP, and the SuperScript First Strand synthesis system for reverse transcription (Life Technologies, Gaithersburg, MD), essentially as described (39).
Positive Selection and Amplification. Equal amounts of biotinylated cDNA prepared from medulla and heart (normalized based on the levels of flaB transcripts) were hybridized separately to Bb-CAL and the biotinylated cDNA-Bb-CAL hybrids bound to streptavidin-coated magnetic beads (Dynal, Lake Success, NY) as shown in Fig. 1B. The Bb-CAL bound to the beads was eluted by boiling and PCR-amplified by using Uniamp primers, as described (39), and schematically outlined in Fig. 1C. The PCR products (Medulla-DECAL and Heart-DECAL) represent B. burgdorferi transcripts expressed in the medulla and heart tissues.
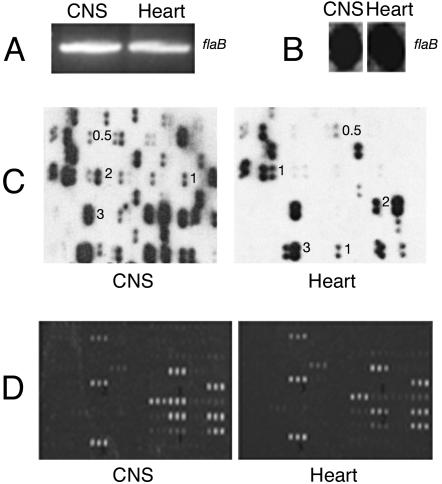
Differential Hybridization Analysis Using Whole-Genome Arrays. Equal amounts of PCR products corresponding to Medulla-DECAL and Heart-DECAL (template DNA normalized based on the level of flaB amplicon obtained by using specific PCR primers; Fig. 2A) were randomly labeled with [α-32P]. The radiolabeled probes were also normalized based on their hybridization intensity to equal amounts of flaB PCR product blotted onto nylon membrane (Fig. 2B). The normalized probes were then used to probe duplicate arrays by using RapidHyb buffer (Amersham Pharmacia–Pharmacia) according to the manufacturer's protocol. Hybridization was scored visually. Spots were given a score from 0 to 3 based on the intensity of hybridization (Fig. 2C) and the results tabulated.
Fig. 2.
Array hybridization. The CNS and Heart Bb-DECAL probes were normalized to the flaB amplicon obtained by PCR (A) and by their hybridization intensity to an equal amount of (25 ng) of flaB PCR product spotted on a nitrocellulose membrane (B). B. burgdorferi whole-genome arrays (C) and glass slide subarrays representing a subset of B. burgdorferi genes (D) were probed with CNS-Bb-DECAL and Heart-Bb-DECAL as described in Materials and Methods. A representative field of each array and the visual scores assigned to the differentially hybridizing elements are shown.
Differential Hybridization Analysis Using Glass Subgenome Arrays. Equal amounts of PCR products corresponding to Medulla-DECAL and Heart-DECAL normalized as described above (Fig. 2A) were labeled with aminoallyl dUTP (Molecular Probes) by using the Nick translation kit (Invitrogen) and the resultant DNA fluorescently labeled by using the Alexa Fluor 647 kit (Molecular Probes). The labeled probes were hybridized to a preblocked B. burgdorferi glass slide subarray and scanned by using an Axon GenePix 4000A array scanner (Axon Instruments, Foster City, CA) essentially as described (31). The intensity of the spot corresponding to flaB was used as a calibration standard in each panel.
Confirmation of Expression by RT-PCR. cDNA, prepared from heart and CNS of steroid-treated N40-infectected NHPs and selectively enriched for B. burgdorferi transcripts by DECAL as described above, was used as template to PCR amplify transcripts corresponding to several genes identified on the nylon whole-genome array and the subgenome glass slide array. The PCR products were electrophoresed and bands visualized under UV light. cDNA prepared from the CNS and heart tissue of steroid-treated NHPs infected with B. burgdorferi strain N40 was also used directly as template without DECAL enrichment to PCR amplify transcripts corresponding to several genes identified by the array analysis. The results obtained by using DECAL-enriched cDNA as template were comparable to those obtained with total cDNA as template. However, DECAL-enriched cDNA templates provided stronger amplicon signals. Because we were limited by the amount of total cDNA available for the studies, technical replicates of the PCR analysis were carried out with DECAL-enriched cDNA templates and are presented in this report.
Bioinformatics. The predicted proteins of selected B. burgdorferi sequences were analyzed by using the psort program (www.psort.org) to assess the probability for the input protein to be localized at the inner membrane (IM), outer membrane (OM), periplasmic, and cytosolic sites. The predicted proteins of selected B. burgdorferi sequences were also analyzed against the PROSITE database (http://ca.expasy.org/prosite) to determine the presence of lipid attachment and processing sites to establish that the input proteins encoded potential lipoproteins.
Results and Discussion
Whole-Genome Array Analysis of B. burgdorferi Genes in the CNS and Heart. Medulla represents a deep tissue of the CNS, distinct from peripheral tissue, and hence was the focus of this study. Heart tissue, a site known to harbor spirochetes (42), was chosen to represent a peripheral tissue. Expression profiles observed in these two sites may be distinct to these tissues and governed by tissue-specific signals. Medulla-DECAL and Heart-DECAL were therefore prepared from B. burgdorferi N40- and JD1-infected NHPs and used to probe duplicate B. burgdorferi strain B31 whole-genome membrane arrays (38). The probes were normalized as described in Materials and Methods. The array hybridizations were performed at least twice, and the results were similar. Only elements that gave reproducible hybridization results in all experiments were included in this study. A sample portion of each array is shown to indicate the differences in spot intensities and the corresponding visual scores assigned to them (Fig. 2C).
A comparison of genes expressed in the CNS and heart was performed and described relative to the expression levels in the heart tissue. A summary of the profiles is presented in Tables 2–4 and graphically presented in Fig. 4, which are published as supporting information on the PNAS web site. Although >1,450 ORFs were expressed by in vitro grown spirochetes (38), detectable hybridization was obtained for 239 chromosomal and 202 plasmid-encoded genes in the CNS of JD1-infected immunocompetent NHPs and for 243 chromosomal and 227 plasmid-encoded genes in the CNS of N40-infected steroid-treated NHPs (Table 1). This may indicate that the in vivo transcriptome of B. burgdorferi is different from the in vitro transcriptome. One cannot rule out the possibility that the inability to detect the expression of several ORFs may be an inherent limitation of the DECAL technique. These results could also be reflective, at least in part, of potential differences in the genomic composition of JD1, N40, and B31. Liang et al. (41) used a B. burgdorferi B31 subgenome array to compare the genomic composition of different B. burgdorferi strains, including N40, 297, Borrelia garinii, and Borrelia afzelii. Based on the hybridization patterns of the different strains to the B31 subgenome array, it was shown that the genomic composition of N40 and 297 was similar to that of B31 (41). Although a similar comparison of the JD1 strain has not been carried out, the genomic composition of the B. burgdorferi sensu stricto strains tested were not vastly different and hybridized to 97% of the subgenome array (41). It was reasonable to presume that JD1 strain would also hybridize to the B31 array elements. Differences in the metabolic and immune status of the steroid-treated and immunocompetent NHPs could further account for the differences in the number of hybridizing elements between the steroid-treated and immunocompetent NHP samples. Because the steroid-treated animals were killed at an earlier time point when compared with immunocompetent animals, the differences in the gene expression profiles between steroid-treated and immunocompetent NHP samples could also relate to the duration of infection.
Table 1. Overall comparison of the expression profiles of chromosomal and plasmid-encoded B. burgdorferi genes in the CNS of steroid-treated and competent (NHP).
| NHP | Total hybridizing elements | Up-regulated in the CNS | Down-regulated in the CNS | Expressed at same levels | Specifically induced in the CNS |
|---|---|---|---|---|---|
| Competent chromosomal | 239 | 99 | 7 | 97 | 36 |
| Competent plasmid | 202 | 58 | 2 | 86 | 56 |
| Steroid-treated chromosomal | 243 | 78 | 10 | 139 | 16 |
| Steroid-treated plasmid | 227 | 36 | 4 | 166 | 21 |
Expression profiles in the CNS are relative to that in the heart tissue of the respective NHPs.
Ninety-seven chromosomal and 86 plasmid-encoded genes were expressed at similar levels in the CNS and heart of JD1-infected immunocompetent NHPs. Of these, 86 chromosomal and 80 plasmid-encoded genes were also expressed at similar levels in the heart and CNS of steroid-treated N40-infected NHPs (Table 2). The expression of 99 chromosomal and 58 plasmid-encoded genes (Table 1) was increased in the CNS of JD1-infected NHPs. The expression of 56 of these 99 chromosomal and 18 of these 58 plasmid-encoded genes was also similarly increased in the CNS of steroid-treated N40-infected NHPs (Table 3, which is published as supporting information on the PNAS web site). The expression of seven chromosomal genes (bb0365, bb0420, bb0419, bb0377, bb0464, bb0070, and bb0580) and two plasmid-encoded genes (bba33 and bba57) was down-regulated in the CNS of JD1-infected NHPs. These genes were also down-regulated in the CNS of N40-infected NHPs (Tables 1 and 3). In addition, at least three other chromosomal (bb0387, bb0376, and bb0715) and two other plasmid-encoded genes (bba66 and bba59) were down-regulated in the CNS of N40-infected NHPs (Table 1 and Table 4, which is published as supporting information on the PNAS web site). Thirty-six chromosomal genes and 56 plasmid-encoded genes were preferentially (no detectable hybridization in the heart tissue) expressed in the CNS of JD1-infected NHPs. Of the 36 chromosomal and 56 CNS-specific plasmid-encoded JD1 genes, 10 chromosomal and 14 plasmid-encoded genes were also preferentially expressed in the steroid-treated N40-infected CNS (Tables 1 and 3).
Subgenome Array Analysis of B. burgdorferi Genes in the CNS and Heart. With the exception of two genes, results obtained using the glass slide subgenome array were consistent with those obtained using the whole-genome membrane array (Table 5). Of the 137 gene elements printed on the glass slide array, detectable hybridization was obtained for 43 elements, of which 21 encoded lipoproteins (Table 5). Detectable hybridization was not obtained for bb0760 and bbi34, both encoding hypothetical proteins, on the whole-genome array. However, glass slide array analysis indicated that bb0760 was up-regulated in the CNS of N40-infected NHPs. Probes prepared from JD1-infected NHPs did not show hybridization to bb0760 on both glass slide and nylon membrane arrays. A similar expression level of bbi34 was observed in the CNS and heart tissues of both immunocompetent and steroid-treated NHPs by using glass slide arrays. Fluorescent-labeled probes are more sensitive than radiolabeled probes, which could account for the detection of bbi34 and bb0760 on the glass slide array.
Genes Expressed at Same Levels in the Heart and CNS Tissues of Immunocompetent and Steroid-Treated NHPs. Eighty-six chromosomal genes and 82 plasmid-encoded genes were expressed at similar levels in the CNS and heart tissues of both immunocompromised and immunocompetent primates (Table 2). At least 75 (87%) chromosomal and 64 (78%) plasmid-encoded genes that were constitutively expressed in the heart and CNS of NHPs were not influenced by changes in temperature in vitro. For example, the expression levels of chromosomal genes such as bb0301 (cell division protein), bb0493 (ribosomal protein L6), bb0633 (exodeoxyribonuclease V), bb0641 (spermidine putrescine ABC transporter), and plasmid-encoded genes, such as bbn23 (pore-forming hemolysin), bbo18, bbn41, and bbg23 encoding hypothetical proteins, were not significantly affected by changes in temperature in vitro (38), nor were they influenced by host milieu (Table 2). It is likely that these gene products serve housekeeping functions important in both environments.
Genes Differentially Expressed in the Heart and CNS Tissues of Immunocompetent and Steroid-Treated NHPs. Regardless of the immune and metabolic status of the animals, the expression profiles of several genes were similar in both the N40- and JD1-infected animals (Tables 3 and 5, which are published as supporting information on the PNAS web site). At least three genes involved in amino acid metabolism [bb0220, bb0738 (Table 3), and bb0372 (Tables 3 and 5) encoding alanyl, valyl, and glutamyl tRNA synthetases, respectively] were up-regulated in the CNS of both immunocompromised and immunocompetent animals. It is plausible that, in comparison to the heart, the CNS tissue may contain these amino acids in abundance and contribute to the increased expression of these genes (44).
Expression of Outer Surface Lipoproteins ospA and ospB in NHPs. The increased expression of two genes, bba15 and bba16, in the CNS of both immunocompetent and steroid-treated NHPs (Table 3) was of particular interest. Expression of bba15 (ospA) could be detected in the CNS and heart tissues (score of 0.5) of both immunocompetent and steroid-treated NHPs; however, the levels of expression were consistently higher in the CNS (score of 1). Although expression of bba16 (ospB) could readily be detected in the CNS samples of these animals (score of 3), no signal for this gene was apparent in heart samples. bba15 and bba16 comprise an operon whose expression has long been assumed to depend on a promoter upstream of bba15 (ospA). The detection of higher levels of bba16 (ospB) than of bba15 (ospA) in the CNS of both immunocompetent and steroid-treated NHPs suggests that bba16 (ospB) may be expressed independently of bba15 (ospA) in vivo. These data are supported by the studies of Liang et al. (22), who reported the expression of ospB independently of ospA in the peripheral tissues of scid mice. The reciprocal expression pattern of OspAB and OspC lipoproteins has long been held as the paradigm for differential gene expression in B. burgdorferi (45, 46). In the unfed tick, OspA apparently serves to tether the spirochete to the tick midgut predominantly via an OspA–tick midgut protein interaction (47), whereas OspC is not expressed at detectable levels in spirochetes within unfed tick midguts. With the onset of feeding, there is a concomitant decrease in OspA expression and an increase in OspC expression (35, 46). The evidence presented in this report for expression of ospA and ospB in NHPs clearly calls this paradigm into question. These findings are consistent, however, with those of Schutzer et al. (43), who reported the presence of IgM antibodies to OspA and OspB specifically in the cerebrospinal fluid (CSF) of patients with early Lyme disease and neurologic involvement. Schutzer et al. (43) also observed the presence of OspC antibodies in both the CSF and sera in 75% of patients with neurologic symptom. These combined data suggest that the coordinate expression of OspA is not entirely restricted to the tick midgut, and that regulation of ospAB and ospC expression in vivo is perhaps more complex than envisaged. This is also consistent with the observations made by Brooks et al. (40) that suggested that bba15 and bba16 were differentially regulated by host-specific signals during cultivation in DMCs.
Dissimilar Expression Profiles of Genes Between the Dexamethasone-Treated and Untreated NHPs. Dexamethasone treatment can affect both the host metabolic and immune system pathways (48, 49), and both are likely to influence B. burgdorferi gene expression. The dichotomy in the expression profiles of several B. burgdorferi genes (Tables 4 and 5) in the dexamethasone-treated and untreated NHPs is, perhaps, reflective of the differences in the metabolic and immune status of the NHPs and is discussed below. The possibility that differences in the genomic composition between the B. burgdorferi strains could contribute to these differences cannot be ruled out until a detailed sequence analysis of these genes is carried out.
(i) Host Metabolic/Nutritional Status on Spirochete Gene Expression. bb0342 and bb0341 encoding the A and B subunits of glutamyl–tRNA amidotransferase, an enzyme involved in the generation of glutamyl-tRNA, were expressed at the same levels in the CNS and heart tissue of dexamethasone-treated NHPs but up-regulated in the CNS of competent NHPs (Table 4). Dexamethasone treatment results in the induction of the vertebrate enzyme glutamine synthetase (50). The increased levels of glutamine in dexamethasone-treated NHPs presumably regulate the levels of the spirochete enzyme that generates the glutamyl-tRNA. In immunocompetent NHPs, the glutamine levels may be preferentially increased in the CNS compared with the heart. The gene bb0380, encoding a magnesium ion transport protein, was expressed at similar levels in the CNS and heart tissue of dexamethasone-treated NHPs (score of 3). In contrast, this gene was up-regulated in the CNS tissue of competent NHPs (Table 4) and is likely to be regulated by differences in magnesium levels in the CNS and heart of immunocompetent NHPs. The ability of B. burgdorferi to change its transcriptome in response to different metabolic cues is perhaps central to its successful dissemination to and persistence in diverse tissue compartments.
It has recently been shown that the oligopeptide permease (opp) genes oppA-I, oppA-II, oppA-III, oppA-IV, and oppA V are differentially expressed under diverse environmental conditions such as carbon and nitrogen availability, pH, temperature, and concentrations of leucine and phosphate, with each gene subject to regulation by its own unique promoter (51). Wang et al. (51) have shown that oppA V is regulated predominantly by temperature. Factors other than temperature also regulate the expression of oppA V in vivo, because expression is apparently increased in unfed ticks and mouse hosts. Wang et al. (51) observed an increase in oppA IV expression in the murine host and a decrease in the tick vector. We found that oppA IV (bbb16) is up-regulated in the CNS in comparison to the hearts of both dexamethasone-treated and untreated NHPs (Tables 3 and 5). The constitutive and higher-level expression of oppA V in the CNS and heart of steroid-treated NHPs (score of 3) may be driven by the metabolic status of the NHPs. OppA IV and V represent paralogs of OppA I–III and appear to have narrower substrate specificities, in contrast to their E. coli orthologs (52). Expression of oppA I–III (bb0328, bb0329, and bb0330, respectively) was not detected at significant levels in either tissue of both steroid-treated and immunocompetent animals. Interestingly, the in vitro expression levels of oppA I–III were higher than those of oppA IV and oppA V in vitro (38). This is in contrast to their in vivo expression profiles. oppB-2 (bb0747), oppC-1 (bb0333), and oppD (bb0334) encode transport polypeptides and ATP-binding proteins that, in conjunction with OppA IV and V, are required to form the peptide transport system (52). The finding that oppB-2, oppC-1, and oppD were up-regulated in the CNS of both groups of NHPs (Table 3) suggests that a functional transporter complex is present within spirochetes during infection.
(ii) Influence of the Immune Status of NHPs on B. burgdorferi Gene Expression. In a mouse model of Lyme disease, several genes were modulated by the host humoral response (31). We observed a similar alteration of the expression profile for the following genes: bb0687 (IM), bb0740 (IM), bb0324 (IM), bbk49 (periplasm or OM), bba15 (OM, lipoprotein), bbg25 (IM) (Tables 2 and 3), bba34 (periplasm or OM, lipoprotein), bbj34 (IM), bb0620 (IM), and bb0652 (IM) (Tables 4 and 5). In scid mice, the expression profiles of these genes were altered in peripheral tissue when exposed to sera from immunocompetent mice (31). In the CNS, where the spirochetes are apparently protected from immune selection pressure, these genes were up-regulated in comparison to the levels in the heart tissue of immunocompetent NHPs. It is conceivable that host humoral responses may influence the expression of bba15 (OspA, OM); bbk49 (conserved hypothetical protein, OM); and bba16 (OspB, OM). However, bb0652 (protein-export membrane protein, IM); bb0740 (conserved hypothetical protein, IM); bbg25 (conserved hypothetical protein IM); and bbj34 (unknown hypothetical protein, IM) encode proteins that are membrane-bound but not surface-exposed. It is difficult to envisage the direct influence of antibodies on the expression of these nonsurface-exposed IM proteins. The observation that the expression of bba16, bba15, bb0740, bbg25, and bbk49 is decreased in the heart of both immunocompetent and steroid-treated NHPs (Tables 3 and 5), however, suggests that tissue-specific factors also modulate the expression of these genes in NHPs, perhaps in conjunction with immune factors. This is consistent with the observations made by Brooks et al. (40) that several OM proteins were differentially regulated within DMCs despite the fact that the DMCs were not permeable to antibodies.
Differential Expression of Paralogous Genes. In both dexamethasone-treated and immunocompetent NHPs, we observed several differentially expressed genes that were members of paralogous families (PFs) and observed similar expression profiles for the genes bb0333, bb0640, bb0217, and bb0747 (PF 41) (Table 3). The expression of all four genes encoding ABC transporter proteins was increased in the CNS compared with the heart tissue. Because the expression of these genes is increased in the CNS of both steroid-treated and immunocompetent NHPs, we presume that the microenvironment of the CNS, rather than the host immune status, drives the increased expression of these genes. Similarly, bbk52 and bba04 (PF 44) were up-regulated in the CNS of both immunocompetent and steroid-treated NHPs (Table 3 and Fig. 3). The differential expression of these paralogous genes in the CNS and heart tissues, wherein temperature is not the variable factor, suggests that a tissue-specific in vivo milieu imposes a level of regulation in addition to that imposed by temperature
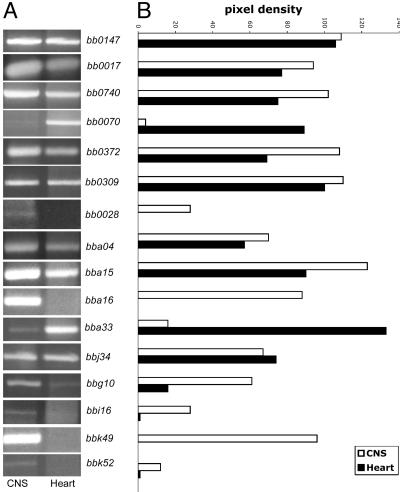
Fig. 3.
Confirmation of the expression profiles. (A) cDNA prepared from steroid treated B. burgdorferi- (N40) infected NHPs and enriched by DECAL was used as template in PCRs by using gene-specific primers, as outlined in Materials and Methods. The amounts of template DNA from CNS and heart tissues in the PCR were normalized against the expression levels of flaB (bb0147). (B) Relative pixel intensities of the amplicons were measured by using scion image, an image processing and analysis software program (www.scioncorp.com).
Consistent with an earlier finding (25) showing that genes within a PF need not be under similar transcriptional regulation at particular growth conditions, we observed dissimilar expression profiles within members of the same PF. Although bbn28 (PF 113) encoding for a putative lipoprotein is expressed at similar levels in the CNS and heart tissue of NHPs (Table 2), its PF members bbm28, bbo28, and bbr28 were apparently not expressed in either tissue. This family is the mlp lipoprotein family (37). The mlp genes tend to be variable, so it is possible that the N40 and JD1 do not have comparable genes. bbn31, bbh27, and bbh07 (PF 50) encoding conserved hypothetical proteins were expressed at the same levels in both heart and CNS tissue of both groups of NHPs (Table 2). The other 18 members of this PF were apparently not expressed in either tissue.
Confirmation of Microarray Data by RT-PCR. Expression levels of select genes (with a focus on lipoproteins and/or membrane proteins) were confirmed by RT-PCR, as described in Materials and Methods. Analysis of the genomic composition of various strains using the subgenome array has suggested that N40 and B31 may be genetically similar (41). Information on the genomic composition of the JD1 is not available. More information on the genomic content of the JD1 strain will be required to design optimal primer pairs to confirm the array analysis by PCR. For this reason, confirmation of the array analysis was limited to the transcriptome of N40 strain in steroid-treated NHPs.
DECAL-enriched cDNA prepared from heart and CNS of steroid-treated N40-infected NHPs was used as template to confirm the profiles determined by array analysis. The expression profiles of the following genes were confirmed by PCR by using gene-specific primers (available on request): bba15, bba16, bba33, bba04, bbi16, bb0028 (Tables 3 and 5); bbj34 (Tables 4 and 5); bb0740, bb0070, bbk49 (Tables 3 and 5); bb0017, bbk52, and bbg10 (Table 3); bb0309 (Tables 2 and 5); and bb0372 (Tables 3 and 5). The expression profiles obtained (Fig. 3) were consistent with the observations made by the whole-genome and subgenome-array analysis. The expression of genes encoding for membrane proteins such as bbk52, bbk49, bbi16, bb0028, bb0740, bba15, and bba16 was specifically induced or up-regulated in the CNS tissue (Fig. 3), consistent with the nylon and glass slide array data. In concurrence with the nylon and glass slide array data, the expression levels of membrane protein-encoding genes such as bb0309 and bbj34 were not significantly different in the CNS and heart tissue in steroid-treated NHPs (Fig. 3). The specific induction of membrane protein encoding genes such as bb0070 and bba33 was apparent in the heart tissue (Fig. 3) of dexamethaone-treated NHPs.
Significance and Implications. The paucibacillary nature of B. burgdorferi in the mammalian host has hampered systematic interrogation of the molecular basis of disease pathogenesis. Recently, several reports have used B. burgdorferi whole-genome microarrays to examine the transcriptome of spirochetes grown in vitro under a variety of conditions (25, 38) and in vivo in DMC (40). Although their observations are consistent with earlier findings, manipulation of in vitro conditions and the DMC model cannot fully replicate the complexities of diverse tissues. The results described in this report iterate the utility of DECAL to address the transcriptome of B. burgdorferi in vivo. Results obtained invoke the predominant role of tissue-specific factors in modulating the gene expression by B. burgdorferi in vivo. Although it is not possible at this juncture to unequivocally prove the functional significance of the genes specifically induced in the CNS, this study has identified target genes that can be examined to understand the interaction of the spirochete with the CNS.
Supplementary Material
Acknowledgments
We thank Benjamin Koski for excellent technical assistance. This work was supported by grants from the National Institutes of Health and American Heart Association. E.F. is the recipient of a Burroughs Wellcome Clinical Scientist Award in Translational Research. This work was supported by National Institutes of Health Grants R01NS34715 and N01A195358 (to A.R.P.), AI42352 and RR00164 (to M.T.P.), AI29375 (to J.D.R. and M.J.C.), AI32947 and AI49200 (to E.F.), and AR46878 (to S.N.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DECAL, differential expression analysis by using a custom-amplified library; NHP, non-human primates; IM, inner membrane; OM, outer membrane; PF, paralogous family; DMC, dialysis membrane chamber.
References
- 1.Barbour, A. G. & Fish, D. (1993) Science 260, 1610-1616. [DOI] [PubMed] [Google Scholar]
- 2.Steere, A. C. (2001) N. Engl. J. Med. 345, 115-125. [DOI] [PubMed] [Google Scholar]
- 3.Pachner, A. R. (2001) Immunol. Rev. 183, 186-204. [DOI] [PubMed] [Google Scholar]
- 4.Philipp, M. T., Aydintug, M. K., Bohm, R. P., Jr., Cogswell, F. B., Dennis, V. A., Lanners, H. N., Lowrie, R. C., Jr., Roberts, E. D., Conway, M. D., Karacorlu, M., et al. (1993) Infect. Immun. 61, 3047-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadavid, D., O'Neill, T., Schaefer, H. & Pachner, A. R. (2000) Lab. Invest 80, 1043-1054. [DOI] [PubMed] [Google Scholar]
- 6.Barthold, S. W., Moody, K. D., Terwilliger, G. A., Jacoby, R. O. & Steere, A. C. (1988) Ann. NY Acad. Sci. 539, 264-273. [DOI] [PubMed] [Google Scholar]
- 7.Philipp, M. T. & Johnson, B. J. (1994) Trends Microbiol. 2, 431-437. [DOI] [PubMed] [Google Scholar]
- 8.Luft, B. J., Steinman, C. R., Neimark, H. C., Muralidhar, B., Rush, T., Finkel, M. F., Kunkel, M. & Dattwyler, R. J. (1992) J. Am. Med. Assoc. 267, 1364-1367. [PubMed] [Google Scholar]
- 9.Pachner, A. R., Delaney, E., O'Neill, T. & Major, E. (1995) Neurology 45, 165-172. [DOI] [PubMed] [Google Scholar]
- 10.Roberts, E. D., Bohm, R. P., Jr., Cogswell, F. B., Lanners, H. N., Lowrie, R. C., Jr., Povinelli, L., Piesman, J. & Philipp, M. T. (1995) Lab. Invest. 72, 146-160. [PubMed] [Google Scholar]
- 11.Pachner, A. R., Cadavid, D., Shu, G., Dail, D., Pachner, S., Hodzic, E. & Barthold, S. W. (2001) Ann. Neurol. 50, 330-338. [PubMed] [Google Scholar]
- 12.Pachner, A. R., Amemiya, K., Bartlett, M., Schaefer, H., Reddy, K. & Zhang, W. F. (2001) Clin. Diag. Lab. Immunol. 8, 225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachner, A. R., Delaney, E. & O'Neill, T. (1995) Ann. Neurol. 38, 667-669. [DOI] [PubMed] [Google Scholar]
- 14.England, J. D., Bohm, R. P., Jr., Roberts, E. D. & Philipp, M. T. (1997) Ann. Neurol. 41, 375-384. [DOI] [PubMed] [Google Scholar]
- 15.Roberts, E. D., Bohm, R. P., Jr., Lowrie, R. C., Jr., Habicht, G., Katona, L., Piesman, J. & Philipp, M. T. (1998) J. Infect. Dis. 178, 722-732. [DOI] [PubMed] [Google Scholar]
- 16.Akins, D. R., Bourell, K. W., Caimano, M. J., Norgard, M. V. & Radolf, J. D. (1998) J. Clin. Invest. 101, 2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babb, K., El-Hage, N., Miller, J. C., Carroll, J. A. & Stevenson, B. (2001) Infect. Immun. 69, 4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll, J. A., Garon, C. F. & Schwan, T. G. (1999) Infect. Immun. 67, 3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fikrig, E., Chen, M., Barthold, S. W., Anguita, J., Feng, W. & Telford, S. R., Flavell, R. A. (1999) Mol. Microbiol. 31, 281-290. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi, J., Piesman, J. & de Silva, A. M. (2001) Proc. Natl. Acad. Sci. USA 98, 670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmore, R. D., Jr., Mbow, M. L. & Stevenson, B. (2001) Microbes Infect. 3, 799-808. [DOI] [PubMed] [Google Scholar]
- 22.Liang, F. T., Jacobs, M. B., Bowers, L. C. & Philipp, M. T. (2002) J. Exp. Med. 195, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, J. R. & Norris, S. J. (1998) Infect. Immun. 66, 3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll, J. A., Cordova, R. M. & Garon, C. F. (2000) Infect. Immun. 68, 6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Revel, A. T., Talaat, A. M. & Norgard, M. V. (2002) Proc. Natl. Acad. Sci. USA 99, 1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, X., Goldberg, M. S., Popova, T. G., Schoeler, G. B., Wikel, S. K., Hagman, K. E. & Norgard, M. V. (2000) Mol. Microbiol. 37, 1470-1479. [DOI] [PubMed] [Google Scholar]
- 27.Ramamoorthy, R. & Scholl-Meeker, D. (2001) Infect. Immun. 69, 2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson, B., Schwan, T. G. & Rosa, P. A. (1995) Infect. Immun. 63, 4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll, J. A., El-Hage, N., Miller, J. C., Babb, K. & Stevenson, B. (2001) Infect. Immun. 69, 5286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obonyo, M., Munderloh, U. G., Fingerle, V., Wilske, B. & Kurtti, T. J. (1999) J. Clin. Microbiol. 37, 2137-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, F. T., Nelson, F. K. & Fikrig, E. (2002) J. Exp. Med. 196, 275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fikrig, E., Feng, W., Aversa, J., Schoen, R. T. & Flavell, R. A. (1998) J. Infect. Dis. 178, 1198-1201. [DOI] [PubMed] [Google Scholar]
- 33.Cassatt, D. R., Patel, N. K., Ulbrandt, N. D. & Hanson, M. S. (1998) Infect. Immun. 66, 5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fikrig, E., Barthold, S. W., Sun, W., Feng, W. & Telford, S. R., Flavell, R. A. (1997) Immunity 6, 531-539. [DOI] [PubMed] [Google Scholar]
- 35.Schwan, T. G. & Piesman, J. (2000) J. Clin. Microbiol. 38, 382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narasimhan, S., Santiago, F., Koski, R. A., Brei, B., Anderson, J. F., Fish, D. & Fikrig, E. (2002) J. Bacteriol. 184, 3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porcella, S. F., Fitzpatrick, C. A. & Bono, J. L. (2000) Infect. Immun. 68, 4992-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojaimi, C., Brooks, C., Akins, D., Casjens, S., Rosa, P., Elias, A., Barbour, A., Jasinskas, A., Benach, J., Katonah, L., et al. (2003) Infect. Immun. 71, 1689-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alland, D., Kramnik, I., Weisbrod, T. R., Otsubo, L., Cerny, R., Miller, L. P., Jacobs, W. R., Jr. & Bloom, B. R. (1998) Proc. Natl. Acad. Sci. USA 95, 13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks, C. S., Hefty, P. S., Jolliff, S. E. & Akins, D. R. (2003) Infect. Immun. 71, 3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, F. T., Nelson, F. K. & Fikrig, E. (2002) Infect. Immun. 70, 3300-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pachner, A. R. (1989) Rev. Infect. Dis. 11, S1482-S1486. [PubMed] [Google Scholar]
- 43.Schutzer, S. E., Coyle, P. K., Krupp, L. B., Deng, Z., Belman, A. L., Dattwyler, R. & Luft, B. J. (1997) J. Clin. Invest. 100, 763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cozzone, A. J. (1980) Biochimie 62, 647-664. [DOI] [PubMed] [Google Scholar]
- 45.Schwan, T. G., Piesman, J., Golde, W. T., Dolan, M. C. & Rosa, P. A. (1995) Proc. Natl. Acad. Sci. USA 92, 2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Silva, A. M. & Fikrig, E. (1997) J. Clin. Invest. 99, 377-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal, U., de Silva, A. M., Montgomery, R. R., Fish, D., Anguita, J., Anderson, J. F., Lobet, Y. & Fikrig, E. (2000) J. Clin. Invest. 106, 561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guarino, N. & Puri, P. (2002) J. Pediatr. Surg. 37, 1563-1567. [DOI] [PubMed] [Google Scholar]
- 49.Oishi, Y., Fu, Z. W., Ohnuki, Y., Kato, H. & Noguchi, T. (2002) Br. J. Dermatol. 147, 859-868. [DOI] [PubMed] [Google Scholar]
- 50.Gaunitz, F., Heise, K., Schumann, R. & Gebhardt, R. (2002) Biochem. Biophys. Res. Commun. 296, 1026-1032. [DOI] [PubMed] [Google Scholar]
- 51.Wang, X. G., Lin, B., Kidder, J. M., Telford, S. & Hu, L. T. (2002) J. Bacteriol. 184, 6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin, B., Short, S. A., Eskildsen, M., Klempner, M. S. & Hu, L. T. (2001) Biochim. Biophys. Acta 1499, 222-231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.