Abstract
Ovarian Clear Cell Carcinoma (OCCC) is an aggressive human cancer that is generally resistant to therapy. To explore the genetic origin of OCCC, we determined the exomic sequences of eight tumors after immunoaffinity purification of cancer cells. Through comparative analyses of normal cells from the same patients, we identified four genes that were mutated in at least two tumors. PIK3CA, which encodes a subunit of phosphatidylinositol-3 kinase, and KRAS, which encodes a well known oncoprotein, had previously been implicated in OCCC. The other two mutated genes were novel: PPP2R1A encodes a regulatory subunit of serine/threonine phosphatase 2 and ARID1A encodes AT-rich interactive domain-containing protein 1A, which participates in chromatin remodeling. The nature and pattern of the mutations suggest that PPP2R1A functions as an oncogene and ARID1A as a tumor suppressor gene. In a total of 42 OCCCs, 7% had mutations in PPP2R1A and 57% had mutations in ARID1A. These results suggest that aberrant chromatin remodeling contributes to the pathogenesis of OCCC.
Ovarian cancers are a heterogeneous group of diseases with distinct clinicopathological and molecular features (1). Among them, OCCCs, which accounts for 10% of epithelial ovarian cancers, is one of the most aggressive types because, unlike the high grade-serous type, it is refractory to standard platinum-based chemotherapy. Previous morphological and molecular studies have indicated that OCCC develops in a stepwise fashion from a common disease progenitor state, in many cases endometriosis, and then proceeds to frank malignancy (2-6). Activating mutations in PIK3CA (7) and genomic amplification of chr20q13.2 (8) are the most common molecular genetic alterations so far identified in OCCC.
To explore the genetic basis of this tumor type, we have determined the sequences of the ∼18,000 protein-encoding genes listed in the RefSeq database in tumors from eight patients (table S1). Because these tumors are composed of a mixture of cancer and stromal cells, we purified the cancer cells using epithelial cell target antibodies attached to magnetic beads (9). Staining of the cells bound to the beads revealed that > 90% of them were OCCC cells. This procedure thereby maximized the sensitivity of the sequencing analyses by eliminating most of the contaminating normal cells (containing normal genomes) from the sample. DNA from the purified cells, as well as from normal cells obtained from the blood or uninvolved tissues of the same patients were used to generate libraries suitable for massively parallel sequencing by synthesis (9). Following capture of the coding sequences of the targeted genes with a SureSelect Enrichment System, the DNA was sequenced using an Illumina GAIIx platform. The average coverage of each base in the targeted regions was 84 fold and 92.7 % of these bases were represented in at least 10 reads (table S2).
Using stringent criteria for analysis of these data (9) we identified 268 somatic mutations in 253 genes among the eight tumors. The range of mutations per tumor was 13 to 125 alterations. Of these, 237 (88%) mutations were confirmed by Sanger sequencing (table S3). The tumor with 125 mutations (OCC06PT) was from a patient with recurrent disease that had previously been treated with chemotherapy. Excluding OCC06PT, there was an average of 20 mutations per tumor (table S2 and S3). The mutation spectrum was enriched for C to T transitions at 5′-CG base pairs, similar to those of other tumors whose exomes have been sequenced (10-14). Only four genes were mutated in more than one of the eight tumors studied: PIK3CA, KRAS, PPP2R1A, and ARID1A. The mutations in each of these four genes, and their somatic nature, were confirmed by Sanger sequencing of the DNA from the tumor and normal tissues of the corresponding patients (examples in Fig. 1). The sequences of these four genes were then determined in the tumor and normal tissues of an additional 34 OCCC cases using PCR amplification and Sanger sequencing with the primers listed in table S4. In total, PIK3CA, KRAS, PPP2R1A, and ARID1A mutations were identified in 40%, 4.7%, 7.1%, and 57% of the 42 tumors, respectively (Table 1).
Figure 1.
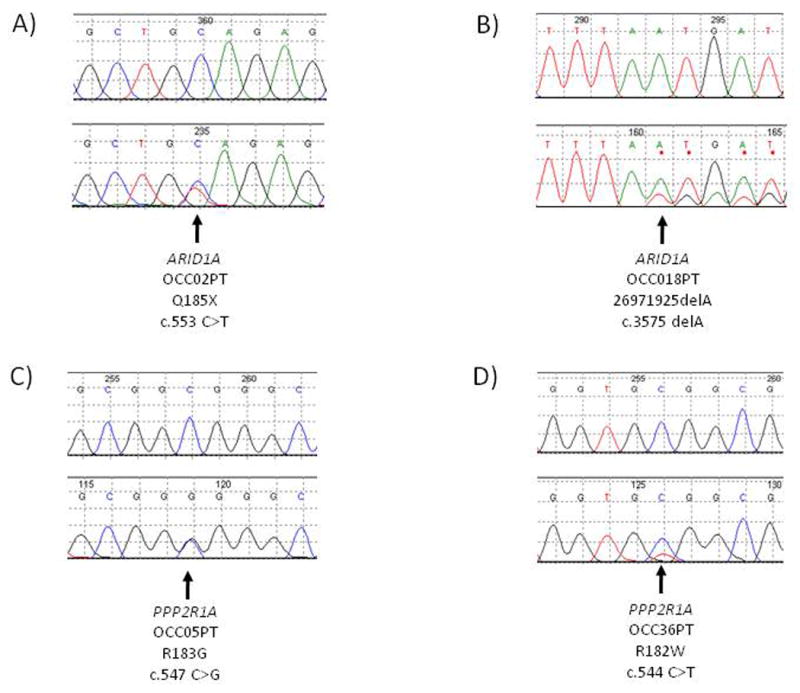
Sequence chromatograms showing somatic ARID1A and PPP2R1A mutations. The lower panels show the tumor and the upper panels show the matched normal control.
Table 1.
Mutations in ARID1A, KRAS, PIK3CA and PPP2R1A in Human Ovarian Clear Cell Carcinomas.
| Sample# | Gene | Transcript Accession | Nucleotide (genomic)* | Nucleotide (cDNA) | Amino acid (protein) | Mutation type |
|---|---|---|---|---|---|---|
| OCC01PT | ARID1A | CCDS285.1 | g.chr1:26972561_26972562insA | c.3854_3855insA | fs | Indel |
| OCC02PT | ARID1A | CCDS285.1 | g.chr1:26896034C>T | c.553C>T | p.Q185X | Nonsense |
| OCC02PT | ARID1A | CCDS285.1 | g.chr1:26978879-26978880dupGT | c.903_904dupGT | fs | Indel |
| OCC03PT | ARID1A | CCDS285.1 | g.chr1:26972009_26972034delTGATGGGGCGCATGTCCTATGAGCCA (hom) | c.3659_3684delTGATGGGGCGCATGTCCTATGAGCCA | fs | Indel |
| OCC07PT | ARID1A | CCDS285.1 | g.chr1:26896066C>A | c.585C>A | p.Y195X | Nonsense |
| OCC08PT | ARID1A | CCDS285.1 | g.chr1:26970389delC | c.3391delC | fs | Indel |
| OCC10PT | ARID1A | CCDS285.1 | g.chr1:26972790_26972792dupGCA (hom) | c.4001_4002dupGCA (hom) | fs | Indel |
| OCC10PT | ARID1A | CCDS285.1 | g.chr1:26979804_26979805delTG (hom) | c.6828_6829delTG(hom) | fs | Indel |
| OCC11PT | ARID1A | CCDS285.1 | g.chr1:26930334_26930335insCCTAC | c.1455_1466insCCTAC | fs | Indel |
| OCC13PT | ARID1A | CCDS285.1 | g.chr1:26974233_26974234insTGGC | c.4926_4927insTGGC | fs | Indel |
| OCC14PT | ARID1A | CCDS285.1 | g.chr1:26972886_26972887_delTT (hom) | c.4011_4012delTT (hom) | fs | Indel |
| OCC15PT | ARID1A | CCDS285.1 | g.chr1:26973940G>A | c.4635G>A | p.W1545X | Nonsense |
| OCC15PT | ARID1A | CCDS285.1 | g.chr1:26978178T>A | c.5202T>A | p.Y1734X | Nonsense |
| OCC16PT | ARID1A | CCDS285.1 | g.chr1:26895967_26895973delCGCCGCC (hom) | c.486_492delCGCCGCC (hom) | fs | Indel |
| OCC18PT | ARID1A | CCDS285.1 | g.chr1:26971925delA | c.3575delA | fs | Indel |
| OCC20PT | ARID1A | CCDS285.1 | g.chr1:26970221delG | c.3223delG | fs | Indel |
| OCC22PT | ARID1A | CCDS285.1 | g.chr1:26979694dupG | c.6718dupG | fs | Indel |
| OCC23PT | ARID1A | CCDS285.1 | g.chr1:26896379_2689637980_insCGTC | c.898_899insCGTC | fs | Indel |
| OCC23PT | ARID1A | CCDS285.1 | g.chr1:26979686_26979687insT | c.6710_6711insT | fs | Indel |
| OCC24PT | ARID1A | CCDS285.1 | g.chr1:26930542C>T | c.1663C>T | p.Q555X | Nonsense |
| OCC27PT | ARID1A | CCDS285.1 | g.chr1:26896263_26896272delCGTCGTCTTC | c.782_791delCGTCGTCTTC | fs | Indel |
| OCC27PT | ARID1A | CCDS285.1 | g.chr1:26971984_26971994delCAGCCCAGTAT | c.3634_3644delCAGCCCAGTAT | fs | Indel |
| OCC30PT | ARID1A | CCDS285.1 | g.chr1:26931823C>T | c.1873C>T | p.Q625X | Nonsense |
| OCC32PT | ARID1A | CCDS285.1 | g.chr1:26960135C>T | c.2122C>T | p.Q708X | Nonsense |
| OCC34PT | ARID1A | CCDS285.1 | g.chr1:26931754G>T | c.1804G>T | p.E602X | Nonsense |
| OCC34PT | ARID1A | CCDS285.1 | g.chr1:26979678delT | c.6702delT | fs | Indel |
| OCC36PT | ARID1A | CCDS285.1 | g.chr1:26928932T>G | c.1341T>G | p.Y447X | Nonsense |
| OCC36PT | ARID1A | CCDS285.1 | g.chr1:26971613delC | c.3442delC | fs | Indel |
| OCC39PT | ARID1A | CCDS285.1 | g.chr1:26896364dupC | c.883dupC | fs | Indel |
| OCC39PT | ARID1A | CCDS285.1 | g.chr1:26965434delC | c.2868delC | fs | Indel |
| OCC41PT | ARID1A | CCDS285.1 | g.chr1:26931831delT | c.1881delT | fs | Indel |
| OCC42PT | ARID1A | CCDS285.1 | g.chr1:26960479_26960488delCGGCCACCCA | c.2179_2188delCGGCCACCCA | fs | Indel |
| OCC04PT | KRAS | CCDS8703.1 | g.chr12:25289551C>T | c.35G>A | p.G12D | Missense |
| OCC05PT | KRAS | CCDS8703.1 | g.chr12:25289551C>G | c.35G>C | p.G12A | Missense |
| OCC01PT | PIK3CA | CCDS43171.1 | g.chr3:180418788C>A | c.1636C>A | p.Q546K | Missense |
| OCC02PT | PIK3CA | CCDS43171.1 | g.chr3:180418776G>A | c.1624G>A | p.E542K | Missense |
| OCC06PT | PIK3CA | CCDS43171.1 | g.chr3:180418785G>A | c.1633G>A | p.E545K | Missense |
| OCC08PT | PIK3CA | CCDS43171.1 | g.chr3:180418785G>A | c.1633G>A | p.E545K | Missense |
| OCC09PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>T | c.3140A>T | p.H1047L | Missense |
| OCC10PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC11PT | PIK3CA | CCDS43171.1 | g.chr3:180418777A>T | c.1625A>T | p.E542V | Missense |
| OCC13PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC15PT | PIK3CA | CCDS43171.1 | g.chr3:180410152C>G | c.1221C>G | p.C407W | Missense |
| OCC20PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC22PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC23PT | PIK3CA | CCDS43171.1 | g.chr3:180399648_180399649insCCTCAA | c.341_342insCCTCAA | fs | Indel |
| OCC27PT | PIK3CA | CCDS43171.1 | g.chr3:180399638A>G | c.331A>G | p.K111E | Missense |
| OCC30PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC35PT | PIK3CA | CCDS43171.1 | g.chr3:180418776G>A | c.1624G>A | p.E542K | Missense |
| OCC36PT | PIK3CA | CCDS43171.1 | g.chr3:180418785G>A | c.1633G>A | p.E545K | Missense |
| OCC42PT | PIK3CA | CCDS43171.1 | g.chr3:180434779A>G | c.3140A>G | p.H1047R | Missense |
| OCC05PT | PPP2R1A | CCDS12849.1 | g.chr19:57407794C>G | c.547C>G | p.R183G | Missense |
| OCC07PT | PPP2R1A | CCDS12849.1 | g.chr19:57407794C>T | c.547C>T | p.R183W | Missense |
| OCC36PT | PPP2R1A | CCDS12849.1 | g.chr19:57407791C>T | c.544C>T | p.R182W | Missense |
Coordinates refer to the human reference genome hg18 release (NCBI 36.1, March 2006).
Samples OCC01 to OCC08 were used for the initial (discovery) screen for mutations
Cancer cell lines with mutations in genes involved in cancer development provide valuable tools for further research. To this end, we extended the analysis of these four genes in seven OCCC cell lines that were derived from tumors independent of those described above. In seven cell lines we identified nine ARID1A mutations in five cell lines, three with PPP2R1A mutations, one with a KRAS mutation and four with PIK3CA, mutations (table S5).
The nature of the somatic mutations in tumors can often be used to classify them as oncogenes or tumor suppressor genes (15). In particular, all bona fide oncogenes are mutated recurrently (that is, at the same codon or clustered at a few adjacent codons in different tumors), and the mutations are nearly always missense. In contrast, all bona fide tumor suppressor genes are mutated at a variety of positions throughout the coding region of the gene and the mutations often truncate the encoded protein through production of a stop codon by a base substitution, an out-of-frame insertion or deletion (indel) or a splice site mutation. Moreover, tumor suppressor gene mutations generally affect both alleles, whereas mutations in oncogenes commonly affect only one allele.
Based on this logic, we can speculate about the likely function of the four genes in OCCC. PIK3CA and KRAS are well-studied oncogenes, and the 19 mutations identified in OCCC were heterozygous and clustered; fourteen of the 17 mutations in PIK3CA were at codons 542, 546, or 1047, while both mutations in KRAS were at codon 12 (Table 1). The three mutations in PPP2R1A were similarly heterozygous and clustered, suggesting that it functions, when mutated, as an oncogene (Table 1). In contrast, the 32 mutations in ARID1A were distributed throughout the coding region and all were predicted to truncate the protein through a base substitution resulting in a stop codon (9 mutations), or an out-of-frame insertion or deletion (23 mutations) (Table 1). In 10 of the 24 tumors with ARID1A mutations, both ARID1A alleles were affected through either a mutation in one allele and loss of heterozygosity of the other allele, or through two mutations which were presumably biallelic. Thus, we hypothesize that ARID1A functions as a tumor suppressor gene and that somatic mutations inactivate the gene product.
The serine/threonine protein phosphatase PP2A represents a family of holoenzymes with various activities. The holoenzyme contains a core composed of a heterodimer of a catalytic subunit (PPP2CA or PPP2CB) and a constant regulatory subunit (PPP2R1A or PPP2R1B). PPP2R1A serves as a scaffold to coordinate the interaction of the core enzyme with one of more than 15 regulatory subunits to form the heterotrimeric holoenzyme (16, 17). Somatic mutations in PPP2R1A are not listed in the Cancer Gene Census of the COSMIC database, although a few alterations in this gene have been previously reported (18). Functional studies have shown that PP2A is involved in the control of cell growth and division. Specifically, this protein is required for proper chromosome segregation through its interactions with Bub1 and Sgo1 (19). The two arginine residues that were somatically mutated in OCCC are highly conserved and reside within one of the HEAT domains of PPP2R1A that are involved in binding regulatory subunits.
The protein encoded by ARID1A can bind to AT-rich DNA sequences and is a component of the ATP-dependent chromatin modeling complex SWI/SNF. The SWI/SNF chromatin-remodeling complex mobilizes nucleosomes and functions as a regulator of gene expression and chromatin dynamics. ARID1A is one of the two mutually exclusive ARID1 subunits of the SWI/SNF complex and is thought to provide specificity to this complex (20). Changes in chromatin can influence the epigenetic regulation of many genes, inducing those that play a role in cancer (20-22). Indeed, functional studies have implicated ARID1A in the ability of the SWI/SNF complex to inhibit cell growth (23). No mutations of ARID1A are listed in the Cancer Gene Census of the COSMIC database, but chromosomal translocations that involve this gene have been identified in a human breast cancer and a human lung cancer cell line (24). Knock-down of ARID1A in a leukemia cell line confers resistance to Fas-mediated apoptosis (25).
The results of this study emphasize two themes in modern cancer genetics. The first is that specific tumor types are characterized by mutations in “communal cancer genes” like KRAS and PIK3CA as well as in “restricted cancer genes” like PPP2R1A and ARID1A. The communal cancer genes are involved in a variety of cancers and have been extensively studied. Restricted cancer genes have been shown to contribute to specific types of leukemias and sarcomas, mainly through translocations (e.g., the ABL oncogene in chronic myelogenous leukemia and EWS fused to an ETS transcription factor family member in Ewing's sarcoma). With the advent of whole exome sequencing, we are beginning to see similar specificity with respect to point mutations (e.g., IDH1 in gliomas (14) and GNAQ in uveal melanomas (26)).
The second theme is that mutations of chromatin-modifying genes are characteristic of certain tumor types. Recent examples include the JARID1C gene, also known as lysine (K)-specific demethylase 5C (KDM5C), in renal cell cancers (27), SMARCA4/BRG1 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4) in lung cancers (28) and now ARID1A in OCCC. Epigenetic changes in cancers, including methylation of deoxycytidine residues in DNA and a variety of covalent modifications of chromatin proteins, have been extensively studied (20-22, 29). Interestingly, however, the reason(s) that DNA methylation and chromatin are different in cancer cells than in normal cells is completely unknown. Similarly, the relationship between the genetic alterations that unequivocally drive tumorigenesis and the epigenetic changes that are so widespread in tumor genomes has not been defined. Discovery of tumor suppressor genes such as ARID1A that are mutated in cancers bridge this gap, as they are likely to directly lead to epigenetic changes in cancer cells through specific modifications of chromatin proteins. They additionally provide a potential approach to determine which of the numerous epigenetic changes in cancers confer a selective growth advantage and which are simply “passengers” that do not play a causal role. The identification of the genes whose chromatin is specifically affected by ARID1A inactivation, and whose expression is thereby modulated, will be the next crucial step in this line of research.
Supplementary Material
Acknowledgments
We thank Y. He for technical advice and M. Whalen, J. Ptak, and N. Silliman for expert technical assistance. Funded by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, AACR Stand Up To Cancer-Dream Team Translational Cancer Research Grant, The Virginia and D. K. Ludwig Fund for Cancer Research, US Department of Defense grant OC0400600 (TLW), and NIH grants CA121113, CA57345, CA129080, CA103937 (IMS), CA122581 (RR).
References
- 1.Cho KR, Shih Ie M. Annu Rev Pathol. 2009;4:287. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erzen M, Rakar S, Klancnik B, Syrjanen K. Gynecol Oncol. 2001;83:100. doi: 10.1006/gyno.2001.6382. [DOI] [PubMed] [Google Scholar]
- 3.Fukunaga M, Nomura K, Ishikawa E, Ushigome S. Histopathology. 1997;30:249. doi: 10.1046/j.1365-2559.1997.d01-592.x. [DOI] [PubMed] [Google Scholar]
- 4.Marquez RT, et al. Clin Cancer Res. 2005;11:6116. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 5.Sato N, et al. Cancer Res. 2000;60:7052. [PubMed] [Google Scholar]
- 6.Veras E, et al. Am J Surg Pathol. 2009;33:844. doi: 10.1097/PAS.0b013e31819c4271. [DOI] [PubMed] [Google Scholar]
- 7.Kuo KT, et al. Am J Pathol. 2009;174:1597. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo KT, et al. Clin Cancer Res. 2010;16:1997. doi: 10.1158/1078-0432.CCR-09-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See supporting material on Science Online.
- 10.Greenman C, et al. Nature. 2007;446:153. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjoblom T, et al. Science. 2006;314:268. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 12.Wood LD, et al. Science. 2007;318:1108. [Google Scholar]
- 13.Jones S, et al. Science. 2008;321:1801. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, et al. Science. 2008;321:1807. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B, Kinzler KW. Nat Med. 2004;10:789. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 16.Eichorn PJ, Creyghton MP, Bernards R. Biochim Biophys Acta. 2009;1795:1. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y. Cell. 2009;139:468. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, et al. Oncogene. 2000;19:1191. doi: 10.1038/sj.onc.1203389. [DOI] [PubMed] [Google Scholar]
- 19.Tang Z, et al. Dev Cell. 2006;10:575. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Wu JI, Lessard J, Crabtree GR. Cell. 2009;136:200. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weissamn B, Knudsen KE. Cancer Res. 2009;69:8223. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banine F, et al. Cancer Res. 2005;65:3542. doi: 10.1158/0008-5472.CAN-04-3554. [DOI] [PubMed] [Google Scholar]
- 23.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Embo J. 2007;26:752. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Zhao YL, Li Y, Fletcher JA, Xiao S. Genes Chromosomes Cancer. 2007;46:745. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 25.Luo B, et al. Proc Natl Acad Sci U S A. 2008 Dec 23;105:20380. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Raamsdonk CD, et al. Nature. 2009;457:599. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalgliesh GL, et al. Nature. 2010;463:360. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina PP, et al. Huma Mutat. 2008;29:617. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. Cell. 2007;128:683. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


