Abstract
Geographic atrophy (GA), an untreatable advanced form of age-related macular degeneration, results from retinal pigmented epithelium (RPE) cell death. Here we show that the microRNA (miRNA)-processing enzyme DICER1 is reduced in the RPE of humans with GA, and that conditional ablation of Dicer1, but not seven other miRNA-processing enzymes, induces RPE degeneration in mice. DICER1 knockdown induces accumulation of Alu RNA in human RPE cells and Alu-like B1 and B2 RNAs in mouse RPE. Alu RNA is increased in the RPE of humans with GA, and this pathogenic RNA induces human RPE cytotoxicity and RPE degeneration in mice. Antisense oligonucleotides targeting Alu/B1/B2 RNAs prevent DICER1 depletion-induced RPE degeneration despite global miRNA downregulation. DICER1 degrades Alu RNA, and this digested Alu RNA cannot induce RPE degeneration in mice. These findings reveal a miRNA-independent cell survival function for DICER1 involving retrotransposon transcript degradation, show that Alu RNA can directly cause human pathology, and identify new targets for a major cause of blindness.
Age-related macular degeneration (AMD), which is as prevalent as cancer in industrialized countries, is a leading cause of blindness1,2. In contrast to neovascular AMD, the more common atrophic form of AMD is without effective therapy3,4. Extensive atrophy of the retinal pigment epithelium (RPE) leads to severe vision loss and is termed GA, whose pathogenesis is unclear. Here, we identify dysregulation of the RNase DICER1 (ref. 5) and the resulting accumulation of transcripts of Alu elements, the most common small interspersed repetitive elements in the human genome6, as a potential cause of GA, and demonstrate strategies to inhibit this pathology in vivo.
DICER1 loss in GA induces RPE death
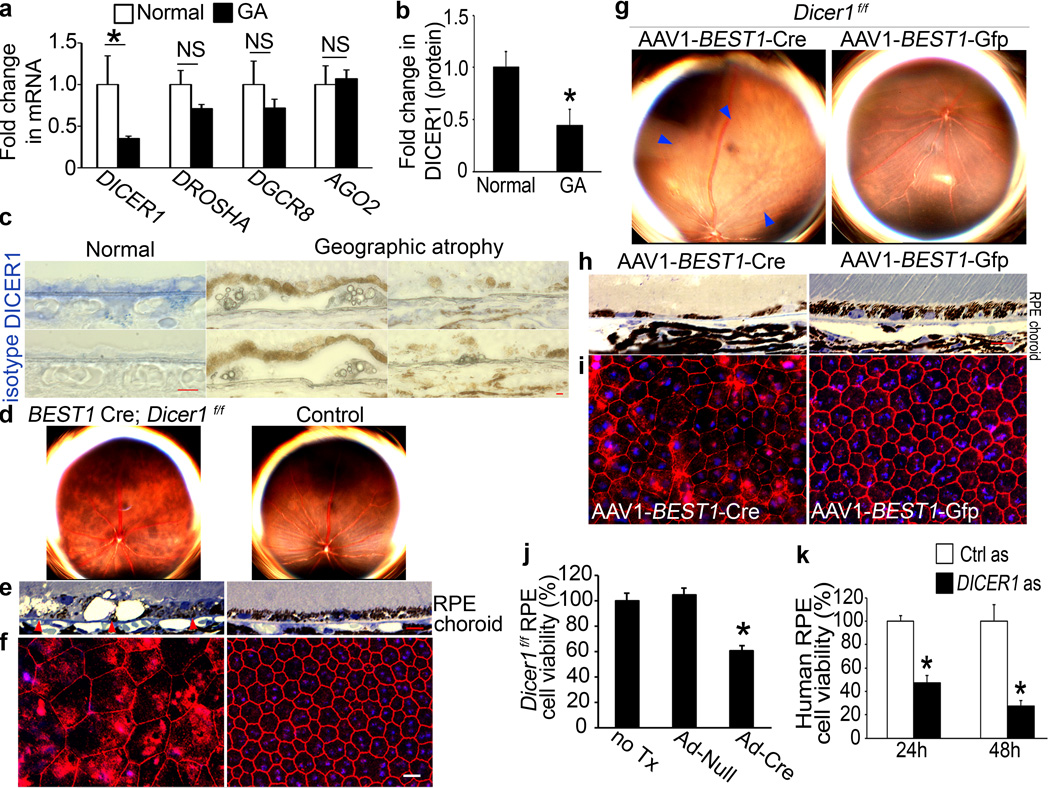
In human donor eyes with GA (n=10), DICER1 mRNA abundance was reduced in the macular RPE by 65±3% (mean±SEM; P=0.0036; Mann-Whitney U test) compared to control eyes (n=11) (Fig. 1a). In contrast, there was no change in the abundance of DROSHA and DGCR8 mRNAs, whose gene products form a complex that processes pri-miRNAs into pre-miRNAs7, or of the gene encoding Argonaute 2 (AGO2, encoded by EIF2C2), the core component of the miRNA effector complex8,9. DICER1 protein expression was reduced in the RPE, but not the neural retina, of eyes with GA compared to controls (Fig. 1b, c and Supplementary Figs. 1 and 2).
Figure 1. DICER1 deficit in GA induces RPE degeneration.
a, DICER1 less abundant in RPE of human eyes with GA (n=10) compared to control RPE (n=11). P = 0.004 by Mann Whitney U test. DROSHA, DGCR8, and EIF2C2 (encoding AGO2) abundance not significantly different (P > 0.11 by Mann Whitney U test). n=10–11. b, DICER1 quantification, assessed by Western blotting (Supplementary Fig. 1), lower in human GA RPE (n=4) compared to control RPE (n=4). P = 0.003 by Student t test. c, Reduced DICER1 (blue) in human GA RPE compared to control eyes. d, e, Fundus photographs (d) and toluidine-blue-stained sections (e) show RPE degeneration in BEST1 Cre; Dicer1f/f mice but not controls. Arrowheads point to basal surface of RPE. f, Flatmounts stained for zonula occludens-1 (ZO-1; red) show RPE disruption in BEST1 Cre; Dicerf/f mice compared to controls. g, h, Fundus photographs (g) and toluidine blue-stained sections (h) show RPE (g, h) and photoreceptor (h) degeneration in Dicer1f/f mice following subretinal injection of AAV1-BEST1-Cre but not AAV1-BEST1-GFP. i, Flatmounts show Dicer1f/f mouse RPE degeneration following subretinal injection of AAV1-BEST1-Cre but not AAV1-BEST1-GFP. Nuclei stained blue with Hoechst 33342. Representative images shown. n=16–32 (d–f); 10–12 (g–i). Scale bars, (c,e,h), 10 µm; (f,i) 20 µm. j, Adenoviral vector coding for Cre recombinase (Ad-Cre) treatment reduces Dicer1f/f mouse RPE cell viability compared to Ad-Null or untreated (no Tx) cells. k, DICER1 antisense (as) reduces human RPE cell viability compared to control antisense (Ctrl as)-treated cells. n=6–8. All error bars indicate mean±s.e.m.
Because DICER1 is downregulated in chemically stressed cells6, we tested whether DICER1 reduction is common to dying retina. DICER1 protein levels were not reduced in the RPE of human eyes with other retinal diseases (vitelliform macular dystrophy, retinitis pigmentosa, retinal detachment; Supplementary Fig. 3). Also, Dicer1 abundance in the RPE was not reduced in numerous mouse models of retinal degeneration including Ccl2−/− Ccr2−/− (refs. 7,8) and Cp−/− Heph−/− mice9 (Supplementary Fig. 3; Supplemental Notes). These data argue that DICER1 depletion in the RPE of eyes with GA is not a generic damage response.
To determine the consequence of DICER1 reduction in the RPE, we interbred Dicer1f/f mice10 with BEST1 Cre mice11, which express Cre recombinase under the control of the RPE cell-specific BEST1 promoter. BEST1 Cre; Dicer1f/f mice uniformly exhibited RPE cell degeneration whereas littermate controls did not (Fig. 1d–f). We also deleted Dicer1 in adult mouse RPE by subretinal injection of an adeno-associated viral vector coding for Cre recombinase under the control of the BEST1 promoter12 (AAV1-BEST1-Cre) in Dicer1f/f mice (Supplementary Fig. 4). These eyes uniformly developed RPE cell degeneration, whereas contralateral eyes that underwent subretinal injection of AAV1-BEST1-GFP and wild-type mouse eyes injected with subretinal AAV1-BEST1-Cre did not (Fig. 1g–i and Supplementary Fig. 4). RPE cell dysmorphology in Dicer1-depleted mice resembled that of human GA eyes (Supplementary Fig. 5). When Dicer1f/f mouse RPE cells were infected with an adenoviral vector coding for Cre recombinase (Ad-Cre), cell viability was reduced (Fig. 1j). Similarly, antisense oligonucleotide mediated knockdown of DICER1 in human RPE cells increased cell death (Fig. 1k). Collectively, these data suggest that DICER1 dysregulation is involved in the pathogenesis of GA.
DICER1 phenotype not due to miRNA dysregulation
We tested whether depleting other miRNA-processing enzymes induces RPE degeneration. Subretinal injection of AAV1-BEST1-Cre in Droshaf/f (ref. 13), Dgcr8f/f (refs. 13,14), or Ago2f/f mice15 did not damage the RPE (Supplementary Fig. 6), suggesting that miRNA imbalances are not responsible for RPE degeneration induced by DICER1 depletion. However, some miRNAs are generated by Dicer1 independent of Drosha and Dgcr8 (refs. 16,17). There is also debate whether Ago2 is essential for miRNA function15,18–21. Mice deficient in Ago1, Ago3, or Ago4 had normal RPE (Supplementary Fig. 7). TRBP (encoded by Tarbp2) recruits DICER1 to the four Argonaute proteins to enable miRNA processing and RNA silencing (ref. 22 and R. Shiekhattar, personal communication); Tarbp2−/− mice too had no RPE degeneration (Supplementary Fig. 7). These data suggest that RPE degeneration induced by Dicer1 ablation involves a mechanism specific to Dicer1 and not to miRNA machinery in general.
To further investigate whether miRNA imbalances might contribute to the DICER1 depletion phenotype, we studied human HCT116 colon cancer cells in which the helicase domain in exon 5 of DICER1 was disrupted. Despite impaired miRNA biogenesis in these HCT-DICER1ex5 cells23, baseline cell viability was not different between HCT-DICER1ex5 and parent HCT116 cells (Supplementary Fig. 8). These findings suggest that the principal biological effect of DICER1 deficit contributing to the development of GA is not miRNA dysregulation, but do not exclude miRNA dysregulation promoting GA through other pathways.
Alu RNA accumulation in GA
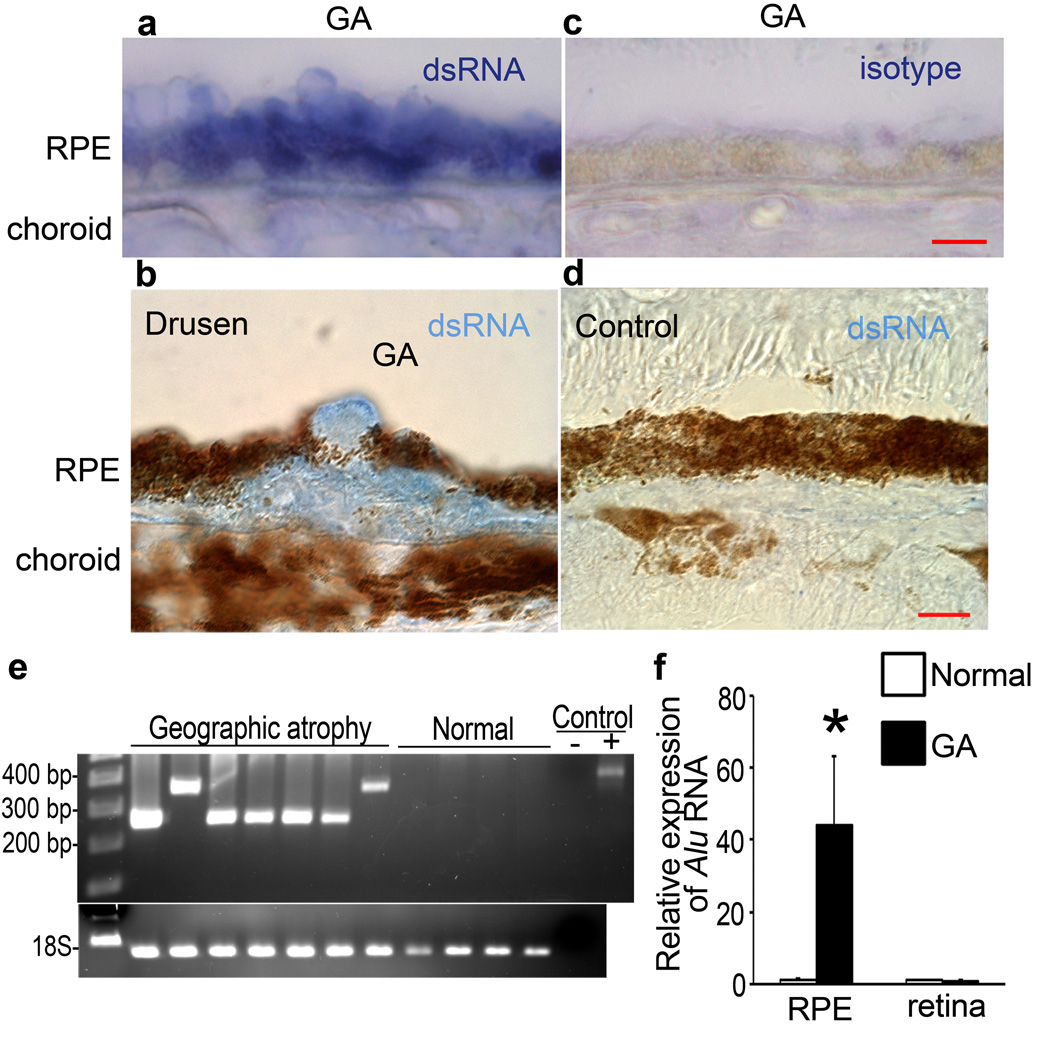
Because miRNA perturbations were not implicated, we speculated that impaired processing of other dsRNAs might be involved. Using an antibody24,25 that recognizes long dsRNA (J2), we detected abundant dsRNA immunoreactivity in the macular RPE of human eyes with GA (n=10; Fig. 2a–c) but not in control eyes (n=10; Fig. 2d). We immunoprecipitated RPE lysates with J2 antibody and then sequenced the dsRNA using a T4 RNA ligase-aided, adaptor-based PCR amplification strategy. Approximately 300-nt long dsRNA species were found in the macular RPE of human eyes with GA (12/12) but not in eyes without GA (0/18) (P=1.210−8 by Fisher’s exact test) (Fig. 2e).
Figure 2. Alu RNA accumulation in GA triggered by DICER reduction.
a, b, dsRNA immunolocalized (blue) in RPE (a, b) and sub-RPE deposits (drusen; b) in human GA. c, d, No staining with isotype antibody in GA RPE (c) and with anti-dsRNA antibody in control eye (d). Scale bars, (a–d), 10 µm. n=10 (a–d) e, PCR amplification of immunoprecipitated dsRNA yielded amplicons with homology to Alu in GA RPE but not normal RPE. Water control (−) showed no amplification and recombinant dsRNA (+) showed predicted amplicon. f, Increased Alu RNA in GA RPE compared to control (n=7). P < 0.05 by Student t test. No significant difference in Alu RNA in neural retina. Values normalized to abundance in normal eyes.
We recovered clones from 8 of the 12 GA eyes and identified two distinct sequences with high homology (E = 3.3×10−103; 1.1×10−76) to Alu RNAs (Supplementary Fig. 9). These sequences showed homology to the Alu Sq subfamily consensus sequence. Alu RNAs were the only dsRNA transcripts identified specifically in the J2-immunoprecipitated GA samples. We confirmed that J2 recognized Alu RNA (Supplementary Fig. 10). There was a dramatic increase in the abundance of Alu RNAs in the RPE of human eyes with GA compared to control eyes (n=7), but not in the neural retina (Fig. 2f, Supplementary Fig. 11). The reference genome did not contain exact matches to these Alu sequences. This could be attributed to genetic variations or regions not represented in the reference genome or to chimeric Alu formation. Further studies should elucidate the genomic origin of and regulatory factors involved in transcription of these Alu RNAs.
DICER1 depletion induces Alu RNA cytotoxicity
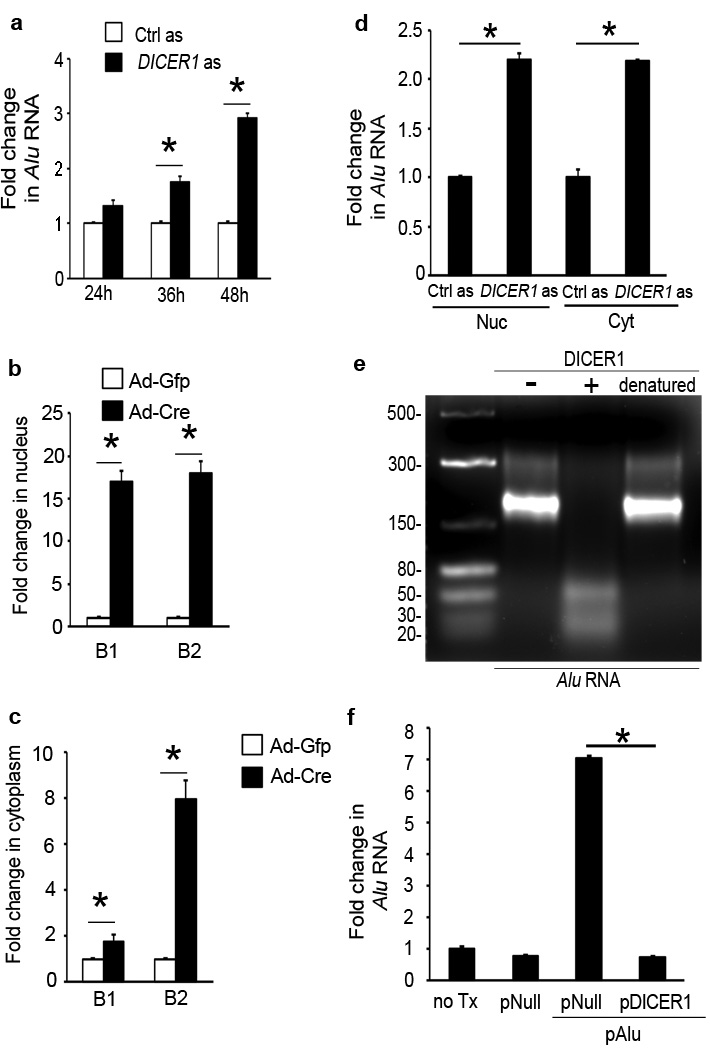
We tested whether Alu RNA accumulation in the RPE of GA was due to deficient DICER1 processing. DICER1 knockdown in human RPE cells using antisense oligonucleotides increased Alu RNA accumulation (Fig. 3a, Supplementary Fig. 12). Ad-Cre infection of Dicer1f/f mouse RPE cells induced accumulation of B1 and B2 RNAs (Fig. 3b, c). DICER1 was expressed in the nucleus and cytoplasm of RPE cells and its depletion induced accumulation of Alu/B1/B2 RNA in both compartments (Fig. 3b–d, Supplementary Fig. 13). Recombinant DICER1, but not heat-denatured DICER1, degraded Alu RNA (Fig. 3e). Enforced expression of DICER1 in human RPE cells reduced the abundance of overexpressed Alu RNA (Fig. 3f), consistent with their degradation by DICER1 in vivo. These data confirm that DICER1 dysregulation can trigger Alu/B1/B2 RNA accumulation.
Figure 3. DICER1 degrades Alu RNA.
a, DICER1 antisense (as) increased Alu RNA in human RPE cells. b, c, Ad-Cre, but not Ad-GFP, increased B1 and B2 RNAs in Dicer1f/f mouse RPE cells in nucleus (b) and cytoplasm (c). d, DICER1 as upregulated Alu RNA in human RPE cell nucleus (Nuc) and cytoplasm (Cyt). e, Agarose gel electrophoresis shows recombinant DICER1 (+), but not heat denatured DICER1, degrades Alu RNA isolated and cloned from human GA RPE. Image representative of 6 experiments. f, Alu RNA in human RPE cells upregulated by plasmid coding for Alu (pAlu) vs. pNull or no treatment (no Tx) at 24 h reduced by pDICER1. * P < 0.05. n=4–8 (a–d, f). Values normalized to control as-treated (for Alu) or Ad-GFP-infected cells (for B elements).
Because cell stresses can induce generalized retrotransposon activation, we wondered whether Alu RNA accumulation in GA might be a generic response in dying retina. However, in the RPE of human eyes with GA and in DICER1-depleted human RPE cells, there was no increase in the abundance of RNAs coded by L1.3, human endogenous retrovirus-W envelope, or hY3 (Supplementary Fig. 14). These data demonstrate that Alu RNA accumulation is a biologically specific response to DICER1 depletion.
Alu RNA upregulation induced by DICER1 knockdown was inhibited by tagetitoxin (an RNA Pol III inhibitor) but not α-amanitin (an RNA Pol II inhibitor) (Supplementary Fig. 15). Northern blotting revealed that Alu RNA from the RPE of human eyes with GA was approximately 300-nt in length, consistent with the length of non-embedded Pol III Alu transcripts. Our Northern probe specifically detected Alu RNA and not 7SL RNA, the evolutionary precursor of Alu. Northern blotting revealed no difference in 7SL RNA abundance between the RPE of GA and control eyes. Real-time RT-PCR analysis showed that 7SL RNA was not dysregulated in the RPE of human eyes with GA or in DICER1-depleted human RPE cells (Supplementary Fig. 16). DICER1 knockdown did not upregulate several Pol II-transcribed genes (ADAR2, NICN, NLRP, SLFN11) containing exon-embedded Alu sequences. These data suggest that Alu RNA in the RPE of human eyes with GA are primary Alu transcripts and not passenger or bystander sequences embedded in other RNAs. Conclusive assignment of these Alu sequences as Pol III transcripts must await precise determination of their transcription start site.
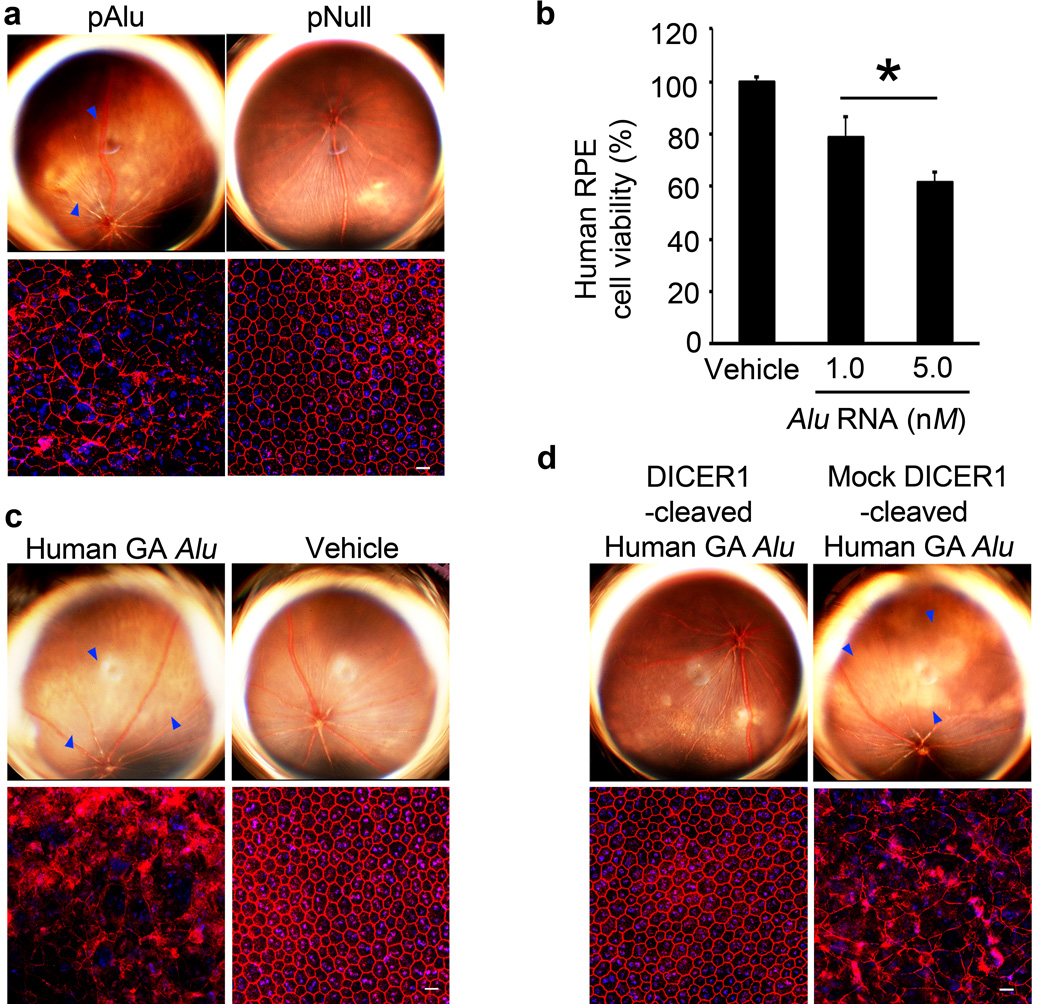
We tested whether Alu RNA accumulation could induce GA. Transfecting human or wild-type mouse RPE cells with a plasmid coding for Alu (pAlu) reduced cell viability (Supplementary Fig. 17). Subretinal transfection of plasmids coding for two different Alu RNAs or for B1 or B2 RNAs induced RPE degeneration in wild-type mice (Fig. 4a, Supplementary Fig. 17, and data not shown). Treating human RPE cells with a recombinant 281-nt long Pol III-derived Alu RNA isolated from a human embryonal carcinoma cell line dose-dependently increased cell death (Fig. 4b), suggesting that endogenous DICER1 degrades small amounts of Alu RNA but can be overwhelmed. Accordingly, DICER1 overexpression blocked pAlu-induced cell death in human RPE cells and RPE degeneration in wild-type mice (Supplementary Fig. 17).
Figure 4. DICER1 protects RPE cells from Alu RNA cytotoxicity.
a, Subretinal pAlu, but not pNull, induced wild-type mouse RPE degeneration (fundus photographs, top row; ZO-1 stained (red) flatmounts, bottom row). b, Alu RNA induced human RPE cytotoxicity. Values normalized to pNull or vehicle. * P < 0.05 by Student t test. n=4–6. c, Subretinal Alu RNA isolated and cloned from human GA RPE induced wild-type mouse RPE degeneration. d, Subretinal injection of this Alu RNA, when cleaved by DICER1, did not induce wild-type mouse RPE degeneration (fundus photographs, top row; flatmounts, bottom row) in contrast to mock-cleaved Alu RNA. Degeneration outlined by blue arrowheads (a,c,d). Scale bars (20 µm). n=10–15.
Subretinal injection delivered Alu RNA to RPE cells in wild-type mice (Supplementary Fig. 18), consistent with the ability of long RNAs with duplex motifs to enter cells26. We cloned a 302-nt long Alu RNA isolated from the RPE of a human eye with GA and transcribed it in vitro to generate partially and completely annealed structures that mimic Alu RNAs transcribed by Pol III and Pol II, respectively. Subretinal injection of either of these Alu RNAs induced RPE degeneration in wild-type mice (Fig. 4f, Supplementary Fig. 19), supporting the assignment of disease causality. In contrast, subretinal injection of these Alu RNAs digested with DICER1 did not induce RPE degeneration (Fig. 4g, Supplementary Fig. 19). When these Alu RNAs were subjected to mock DICER1 digestion, they induced RPE degeneration, suggesting a role for DICER1 in protecting against Alu RNA-induced degeneration.
In contrast, subretinal transfection of transfer RNA or plasmids coding for 7SL RNA or two different primary miRNAs did not induce RPE degeneration in wild-type mice (Supplementary Fig. 20). Chemically synthesized dsRNAs that mimic viral dsRNA can induce RPE degeneration by activating toll like receptor-3 (TLR3)27; however, pAlu transfection did not induce TLR3 phosphorylation in human RPE cells and did induce RPE degeneration in Tlr3−/− mice (Supplementary Fig. 21). Therefore, Alu RNA-induced RPE degeneration cannot be attributed solely to its repetitive or double-stranded nature, as it exerted effects distinct from other structured dsRNAs of similar length.
The mechanism of RPE cell death in GA is undefined. DNA fragmentation has been identified in RPE cells in human eyes with GA28, and Dicer1 knockdown has been associated with induction of apoptosis in diverse tissues10,29. We now provide evidence of caspase-3 cleavage in regions of RPE degeneration in human eyes with GA (Supplementary Fig. 22). Caspase-3 cleavage was also observed in the RPE cells of BEST1 Cre; Dicer1f/f mice and in Alu RNA-stimulated or -overexpressing human RPE cells. These data suggest a role for Alu RNA-induced RPE cell apoptosis triggered by DICER1 dysregulation in GA.
To study whether an imbalance in small RNA species produced from long Alu RNAs could contribute to RPE degeneration, we exposed human RPE cells or wild-type mice to DICER1 cleavage fragments of Alu RNA. Subretinal transfection of these fragments did not damage wild-type mouse RPE cell, and co-administering these fragments did not prevent RPE cell degeneration induced by pAlu (Supplementary Fig. 23). Similarly, these fragments did not prevent human RPE cell death induced by Alu RNA overexpression. These data suggest that upregulation of long Alu RNA rather than imbalance in Alu RNA-derived small RNA fragments is responsible for RPE degeneration induced by DICER1 reduction.
To dissect the contribution of Alu RNA accumulation versus that of miRNA dysregulation to RPE degeneration in the context of DICER1 deficit, we re-examined HCT-DICER1ex5 cells in which miRNA biogenesis is impaired but long dsRNA cleavage is preserved due to the intact RNase III domains. Alu RNA levels were not different between HCT-DICER1ex5 and parent HCT116 cells (Supplementary Fig. 24). In contrast, DICER1 knockdown in HCT116 cells upregulated Alu RNA. Also, Alu RNA induces similar levels of cytotoxicity in HCT-DICER1ex5 and parent HCT116 cells, suggesting that coexisting miRNA expression deficits do not augment Alu RNA-induced RPE degeneration. In conjunction with the discordance in the RPE degeneration phenotype between ablation of Dicer1 and that of various other small RNA biogenesis pathway genes in mice, our findings suggest that Alu RNA accumulation is critical to DICER1 reduction-induced cytotoxicity.
RPE degeneration blocked by Alu RNA inhibition
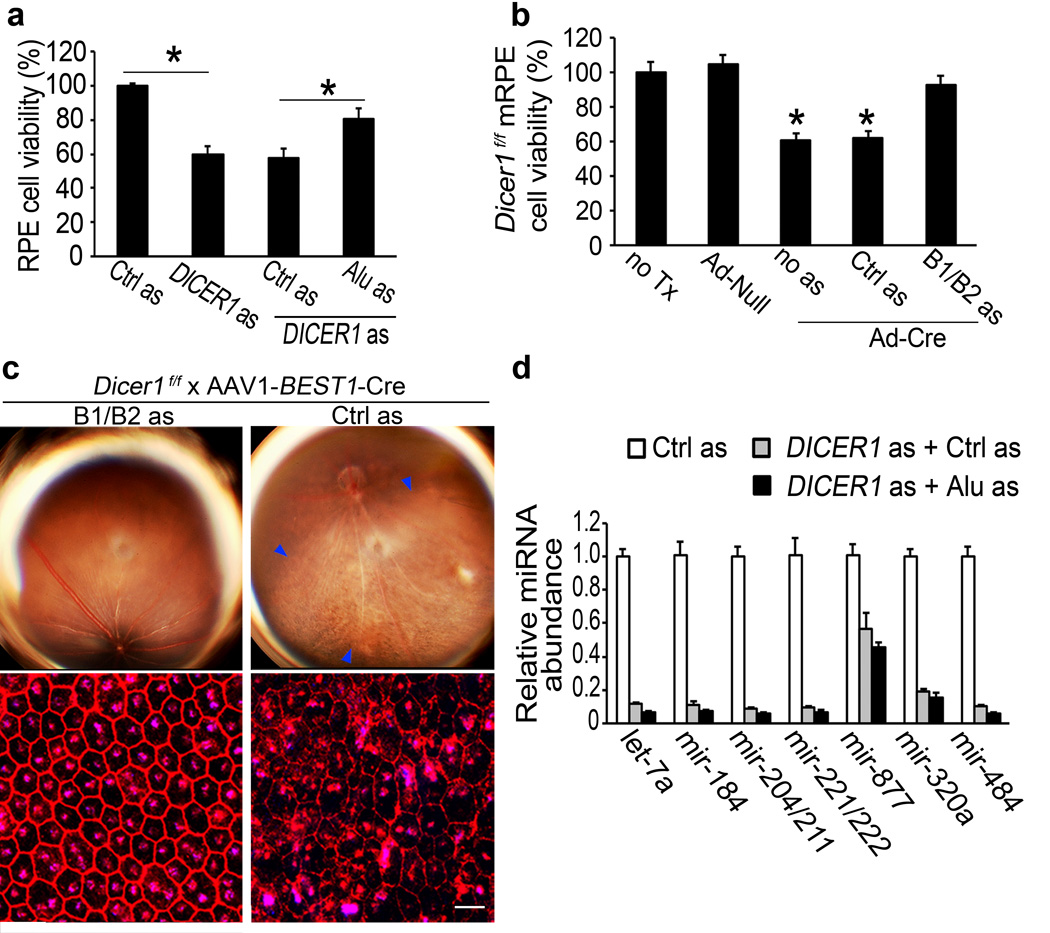
We tested whether DICER1 reduction-induced cytotoxicity is due to Alu RNA accumulation. DICER1 knockdown-induced human RPE cytotoxicity was inhibited by antisense oligonucleotides targeting Alu RNA sequences but not by scrambled antisense control (Fig. 5a and Supplementary Fig. 25). Ad-Cre infection of Dicer1f/f mouse RPE cells reduced cell viability, and this was blocked by antisense oligonucleotides targeting B1/B2 RNAs but not by scrambled antisense control (Fig. 5b and Supplementary Fig. 25). Subretinal administration of antisense oligonucleotides that reduced accumulation of B1/B2 RNAs inhibited RPE degeneration in AAV1-BEST1-Cre-treated Dicer1f/f mice (Fig. 5c and Supplementary Fig. 25), providing evidence of in vivo functional rescue.
Figure 5. DICER1 dyregulation induces RPE cell death via Alu RNA accumulation.
a, Human RPE cytotoxicity induced by DICER1 as rescued by Alu RNA as. Values normalized or compared to control (Ctrl) as. b, Ad-Cre but not Ad-Null induced Dicer1f/f mouse RPE cytotoxicity. B1/B2 RNA as, but not control (Ctrl) as, rescued viability. Values normalized to untreated cells (no Tx). * P < 0.05 by Student t test. n=4–6 (a,b). c, Subretinal AAV-BEST1-Cre induced RPE degeneration (blue arrowheads in fundus photograph, top row; ZO-1 stained (red) flatmounts, bottom row) in Dicer1f/f mice 20 days after injection was inhibited by subretinal cholesterol-conjugated B1/B2 as, but not cholesterol-conjugated Ctrl as, 10 days after AAV-BEST1-Cre injection. Values normalized to Ctrl as-treatment. n=8. Scale bar, 20 µm. d, DICER1 as induced global miRNA expression deficits in human RPE cells compared to Ctrl as. No significant difference in miRNA abundance between Alu as and Ctrl as-treated DICER1 depleted cells. n=3.
We tested whether Alu inhibition also rescued miRNA expression deficits as a potential explanation for the functional rescue of DICER1 depletion-induced RPE degeneration. As expected, DICER1 knockdown in human RPE cells reduced the abundance of numerous miRNAs (Fig. 5d). However, inhibition of Alu RNA did not recover miRNA expression. Thus, rescue of RPE cell viability by Alu RNA inhibition despite persistent global miRNA expression deficits argues that RPE degeneration induced by DICER1 deficit is due to Alu RNA accumulation and not miRNA dysregulation.
Collectively, these data support a model in which primary Alu transcripts are responsible for RPE degeneration. Whether similar pathology can also result from upregulation of as yet undefined Pol II transcripts with embedded Alu sequences is an intriguing possibility that requires further study. Importantly, we demonstrated that primary Alu transcripts are elevated in human disease, that Alu transcripts recapitulate disease in relevant experimental models, and that targeted suppression of Alu transcripts successfully inhibits this pathology. These observations have direct relevance for clinical strategies to prevent and treat GA.
Discussion
Our findings elucidate a critical cell survival function for DICER1 by functional silencing of toxic Alu transcripts. This unexpected function suggests that RNAi-independent mechanisms should be considered in interpreting the phenotypes of systems in which Dicer1 is dysregulated. For example, it would be interesting to test whether Dicer1 ablation induced cytotoxicity in mouse neural retina30 and heart31 might also involve B1/B2 RNA accumulation. More broadly, recognition of DICER1’s hitherto unidentified function as an important controller of transcripts derived from the most abundant genomic repetitive elements can illuminate new functions for RNases in cytoprotective surveillance. DICER1 expression is reduced in GA and partial loss of DICER promotes RPE degeneration; thus loss of heterozygosity in DICER1 may underlie the development of GA, similar to its function as a haploinsufficient tumor suppressor32–34.
This also is, to our knowledge, the first example of how Alu could cause a human disease via direct RNA cytotoxicity rather than by inducing chromosomal DNA rearrangements or insertional mutagenesis through retrotransposition, which have been implicated in diseases such as α-thalassemia35, colon cancer36, hypercholesterolemia37,38, and neurofibromatosis39. Future studies should determine the precise chromosomal locus of the Alu RNA elements that accumulate in GA and the nature of transcriptional and post-transcriptional machinery that enable their biogenesis.
In addition to processing miRNAs5, DICER1 has been implicated in heterochromatin assembly40,41. Since Alu elements are abundant within heterochromatin42, whether perturbations in centromeric silencing underlie the pathogenesis of GA warrants study. The finding that chromatin remodelling at Alu repeats can regulate miRNA expression43 raises the intriguing possibility of other regulatory intersections between DICER1 and Alu. It also remains to be investigated whether centromeric satellite repeats that accumulate in Dicer1-null mouse embryonic stem cells44,45 might be involved in the pathogenesis of GA.
In the mouse germline, Dicer1 has been implicated in generating endogenous small interfering RNAs (endo-siRNAs) from repeat elements46,47. If this process is conserved in mammalian somatic tissues, it would be interesting to learn whether endo-siRNAs serve a homeostatic function in preventing the development of GA. Given that caspases can cleave Dicer1 and convert it into a DNase that promotes apoptosis in nematodes48, our finding that Alu RNA induces caspase activation introduces the possibility of bidirectional regulation between DICER1 and Alu that triggers feed-forward disease-amplifying loops.
The inciting events that trigger an RPE-specific reduction of DICER1 in patients with GA are unknown. Potential culprits could include oxidative stress, which is postulated to underlie AMD pathogenesis4, as we found that hydrogen peroxide downregulates DICER1 in human RPE cells (Supplementary Fig. 26). While upstream triggers of DICER1 dysregulation and the role of other DICER-dependent, DROSHA/DGCR8-independent small RNAs in GA await clarification, the ability of Alu RNA antisense oligonucleotides to inhibit induced by DICER1 depletion-induced RPE cytotoxicity provides a rationale to investigate Alu RNA inhibition or DICER1 augmentation as potential therapies for GA.
METHODS SUMMARY
Subretinal injections (1 µL) were performed using a Pico-Injector (PLI-100, Harvard Apparatus). Plasmids were transfection in vivo using 10% Neuroporter (Genlantis). Immunolabeling was performed using antibodies against dsRNA (clone J2, English & Scientific Consulting), DICER1 (Santa Cruz Biotechnology), zonula occludens-1 (Invitrogen), Cre recombinase (EMD4Biosciences), or cleaved caspase-3 (Cell Signaling). dsRNA was isolated by immunoprecipitating homogenized tissue lysates with 40 µg of J2 for 16 h at 4 °C. Purified dsRNA was ligated to an anchor primer and purified by MinElute Gel extraction columns (Qiagen). Ligated dsRNA was denatured, reverse transcribed, and amplified by PCR. Amplified cDNA products were cloned into PCRII TOPO vector (Invitrogen) and sequenced. Homology to Alu consensus sequences was determined using CENSOR. Cell viability was assessed using CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Total RNA (1 µg) was reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences) and amplified by real-time quantitative PCR (Applied Biosystems 7900 HT) with Power SYBR green Master Mix. Relative expressions were determined by the 2−ΔΔCt method. miRNA abundance was quantified using All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Human tissue
Donor eyes or ocular tissues from patients with GA due to AMD or patients without AMD were obtained from various eye banks in Australia and the United States of America. These diagnoses were confirmed by dilated ophthalmic examination prior to acquisition of the tissues or eyes or upon examination of the eye globes post mortem. The study followed the guidelines of the Declaration of Helsinki. Institutional review boards granted approval for allocation and histological analysis of specimens.
Animals
All animal experiments were in accordance with the guidelines of the University of Kentucky Institutional Animal Care and Use Committee and the Association for Research in Vision and Ophthalmology.
Immunolabeling and Histology
Fixed human tissue was stained with the antibodies against dsRNA (clone J2, English & Scientific Consulting) or human DICER1 (Santa Cruz Biotechnology). Bound antibody was detected with alkaline phosphatase streptavidin solution (Invitrogen) and the enzyme complex was visualized by Vector Blue (Vector Laboratories). Mouse RPE/choroid flat mounts were fixed with 4% paraformaldehyde or 100% methanol and stained with rabbit antibodies against human zonula occludens-1 (Invitrogen), Cre recombinase (EMD4Biosciences), or human cleaved caspase-3 (Cell Signaling) and visualized with Alexa594- or Cy5-conjugated secondary antibodies. Fixed primary human RPE cells were stained with antibodies against dsRNA or human DICER1 and visualized with Alexa Fluor - conjugated secondary antibodies. Nuclei were visualized with DAPI counterstaining.
Subretinal injections
Subretinal injections (1 µL) in mice were performed using a Pico-Injector (PLI-100, Harvard Apparatus). In vivo transfection of plasmids was achieved using 10% Neuroporter (Genlantis). AAV1-BEST1-Cre12 or AAV1-BEST1-GFP were injected at 1.0×1011 pfu/mL and recombinant Alu RNAs were injected at 0.3 mg/mL. Cell-permeating cholesterol conjugated-B1/B2 antisense oligonucleotides or cholesterol conjugated-control antisense (both from Integrated DNA Technologies) were injected (2 µg in 1 µL) 10 days after AAV1-BEST1-Cre was injected in Dicer1f/f mice.
dsRNA isolation
Human eyes were stored in RNAlater (Ambion). Homogenized lysates were immunoprecipited with 40 µg of mouse antibody against dsRNA (clone J2) for 16 h at 4 °C. Immunocomplexes were collected on protein A/G agarose (Thermoscientific) and dsRNA species were separated and isolated using Trizol (Invitrogen) according to the manufacturer’s instructions. Purified dsRNA was then ligated to an anchor primer and purified by MinElute Gel extraction columns (Qiagen). Ligated dsRNA was then denatured, reverse transcribed into cDNA, and amplified by PCR. Amplified cDNA products were cloned into PCRII TOPO vector (Invitrogen) and sequenced at the University of Kentucky Advanced Genetic Technologies Center. The homology of isolated cDNA sequences to known Alu consensus sequences was determined using the CENSOR server49.
Cell culture
All cell lines were cultured at 37 °C and 5% CO2. Primary mouse RPE cells were isolated as previously described50 and grown in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% FBS and standard antibiotics concentrations. Primary human RPE cells were isolated as previously described27 and maintained in DMEM supplemented with 20% FBS and antibiotics. Parental HCT116 and isogenic Dicerex5 cells23 were cultured in McCoy’s 5A medium supplemented with 10% FBS. Transient tranfections of plasmid and antisense oligonucleotides were performed with Lipofectamine2000 (Invitrogen) and Oligofectamine (Invitrogen) respectively. Cell viability measurements were performed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) in according to the manufacturer’s instructions.
Alu RNA Synthesis
Two Alu RNAs were synthesized: 281 nt Alu sequence originating from the cDNA clone TS 103 which is known to be expressed in human cells51 and a 302 nt Alu sequence isolated from the RPE of a human eye with GA. Alu RNAs were synthesized using a RNA polymerase T7 promoter and runoff transcription followed by gel purification as previously described52, yielding ssRNAs that fold into a defined secondary structure identical to Pol III derived transcripts. We also synthesized a fully complementary dsRNA form (resembling a Pol II derived transcript) of the 302 nt human GA Alu using linearized PCRII TOPO plasmid templates using T7 or SP6 RNA polymerases (MegaScript, Ambion) according to the manufacturer’s recommendations. After purification, equal molar amount of each transcript were combined and heated at 95 °C for 1 min, cooled and then annealed at room temperature for 24 h. The Alu dsRNA was precipitated, suspended in water and analyzed on 1.4% non-denaturing agarose gel using the single-stranded complementary strands as controls.
Real-time PCR
Total RNA was extracted from tissues or cells using Trizol reagent (Invitrogen) according to manufacturer’s recommendations and were treated with RNase free DNase (Ambion). Total RNA (1 µg) was reverse transcribed as previously described2 using qScript cDNA SuperMix (Quanta Biosciences). The RT products (cDNA) were amplified by real-time quantitative PCR (Applied Biosystems 7900 HT Fast Real-Time PCR system) with Power SYBR green Master Mix. Oligonucleotide primers specific for DICER1 (forward 5'-CCCGGCTGAGAGAACTTACG-3' and reverse 5'-CTGTAACTTCGACCAACACCTTTAAA-3'), DROSHA (forward 5'-GAACAGTTCAACCCCGATGTG-3' and reverse 5'-CTCAACTGTGCAGGGCGTATC-3'), DGCR8 (forward 5'-TCTGCTCCTTAGCCCTGTCAGT-3' and reverse 5'-CCAACACTCCCGCCAAAG-3'), EIF2C2 (forward 5'-GCACGGAAGTCCATCTGAAGTC-3' and reverse 5'-CCGGCGTCTCTCGAGATCT-3'), human 18S rRNA (forward 5'-CGCAGCTAGGAATAATGGAATAGG-3' and reverse 5'-GCCTCAGTTCCGAAAACCAA-3'), Alu (forward 5'- CAACATAGTGAAACCCCGTCTCT-3’ and reverse 5'-GCCTCAGCCTCCCGAGTAG-3'), LINE L1.3 (ORF2) (forward 5'-CGGTGATTTCTGCATTTCCA-3' and reverse 5'-TGTCTGGCACTCCCTAGTGAGA-3'), HERV-WE1 (forward 5'- GCCGCTGTATGACCAGTAGCT-3' and reverse 5'-GGGACGCTGCATTCTCCAT-3'), human Ro-associated Y3 (hY3) (forward 5'-CCGAGTGCAGTGGTGTTTACA-3' and reverse 5'-GGAGTGGAGAAGGAACAAAGAAATC-3'), 7SL (forward 5'-CGGCATCAATATGGTGACCT-3' and reverse 5'-CTGATCAGCACGGGAGTTTT-3'), B1 (forward 5'-TGCCTTTAATCCCAGCACTT-3' and reverse 5'-GCTGCTCACACAAGGTTGAA-3'), B2 (forward 5'-GAGTTCAAATCCCAGCAACCA-3' and reverse 5'-AAGAGGGTCTCAGATCTTGTTACAGA-3'), Dicer1 (forward 5'-CCCACCGAGGTGCATGTT-3' and reverse 5'-TAGTGGTAGGAGGCGTGTGTAAAA-3'), mouse 18S rRNA (forward 5'- TTCGTATTGCGCCGCTAGA-3' and reverse 5'- CTTTCGCTCTGGTCCGTCTT-3') were used. The QPCR cycling conditions were 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles of a two-step amplification program (95 °C for 15 s and 58 °C for 1 min). At the end of the amplification, melting curve analysis was applied using the dissociation protocol from the Sequence Detection system to exclude contamination with unspecific PCR products. The PCR products were also confirmed by agarose gel and showed only one specific band of the predicted size. For negative controls, no RT products were used as templates in the QPCR and verified by the absence of gel-detected bands. Relative expressions of target genes were determined by the 2−ΔΔCt method.
miRNA PCR
miRNA abundance was quantified using the All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia). Briefly, total RNA was polyadenylated and reverse transcribed using a poly dT-adaptor primer. Quantitative RT-PCR was carried out using a miRNA-specific forward primer and universal reverse primer. PCR products were subjected to dissociation curve and gel electrophoresis analyses to ensure that single, mature miRNA products were amplified. Data were normalized to ACTB levels. The forward primers for the miRNAs were as follows: miR-184 (5'-TGGACGGAGAACTGATAAGGGT-3'); miR-221/222 (5'-AGCTACATCTGGCTACTGGGT-3'); miR-204/211 (5'-TTCCCTTTGTCATCCTTCGCCT-3'); miR-877 (5'-GTAGAGGAGATGGCGCAGGG-3'); miR-320a (5'-AAAAGCTGGGTTGAGAGGGCGA-3'); miR-484 (5'-TCAGGCTCAGTCCCCTCCCGAT-3'); let-7a (5'-TGAGGTAGTAGGTTGTATAGTT-3'). The reverse primers were proprietary (Genecopoeia). The primers for ACTB were forward (5'-TGGATCAGCAAGCAGGAGTATG-3') and reverse (5'-GCATTTGCGGTGGACGAT-3').
Western Blot
Tissues were homogenized in lysis buffer (10 mM Tris base, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NP-40, protease and phosphatase inhibitor cocktail (Roche)). Protein concentrations were determined using a Bradford assay kit (Bio-Rad) with bovine serum albumin as a standard. Proteins (40–100 µg) were run on 4–12% Novex Bis-Tris gels (Invitrogen). The transferred membranes were blocked for 1 h at RT and incubated with antibodies against DICER1 (1:1,000, ref. 45; or 1:200, Santa Cruz Biotechnology) at 4 °C overnight. Protein loading was assessed by immunoblotting using an anti-Tubulin antibody (1:1,000; Sigma-Aldrich). The secondary antibodies were used (1:5,000) for 1 h at RT. The signal was visualized by enhanced chemiluminescence (ECL Plus) and captured by VisionWorksLS Image Acquisition and Analysis software (Version 6.7.2, UVP, LLC). Densitometry analysis was performed using ImageJ (NIH). The value of 1 was arbitrarily assigned for normal eye samples.
RNA polymerase inhibition
Human RPE cells were transfected with DICER1 or control antisense oligonucleotides using Lipofectamine 2000. After a change of medium at 6 h, the cells were incubated with 45 µM tagetitoxin (Epicentre Technologies, Tagetin) or 10 µg/ml aamanitin (Sigma-Aldrich) and the total RNA was collected after 24 h.
Cell viability
MTS assays were performed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) in according to the manufacturer’s instructions.
Supplementary Material
Acknowledgements
We thank M. Chrenek, J. Garcia-Perez, T. Heidmann, C. Kanellopoulou, D.M. Livingston, J.V Moran, R.F. Mullins, J.M. Nickerson, E.A. Pearce, A. Tarakhovsky, B. Vogelstein, V.E. Velculescu, and D.J. Zack for providing mice, reagents, or tissues; R. King, L. Xu, M. McConnell, C. Payne, G. R. Pattison, G.J. Jaffe, S. Medearis, and C. Spee for technical assistance; and A. Sinai, R. Mohan, T.S. Khurana, R.A. Brekken, P.L. Deininger, S. Bondada, P.A. Pearson, A.M. Rao, G.S. Rao and K. Ambati for discussions. J.A. was supported by National Eye Institute (NEI)/National Institutes of Health (NIH) grants, the Doris Duke Distinguished Clinical Scientist Award, the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research, the Dr. E. Vernon Smith and Eloise C. Smith Macular Degeneration Endowed Chair, the Senior Scientist Investigator Award (Research to Prevent Blindness, RPB), and a departmental unrestricted grant from the RPB. J.Z.B. was supported by the University of Kentucky Physician Scientist Award, the International Retinal Research Foundation, and the American Health Assistance Foundation. M.E.K. and S.B. were supported by NIH T32 grants. B.K.A. was supported by NEI/NIH grants, the VA Merit Award and the Department of Defense. P.P. is a Senior Scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). DKL acknowledges support from Global Research Laboratory program by MEST, Korea.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.K., S.D., V.T., W.C., B.J. F., M.E.K., S.L.P., J.K., J.A.G., K.K., N.L.J., B.D.G., Y.H., R.J.C.A., A.D.B., S.B, J.W., M.H, Y.S., and J.Z.B. performed experiments. W.W.H., V.A.C, D-K. L, J.W.Y., C.M.R, D.R.H., H.E.G., Q.Z., J.M.P., M.C.M., A.H.M., M.M.W.C., D.R.L., E.F., E.A.P., P.P., F.W.B., R.E.B., S.M., G.C., and J.L.D. provided tissues or reagents. J.A. conceived and directed the project, and wrote the paper with assistance from P.P., C.M.R., K.K., J.F.K., J.A.G., E.F., M.M.W.C., B.J.F., B.D.G., and B.K.A. All authors had the opportunity to discuss the results and comment on the manuscript.
Author Information The Alu sequences have been deposited in GenBank under the accession numbers HN176584 and HN176585. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda A, et al. CCR3 is a target for age-related macular degeneration diagnosis and therapy. Nature. 2009;460:225–230. doi: 10.1038/nature08151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrara N. Vascular endothelial growth factor and age-related macular degeneration: from basic science to therapy. Nat Med. 2010;16:1107–1111. doi: 10.1038/nm1010-1107. [DOI] [PubMed] [Google Scholar]
- 4.Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 9.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacovelli J, et al. Generation of cre transgenic mice with postnatal RPE-specific ocular expression. Invest Ophthalmol Vis Sci. 2011 Jan 6; doi: 10.1167/iovs.10-6347. published ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander JJ, Hauswirth WW. Adeno-associated viral vectors and the retina. Adv Exp Med Biol. 2008;613:121–128. doi: 10.1007/978-0-387-74904-4_13. [DOI] [PubMed] [Google Scholar]
- 13.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong MM, et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer A, et al. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med. 2010;207:1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Kaneda M, Tang F, O'Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schonborn J, et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, et al. Toll-like receptor 3 and geographic atrophy in age-related macular degeneration. N Engl J Med. 2008;359:1456–1463. doi: 10.1056/NEJMoa0802437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunaief JL, Dentchev T, Ying GS, Milam AH. The role of apoptosis in age-related macular degeneration. Arch Ophthalmol. 2002;120:1435–1442. doi: 10.1001/archopht.120.11.1435. [DOI] [PubMed] [Google Scholar]
- 29.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damiani D, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JF, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill DA, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls RD, Fischel-Ghodsian N, Higgs DR. Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell. 1987;49:369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 36.Nystrom-Lahti M, et al. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- 37.Lehrman MA, et al. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehrman MA, Goldstein JL, Russell DW, Brown MS. Duplication of seven exons in LDL receptor gene caused by Alu-Alu recombination in a subject with familial hypercholesterolemia. Cell. 1987;48:827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- 39.Wallace MR, et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- 40.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 41.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 42.Prades C, Laurent AM, Puechberty J, Yurov Y, Roizes G. SINE and LINE within human centromeres. J Mol Evol. 1996;42:37–43. doi: 10.1007/BF00163209. [DOI] [PubMed] [Google Scholar]
- 43.Saito Y, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 44.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang P, Tyrrell J, Han I, Jaffe GJ. Expression and modulation of RPE cell membrane complement regulatory proteins. Invest Ophthalmol Vis Sci. 2009;50:3473–3481. doi: 10.1167/iovs.08-3202. [DOI] [PubMed] [Google Scholar]
- 51.Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 52.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.