Summary
The anti-phagocytic polysaccharide capsule of the human fungal pathogen Cryptococcus neoformans is a major virulence attribute. However, previous studies of the pleiotropic virulence determinant Gat201, a GATA-family transcription factor, suggested that capsule-independent antiphagocytic mechanisms exist. We have determined that Gat201 controls the mRNA levels of ~1100 genes (16% of the genome) and binds the upstream regions of ~130 genes. Seven Gat201-bound genes encode for putative and known transcription factors—including two previously implicated in virulence—suggesting an extensive regulatory network. Systematic analysis pinpointed two critical Gat201-bound genes, GAT204 (a transcription factor) and BLP1, which account for much of the capsule-independent antiphagocytic function of Gat201. A strong correlation was observed between the quantitative effects of single and double mutants on phagocytosis in vitro and on host colonization in vivo. This genetic dissection provides evidence that capsule-independent anti-phagocytic mechanisms are pivotal for successful mammalian infection by C. neoformans.
INTRODUCTION
The fungus Cryptococcus neoformans is a leading cause of morbidity and mortality in AIDS patients, resulting in one million cases and 600,000 deaths annually worldwide (Park et al., 2009). Although cryptococcosis is typically associated with immunodeficient patients, a recent outbreak caused by the sister species Cryptococcus gattii in the Pacific Northwest has resulted in the deaths of numerous immunocompetent individuals (Datta et al., 2009). It is thus critical to understand the interactions of the Cryptococcus species complex with the mammalian immune system.
C. neoformans is thought to be acquired through the inhalation of spores or yeast (Botts and Hull, 2010), and alveolar macrophages are considered the first line of defense against cryptococcal infection. Experimental evidence supports this, particularly early in infection (Monga, 1981; Osterholzer et al., 2009). C. neoformans can also survive and proliferate within the phagolysosome, as well as exit macrophages by lytic and nonlytic pathways, all of which might be important for pathogenesis (Voelz and May, 2010). Macrophage activation has been shown to be important for T-cell proliferation, the development of the T-cell response, and chemokine-mediated recruitment of neutrophils and monocytes into the tissue (Monari et al., 2006a).
C. neoformans is rarely taken up by macrophages in the absence of opsonizing agents such as complement or antibodies, even after 24 hours of co-incubation (Levitz and DiBenedetto, 1989; Liu et al., 2008). This is in striking contrast to other yeasts: unopsonized Saccharomyces cerevisiae or Candida albicans are phagocytosed after less than an hour of co-incubation (Lohse and Johnson, 2008; Tejle et al., 2002). Opsonizing agents are likely present in lower levels early in infection. Within the lungs, complement is not present at constitutively high levels, but is synthesized in response to infection by many different cell types within the lungs and other tissues in a process that is estimated to take up to a week (Blackstock and Murphy, 1997; Rothman et al., 1989). Antibody generation is thought to take two to five weeks for peak production (Nussbaum et al., 1999). Therefore, cell-mediated killing by macrophages during the initial infection, prior to complement- and antibody-generation, is likely important for limiting proliferation of the invading pathogen. Studies have shown that AIDS patients generate less anti-C. neoformans antibody than healthy patients, and the severe prognosis for cryptococcosis in AIDS patients may be linked to this defect (Subramaniam et al., 2009). Antibodies generated by infected AIDS patients may be less protective against C. neoformans infection, produced at titers too low for effective opsonization, or production could be critically delayed early in infection (Dromer et al., 1988; Dromer et al., 1995). It is thus possible that yeast evasion of phagocytosis by macrophages plays an important role in the development of cryptococcosis in immunocompromised individuals.
The characteristic polysaccharide capsule of C. neoformans is considered one of its chief virulence traits. Its primary components are glucuronoxylomannan (GXM) and galactoxylomannan (GalXM), two high molecular mass polymers that have immunomodulatory properties, including suppression of both adaptive and innate immune mechanisms (Monari et al., 2006a; Monari et al., 2006b) The production of capsule by C. neoformans has been previously associated with inhibition of phagocytosis, although this correlation has been reported predominantly in the context of C. neoformans cells opsonized with serum (Del Poeta, 2004). It remains unclear how the capsular polysaccharides GXM and GalXM contribute to inhibition of unopsonized phagocytosis, which is presumably relevant to the control of disease early in the infection process. While the capsule is clearly an important virulence trait, clinical studies have suggested for decades the existence of important capsule-independent mechanisms: cryptococcosis cases produced by capsule-deficient strains exhibit a similar clinical course as infections produced by capsule-proficient strains (Torres et al., 2005).
Previously, our group described the first large-scale systematic genetic screen for virulence determinants of C. neoformans (Liu et al., 2008). We constructed and analyzed ~1200 gene knockout strains in the background of the H99 clinical isolate of C. neoformans var grubii (serotype A). A signature-tagged mutagenesis screen led to the identification of numerous factors required for the expression of known virulence factors as well as dozens of genes who contributed to virulence independently of known mechanisms. Among the mutants that produced the strongest defect in infectivity but did not affect proliferation rate at body temperature was a deletion in the GAT201 gene (Liu et al., 2008). GAT201 encodes a member of the GATA family of zinc finger transcriptional regulators that is conserved from yeast to man (Teakle and Gilmartin, 1998). A GAT201 knockout mutant shows reduced capsule size and dramatic attenuation in virulence in a mouse inhalation model of infection. Significantly, we found that gat201Δ mutants are robustly taken up by macrophages in unopsonized conditions. In contrast, less than 1% of macrophages are associated with yeast when infected with wild type C. neoformans for the same period. The defect in phagocytosis evasion by gat201Δ cells cannot be explained by the capsule size defect exhibited by gat201Δ cells: cap10Δ, cap60Δ, and cap64Δ cells, which are devoid of capsule, display a much more modest increase in association with macrophages than a gat201Δ mutant. More importantly, when CAP genes are knocked out in combination with GAT201, the resulting capΔgat201Δ strains still show dramatically increased percentages of macrophages with associated yeast cells over that displayed by the single cap mutants. These data suggested that Gat201 inhibits phagocytosis via both capsule-dependent and capsule-independent mechanisms (Liu et al., 2008). However, the relative contribution of these two functions to the virulence function of Gat201 remained to be addressed.
Given that Gat201 is predicted to be a transcriptional regulator, understanding its role in virulence requires identification and characterization of its direct targets. ChIP-chip and ChIP-seq are ideal methods for identifying direct targets of transcription factors, but the application of these methods to C. neoformans (whose polysaccharide capsule thickness can exceed the diameter of the cell) is nontrivial. In this study, we report the successful development of ChIP-chip methods for capsule-bearing C. neoformans, the identification of the direct targets of Gat201, and detailed analysis of these targets by systematic gene disruption and phenotypic characterization. We demonstrate that two targets, GAT204 and BLP1, together explain a bulk of the capsule-independent phagocytosis evasion (“anti-phagocytosis”) activity of Gat201, and that they are critical for C. neoformans’s ability to colonize the mammalian lung early in the infection process.
RESULTS
Gat201 controls the expression of ~1100 genes in response to environmental cues
Our previous work (Liu et al., 2008) indicated that Gat201 is a key virulence regulator, possibly through its inhibition of phagocytosis by macrophages. As Gat201 is a GATA-family transcription factor, we used microarray-based expression profiling to identify genes that are transcriptionally-regulated by Gat201 (Figure 1A). When we compared the transcript profiles of wild type cells to gat201Δ cells grown in standard yeast culture conditions (30°C, YPAD medium, atmospheric CO2), we observed few changes in mRNA accumulation as determined using Statistical Analysis of Microarrays (SAM) analysis (Tusher et al., 2001). In comparison, when the strains were cultured in tissue culture conditions (37°C, DMEM, 5% CO2), the same conditions used for culturing macrophages, we saw a dramatic Gat201-dependent transcriptional program (Figure 1B). Over 1,100 genes (~16% of the genome) showed a ≥2-fold difference in transcript levels and were identified by SAM analysis as differentially up- or down-regulated in a Gat201-dependent manner. We performed time course experiments utilizing RT-qPCR to examine gene expression of two representative genes from the up-regulated set (MEU1 and CNJ1810) following transfer of cells from yeast to tissue culture conditions. Through the time course, we observed steady increases in transcript levels, with maximal expression occurring between 8 and 24 hours after transfer (Figure 1C). Interestingly, GAT201 RNA levels also increased dramatically under these conditions, suggesting a positive feedback loop.
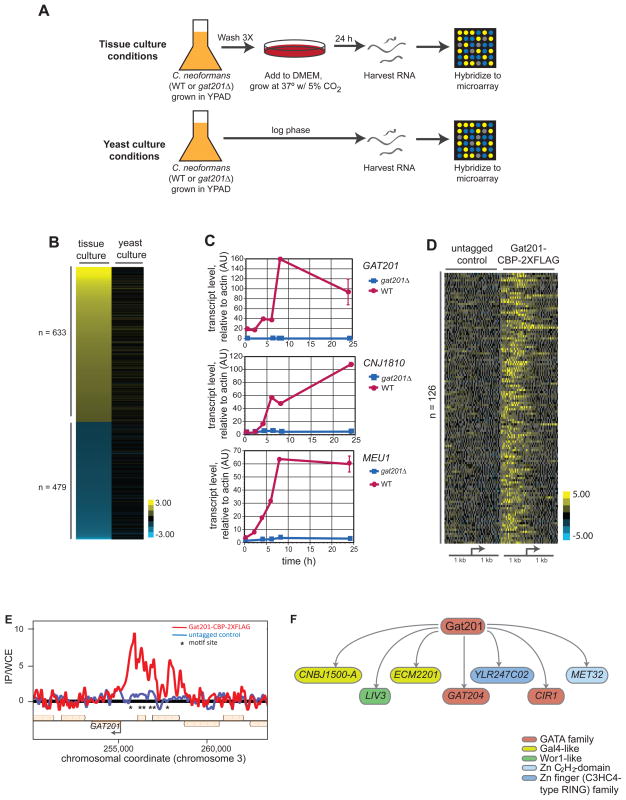
Figure 1. Identification of Gat201 transcriptional program and direct Gat201 targets.
(A) Schematic of growth conditions used for transcriptional microarrays of yeast cells grown in tissue culture conditions (DMEM, 37°C, 5% CO2) or yeast culture conditions (YPAD, 30°C, atmospheric CO2).
(B) Transcript profiling using custom whole-genome DNA microarrays was performed comparing gat201Δ versus wild type cells grown in tissue culture and in yeast culture conditions. Genes shown are those with both a minimum of a two-fold change and a statistically significant difference (using SAM analysis) between expression profiles in tissue culture and yeast media conditions. Values are plotted as the average ratio of transcript level in gat201Δ cells vs. wild type cells (log2) from four arrays per growth condition.
(C) RT-qPCR analysis of the indicated transcripts following transfer from yeast culture conditions into tissue culture conditions. Samples were harvested at the indicated time points following transfer for RNA isolation and cDNA synthesis. Error bars at the 24 hour time point are the standard deviation of two samples.
(D) Global summary of ChIP-chip results. Genes with enriched for Gat201-CBP-2XFLAG bound in their promoter regions were identified by comparing signals from replicate experiments performed with untagged and tagged strains. Genes are displayed which the largest difference between tagged and untagged genotypes in median value of the top five probe values among those within 1 kb of the predicted translational start site. Depicted are the ratios (log2) of immunoprecipitated DNA (IP) to whole cell extract (WSE) for each probe.
(E) Example of ChIP-chip data. Ratios (log2) of IP DNA to WCE were plotted for each probe of the two tiling arrays at the chromosomal coordinates indicated. Asterisks indicated locations of motif in the chromosomal sequence shown.
(F) Gat201 directly binds the regulatory regions of set of known and putative transcription factors.
See also Figure S2.
Gat201 binds the promoters of 126 genes and controls mRNA levels of 62 of these genes
To identify genes directly bound by Gat201, we used a chromatin immunoprecipitation-microarray hybridization (ChIP-chip) approach (Nobile et al., 2009). We tagged the endogenous GAT201 gene with a C-terminal calmodulin-binding peptide-2X FLAG (CBP-2XFLAG) epitope. For these studies we developed a ChIP protocol for encapsulated cells (see Supplemental Procedures). Immunoprecipitated DNA (IP) and reference DNA (WCE) from a Gat201-CBP-2XFLAG-expressing and an untagged parent strain were amplified and hybridized against a custom microarray spanning the entire genome of the strain H99. We identified 126 genes that show enrichment of Gat201-CBP-2XFLAG in their promoter regions when compared to the reference DNA and the untagged control strain (Figure 1D). As predicted from our time course analysis, Gat201-CBP-2XFLAG binds in the promoter of GAT201 itself (Figure 1E).
Having identified genes with Gat201-CBP-2XFLAG bound in their promoters, we next sought to determine if their transcription was Gat201-dependent (Figure S1). We used RT-qPCR to quantify the mRNA levels for 126 genes with the strongest enrichment for Gat201-CBP-2XFLAG in their promoters, comparing wild type and gat201Δ cells grown in yeast culture and tissue culture conditions. Transcript levels were normalized to the levels found in wild type cells grown in yeast culture conditions. We identified a set of 62 Gat201-bound genes that demonstrated Gat201-dependent transcription induction in tissue culture conditions (Table 1). The promoter regions of these genes are enriched for a seven-nucleotide motif when compared to the promoters of all genes in the H99 C. neoformans genome (Figure S2A). Strikingly, Gat201 activates the transcription of genes encoding seven known and putative transcription factors: CNBJ1500-A, GAT204, ECM2201, CIR1, LIV3, YLR236C02, and MET32. These results suggest that Gat201 controls its remarkably large transcriptional regulon in part by activating the transcription of genes coding for downstream regulators (Figure 1F).
Table 1. Gat201 target genes.
Genes with Gat201-CBP-2XFLAG bound in their promoter with Gat201-dependent induction in tissue culture conditions. CNAG numbers are gene identifiers from the Broad Institute’s annotation of H99. CDS numbers are gene identifiers from our annotation. “WT/gat201Δ” refers to the ratio of transcript level for a given gene in wild type cells over that in gat201Δ cells in tissue culture conditions. “DMEM induction” is the amount of transcript for a given gene in wild type cells grown in tissue culture conditions relative to the transcript level in wild type cells grown in yeast culture conditions. When previously characterized in the literature, the name for a gene is given. If there is no prior reference to the gene, the name assigned in our annotation is given, if available. Annotations were assigned by BLASTP and SMART analysis. Italics indicate those genes that were not successfully knocked out
| CNAG # | CDS # | gat201Δ/WT | DMEM Induction | Name | Annotation |
|---|---|---|---|---|---|
| CNAG_06517 | CDS_4922 | 144.51 | 117.50 | YOR296W02 | C2 domain-containing protein |
| CNAG_04736 | CDS_5892 | 103.76 | 153.95 | CNJ1800 | unknown |
| CNAG_06186 | CDS_1099 | 37.53 | 103.95 | CNM1740 | sugar transport |
| CNAG_01553 | CDS_3244 | 35.87 | 36.20 | CNC0890 | CHORD-containing protein |
| CNAG_05867 | CDS_3724 | 32.25 | 239.48 | CNF3650 | fucose permease |
| CNAG_01601 | CDS_2811 | 31.95 | 195.53 | ATG15 | potential vacuolar triglyceride lipase |
| CNAG_06389 | CDS_7034 | 31.10 | 20.86 | unknown | |
| CNAG_05147 | CDS_5668 | 23.64 | 51.11 | CNI2090 | unknown |
| CNAG_04735 | CDS_5889 | 23.00 | 69.96 | CNJ1810 | metalloprotease |
| CNAG_00165 | CDS_4036 | 18.59 | 39.71 | MEU1 | phosphorylase |
| CNAG_04756 | CDS_5770 | 15.69 | 72.05 | CNJ1610 | ribonuclease H-related protein |
| CNAG_00331 | CDS_4152 | 8.49 | 22.77 | YMR210W01 | alpha/beta hydrolase fold |
| CNAG_02553 | CDS_1762 | 8.31 | 17.90 | SPS1904 | putative oxidoreductase |
| CNAG_05821 | ND | 8.20 | 20.59 | unknown | |
| CNAG_03848 | CDS_6469 | 8.07 | 22.97 | CNB2310-B | putative glutaredoxin |
| CNAG_05312 | CDS_5280 | 7.93 | 3.45 | CNI3590 | macrophage activating glycoprotein |
| CNAG_00456 | ND | 6.93 | 24.06 | unknown | |
| CNAG_00374 | CDS_6603 | 6.27 | 10.73 | unknown | |
| CNAG_01777 | CDS_2931 | 6.19 | 1.47 | YNL274C03 | Glyoxylate reductase/ |
| CNAG_00919 | CDS_669 | 5.67 | 3.05 | KEX101 | carboxypeptidase S3 |
| CNAG_06762 | CDS_5944 | 5.57 | 36.16 | GAT204 | GATA-family transcription factor |
| CNAG_01552 | CDS_3426 | 5.38 | 44.66 | BET1 | T-SNARE membrane protein |
| CNAG_06346 | ND | 5.04 | 5.14 | BLP1 | Barwin-like domain |
| CNAG_05640 | CDS_5612 | 5.02 | 7.19 | SMF1 | NRAMP manganese transporter |
| CNAG_06493 | CDS_4944 | 4.81 | 14.99 | CNN2220 | unknown |
| CNAG_02189 | CDS_1614 | 4.68 | 7.46 | YJL216C | putative alpha-amylase |
| CNAG_03012 | CDS_2777 | 4.52 | 2.07 | QSP1 | quorum sensing peptide |
| CNAG_02777 | CDS_3007 | 4.49 | 158.67 | PHO840 | putative phosphate transporter |
| CNAG_05664 | CDS_5254 | 2.17 | 1.22 | BAT1 | branched amino-acid transaminase |
| CNAG_03136 | CDS_2688 | 4.29 | 20.43 | CNG0410 | putative fatty acid synthase |
| CNAG_04768 | CDS_5907 | 4.14 | 6.35 | CNBJ1500-A | Gal4-like DNA binding domain |
| CNAG_06098 | CDS_1286 | 4.09 | 5.40 | CNM0910 | glucosamine-6-phosphate |
| CNAG_03122 | ND | 3.82 | 772.37 | unknown | |
| CNAG_06200 | CDS_1088 | 3.70 | 3.92 | CNM1860 | PAS domain-containing protein |
| CNAG_06242 | CDS_1107 | 3.56 | 8.20 | CFT1 | high-affinity iron permease |
| CNAG_06108 | ND | 3.40 | 2.83 | unknown | |
| CNAG_01778 | CDS_3364 | 3.30 | 6.15 | CNC4450 | mitochondrial carrier protein-like |
| CNAG_04737 | CDS_5893 | 3.25 | 1.50 | CNJ1790 | potential methyltransferase |
| CNAG_04517 | ND | 3.21 | 3.64 | unknown | |
| CNAG_00883 | CDS_781 | 3.00 | 11.16 | ECM2201 | Gal4-like DNA binding |
| CNAG_05229 | CDS_5329 | 2.86 | 19.11 | CNI2770 | stomatin-like protein |
| CNAG_00884 | CDS_775 | 2.74 | 5.74 | SEC10 | unknown |
| CNAG_03413 | CDS_2415 | 2.73 | 9.52 | CNG3070 | alginate lyase |
| CNAG_04312 | CDS_112 | 2.43 | 2.88 | PMI4001 | mannose-6-phosphate isomerase |
| CNAG_05835 | CDS_3521 | 2.35 | 3.21 | LIV3 | Wor1-like transcription factor |
| CNAG_03915 | CDS_6280 | 2.32 | 9.78 | CNB1700 | unknown |
| CNAG_04140 | CDS_43 | 2.20 | 4.99 | CNH2650 | unknown |
| CNAG_02856 | CDS_3358 | 2.20 | 2.05 | TRK1 | putative potassium transporter |
| CNAG_03412 | CDS_2417 | 2.16 | 3.97 | CTS202 | chitinase |
| CNAG_02219 | CDS_1876 | 2.08 | 1.71 | YLR247C02 | zinc finger |
| CNAG_03847 | CDS_6346 | 2.08 | 7.85 | CNB2320 | unknown |
| CNAG_01681 | CDS_2877 | 2.07 | 10.13 | FCY2201 | putative purine-cytosine permease |
| CNAG_06763 | CDS_5942 | 2.05 | 6.61 | DIA4 | putative seryl-tRNA synthetase |
| CNAG_04864 | CDS_5991 | 2.01 | 3.22 | CIR1 | GATA-type transcription factor |
| CNAG_04436 | CDS_146 | 1.99 | 12.71 | CNH1150 | unknown |
| CNAG_04435 | ND | 1.97 | 6.83 | ferric reductase | |
| CNAG_00184 | CDS_4067 | 1.84 | 1.87 | PCF11 | Pre-mRNA cleavage complex II |
| CNAG_00068 | CDS_4631 | 1.81 | 7.46 | MET32 | Zn C2H2-domain containing protein |
| CNAG_06187 | ND | 2.54 | 3.39 | STR1 | streptomycin biosynthesis protein |
| CNAG_01040 | CDS_703 | 24.63 | 10.64 | KEX102 | carboxypeptidase S3 |
| CNAG_05159 | ND | 18.83 | 6.09 | unknown | |
| CNAG_00546 | CDS_4340 | 2.95 | 5.68 | CHS301 | chitin synthase |
Identification of key Gat201-bound genes
We considered the 62 direct downstream targets of Gat201 to be likely effectors of Gat201-mediated phagocytosis inhibition. We thus targeted each of these genes for deletion by homologous recombination (Figure S1A). We successfully knocked out 46 of 62 genes, then tested each of the mutant strains for phagocytosis defects by co-incubation with RAW264.7 macrophages. Two genes emerged as having a role in phagocytosis inhibition: CNAG_06346 and CNAG_06762. CNAG_06346 encodes a protein with a highly conserved Barwin-like domain that we named Barwin-like protein 1 (Blp1). CNAG_06762 encodes for the GATA-family transcription factor that we previously annotated as Gat204 (Chow et al., 2007). BLP1 and GAT204 show enhanced binding of Gat201-CBP-2XFLAG in their promoter regions (Figure S2B) and their expression is Gat201-dependent in tissue culture conditions (Figure 2A).
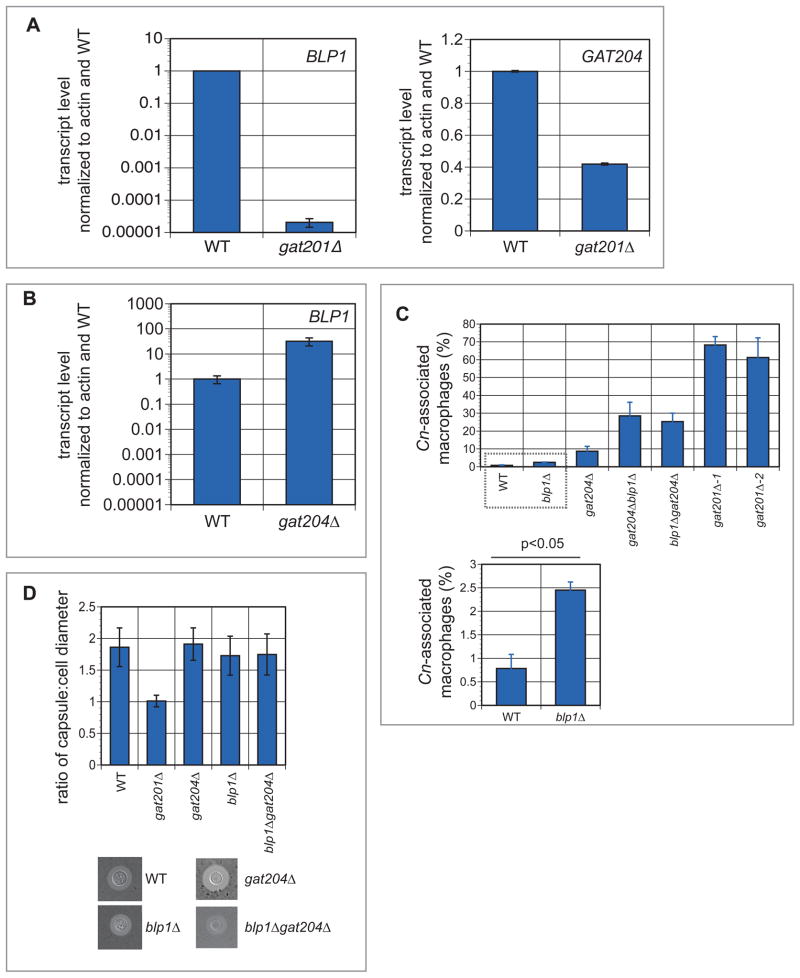
Figure 2. In vitro analysis of Gat204 and Blp1.
(A) Activation of BLP1 and GAT204 by Gat201. BLP1 (left panel) and GAT204 (right panel) transcript levels were assessed in wild type and gat201Δ cells grown in tissue culture conditions for 24 hours. Error bars are standard deviation (SD) of 2–4 samples.
(B) Repression of BLP1 by Gat204. BLP1 transcript levels were assessed in wild type and gat204Δ cells grown in tissue culture conditions for 24 hours. Error bars are SD of two samples.
(C) Quantification of phagocytosis defects. RAW264.7 macrophages were co-incubated with the designated C. neoformans strains for 24 hours then washed three times with PBS to remove unphagocytosed yeast. The macrophages were then counted to assay the number of macrophages with yeast associated. At least 200 macrophages were counted per well and each strain was assayed in triplicate. By Student’s t-test, the difference between blp1Δ and wild type association with macrophages was determined to be statistically significant.
(D) Quantitative measurements of capsule size. Capsule was induced in the indicated strains by incubation in tissue culture conditions for 24 hours. Capsule and cell size were measured for at least 30 cells per strain, and the ratio of capsule to cell diameter calculated and averaged for each strain. Error bars denote SD.
See also Figure S1.
As Gat204 is itself a transcription factor, we examined BLP1 transcription in gat204 cells. BLP1 expression is increased in gat204 cells in tissue culture conditions (Figure 2B), suggesting that Gat204 may negatively regulate BLP1 transcription, or that BLP1 transcription is up-regulated as a compensatory mechanism when Gat204 is lost.
Gat201-bound genes mediate its anti-phagocytic function
Following 24 hour co-incubation with blp1Δ cells, approximately 2.5% of RAW264.7 macrophages were associated with yeast, a modest but statistically significant (p<0.05) increase over the 0.8% of macrophages associated with wild type yeast (Figure 2C). Co-incubation with gat204Δ cells resulted in yeast-association in ~9% of RAW264.7 cells. Strikingly, two independently-derived double knockout strains in GAT204 and BLP1 showed 25–30% of RAW264.7 macrophages associated with C. neoformans, indicating a functional synergy in their mechanism of phagocytosis inhibition, and recapitulating almost 40% of the original Gat201-dependent phenotype. We also observed this synergistic increase in phagocytosis in bone marrow-derived macrophages (BMDMs) (Figure S3A). Finally, we distinguished between macrophage-internalized and -externally associated yeast cells using differential staining techniques. We confirmed that the gat204Δ and blp1Δ mutations acted synergistically to produce increases in actual uptake. We also documented an increase in external association of gat204Δ, blp1Δ, and gat204Δblp1Δ strains (Figure S3B,C), which suggests that recognition and adherence may be what is triggering phagocytosis in the mutants.
Agrobacterium T-DNA insertion screen independently identifies GAT201 and GAT204
Our original knockout library only covered a portion of the C. neoformans genome, so we sought a more global approach to identify anti-phagocytic factors. We therefore used the Agrobacterium tumefaciens T-DNA system (McClelland et al., 2005) to produce insertional mutants across the C. neoformans genome and then screened this mutant library for anti-phagocytosic defects (Figure S1B). A library of ~30,000 insertions was generated, from which 2×108 yeast cells from overnight YPAD culture were added to plated RAW264.7 macrophages. Following rounds of selection for phagocytosed yeast cells, the yeast culture was enriched for strains mutated in genes important for phagocytosis inhibition. The sites of insertion were mapped for strains that individually demonstrated increased uptake by macrophages. Strikingly, this global approach identified three independent insertional events each into GAT201 and GAT204 (Figure S1C). We also identified the copper transporter CTR2, a gene not regulated by Gat201 that causes capsule defects when mutated and is thus presented elsewhere (Chun and Madhani, 2010). We did not identify strains with mutations in BLP1 in this screen, which is not surprising given the mild phenotype of the blp1Δ single mutant. We also did not identify capsule gene mutants; however, these strains exhibit a growth defect (Chun and Madhani, unpublished observation) and are likely out-competed during the selection process. Another gene known to be involved in capsule-independent phagocytosis, APP1, acts only in opsonized cells (Luberto et al., 2003), and was not identified in our screen for unopsonized phagocytosis. These data underline the importance of Gat201 and Gat204 in limiting phagocytosis of C. neoformans.
Gat204 and Blp1 are dispensable for capsule formation
Interestingly, the level of capsule, as detected by India ink staining (Figure 2D), was unchanged in these mutants, unlike in the gat201Δ strain (Liu et al., 2008). gat204Δ, blp1Δ, and gat204Δblp1Δ mutants were also stained for β-glucan, chitin, and mannoprotein but did not show any changes relative to wild-type cells (data not shown), demonstrating that gross cell surface architecture is unchanged in these mutants. Growth at 37°C was unchanged in the gat204Δ and blp1Δ mutant strains (Figure 3A). None of the mutants were deficient in the production of melanin, a pigmented compound thought to be important for virulence in the host (Figure 3B).
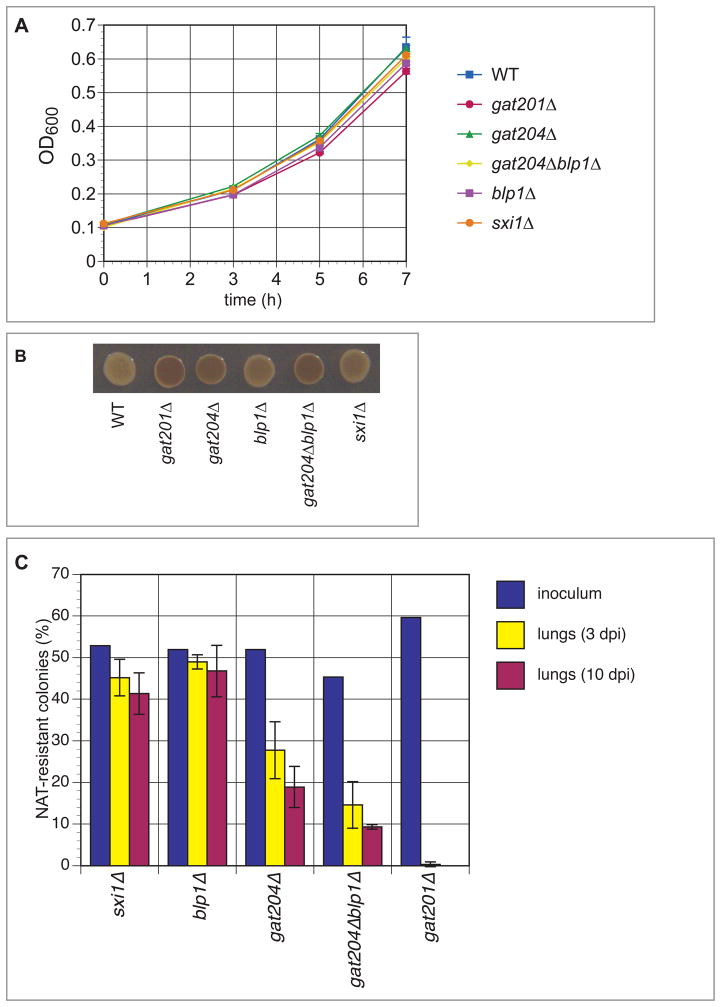
Figure 3. Anti-phagocytosis mutants show reduced fitness during infection but normal rates in vitro.
(A) Growth rate measurements. Log-phase wild type and mutant strains were grown for seven hours at 37°C in YPAD medium, and OD600 was measured every 2–3 hours. Error bars denote SD for cultures grown in triplicate.
(B) Melanin assays. Melanin assay on L-DOPA plates demonstrates that none of the strains are hypomelanized. gat201Δ is hypermelanized, as previously noted in Liu et al (2008). gat204Δ is also hypermelanized, as is the gat204Δblp1Δ mutant.
(C) Mouse infection data. Approximately 1:1 mixtures of H99 and the indicated mutant (marked with a nourseothricin [NAT] resistance gene) were inoculated intranasally into mice (A/J) (5 × 105 total cells/mouse). The actual proportion of mutant cells in each inoculum were determined by plating a dilution of the inoculum on non-selective medium and then assaying 100–200 individual colonies for NAT resistance. At three and ten days post-infection, animals were sacrificed and the lungs, brains, and spleen from each were homogenized and serial dilutions were plated. 100–200 colonies per organ were assayed for NAT resistance to determine the percentage of mutant cells. Error bars represent the SD from three mice per inoculum. See also Figure S3.
Blp1 is part of a six-gene family in C. neoformans
SMART protein sequence analysis of Blp1 identified a conserved Barwin-like domain (Figure S2C). This domain is found in a number of protein families in plants, fungi, and bacteria, several members of which have antifungal properties (Hejgaard et al., 1992). Analysis of predicted protein sequence by SignalP reveals a high probability (1.000) of a signal peptide. BLP1 appears to be part of a six-member family in C. neoformans, all of which contain the Barwin-like domain and a signal sequence.
Inhibition of phagocytosis correlates with effective colonization of the lungs
To determine whether inhibition of phagocytosis by C. neoformans correlates with its ability to colonize the lung, we performed competitive infections. Wild type cells were mixed in equal proportion with gat201Δ, gat204Δ, blp1Δ, or gat204Δblp1Δ cells. As a control strain, we used a knockout mutant in SXI1, which is not expressed in non-mating cells and is dispensable for virulence (Hull et al., 2004). Inocula were administered to mice through an inhalation model of infection. The lungs were harvested after three or ten days and the proportion of mutant to wild type cells assessed. Strikingly, we observed a correlation between the degree of phagocytosis inhibition in a given strain and the relative ability of that strain to colonize the lung tissue (Figure 3C). While the mutant cells made up ~50% of each inoculum, their prevalence after three or ten days in the lungs correlated with their anti-phagocytic abilities. The gat201Δ strain, which showed the least phagocytic inhibition of all the strains tested, was entirely absent from lung tissue homogenate-derived colonies. The gat204Δ strain, which showed decreased phagocytic inhibition compared to wild type, also displayed decreased representation in the lungs (~27% at 3dpi, ~19% at 10dpi). gat204Δblp1Δ, which displayed a level of phagocytic inhibition between that of gat201Δ and gat204Δ, showed a level of representation in the lungs also intermediate to the two strains. The blp1Δ mutant, which displayed the weakest phagocytosis phenotype, and the control sxi1Δ mutant represented ~50% of the cells recovered from the lungs.
DISCUSSION
Cryptococcus species produce a polysaccharide capsule, which numerous studies have shown to be a major virulence factor and an important mechanism of immune modulation. In particular, capsule inhibits phagocytosis of C. neoformans by macrophages (Monari et al., 2006a). However, our previous work demonstrates that the pleiotropic transcriptional activator Gat201 controls a capsule-independent mechanism by which C. neoformans evades phagocytosis but also regulates capsule synthesis (Liu et al., 2008). Here, we describe a regulatory circuit dissection approach to further illuminate capsule-independent antiphagocytic mechanisms and to test their roles in infection. The work described in this paper demonstrates that at least two such mechanisms exist, begins to identify potential end-effectors of this regulatory circuit, and—importantly—provides strong evidence that such mechanisms are important for successful mammalian infection by C. neoformans.
An integrative approach identifies the direct targets of the Gat201 virulence regulator
We found that over one thousand genes, or approximately 1/6 of the C. neoformans genome, showed statistically significant changes in expression in gat201Δ cells compared to wild type. As expression of GAT201 itself was induced in these conditions, Gat201 could function downstream of a signaling pathway(s) that senses environmental changes and modulates a massive reprogramming of the cellular transcriptome.
To identify the direct targets of Gat201, we developed ChIP-chip methods. Through this approach we identified 126 genes that demonstrate robust binding of Gat201 in their promoter regions, 62 of which depend on Gat201 for transcript accumulation in tissue culture conditions. Significantly, seven targets of Gat201 themselves encode known or predicted transcription factors, two of which, Liv3 and Cir1, have been previously implicated in pathogenicity (Jung et al., 2006; Liu et al., 2008). The results reported here implicate Gat204 in infectivity. Thus, it appears that Gat201 is part of a critical large virulence transcriptional regulatory network that controls pathogenicity in response to environmental signals.
Systematic analysis reveals the existence of multiple capsule-independent anti-phagocytosis mechanisms
Through a systematic gene disruption approach, we identified two genes, BLP1 and GAT204, which affect the ability of C. neoformans to evade unopsonized phagocytosis by macrophages. Together, these two genes recapitulate a significant portion of the phenotype of the gat201Δ mutant strain. These observations suggest that at least three mechanisms exist to inhibit phagocytosis: GXM, those dependent on Blp1 and Gat204, and at least one mechanism that is Gat201-dependent but Blp1/Gat204-independent.
Capsule-independent phagocytosis inhibition and mammalian infectivity of Cryptococcus neoformans
Finally, we have demonstrated that the relative ability of a given strain to evade or inhibit phagocytosis correlates with its ability to survive and proliferate within the lung tissue. We examined time points relatively early in infection, when we expect alveolar macrophages to play a crucial role in the initial host response, but before a full adaptive immune response develops. Cells lacking Gat201 are cleared from the lungs of infected mice within three days, demonstrating an early and pivotal role for this transcription factor in pathogen success. Significantly, the analysis of single and double mutants demonstrated that the roles of Gat201, Gat204 and Blp1 in survival within the murine lung are proportional to their impact on phagocytosis in vitro. The correlation between the in vitro and in vivo data strongly indicates that multiple capsule-independent anti-phagocytosis mechanisms are critical for infection of the mammalian host. Thus, while a major focus of C. neoformans research has justifiably been its polysaccharide capsule, this work demonstrates capsule-independent anti-phagocytic mechanisms are important for successful host colonization.
EXPERIMENTAL PROCEDURES
Gene nomenclature
Genes were identified using annotation from the H99 sequence from the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) and from our own annotation of the H99 sequence (http://cryptogenome.ucsf.edu). Gene annotations from the Broad are designated by their nomenclature “CNAG_#”, while our own annotations are designated “CDS_#”.
Strains and media
C. neoformans growth conditions were as previously described (Chun et al., 2007; Chun and Madhani, 2010; Liu et al., 2008), with minor modifications found in the Supplemental Procedures.
Phagocytois assay
RAW264.7 macrophage assays were performed as previously described (Chun and Madhani, 2010; Liu et al., 2008). Bone marrow-derived macrophage experiments were performed in much the same manner with minor modifications described in Supplemental Procedures.
Expression profiling
Expression profiling was performed as previously described (Liu et al., 2008) with minor modifications described in Supplemental Procedures. Analysis was performed as described in Chun et al., 2007.
ChIP-on-chip
The ChIP-chip tiling arrays were designed on 244,000 probes of 60-bp length, averaging 80-bp between probes, across the entire H99 sequence as it was published by the Broad Institute, current to 2007. Experiments were performed as previously described (Nobile et al., 2009) with C. neoformans-specific modifications described in the Supplemental Procedures.
RT-qPCR
Gene expression differences observed using microarray-based transcript profiling were confirmed utilizing RT-qPCR as previously described (Chun et al., 2007) with minor modifications (see Supplemental Procedures).
A. tumefaciens-mediated transformation of C. neoformans
These experiments were performed as described in McClelland et al., 2005 (McClelland et al., 2005) with minor modifications as described in Chun and Madhani, 2010.
Phagocytosis Screen with Insertional Mutant Library
This procedure was performed as previously described (Chun and Madhani, 2010).
Capsule assay
Experiments were performed as previously described (Liu et al., 2008). See Supplemental Procedures for more details.
Growth curve
Overnight YPAD cultures grown at 37°C were diluted back to OD600=0.1 in YPAD, in triplicate. Cultures were grown at 37°C for 7 hours, and the OD600 was measured every 2–3 hours.
Melanin assay
From overnight culture, C. neoformans cells were diluted to OD600 = 0.6 in water then spotted onto melanin-inducing plates containing L-DOPA (L-dihydroxyphenylalanine, Sigma, 100 mg l-1) and grown for three days at 37°C.
Intranasal co-infection experiments
Experiments were performed as described previously (Liu et al., 2008).
Supplementary Material
Acknowledgments
We thank Paolo Manzanillo and Jeffery S. Cox for the kind gift of bone marrow-derived macrophages. This work was supported by grant R01AI065519 from the National Institute of Allergy and Infectious Disease (NIAID) to HDM. CDC and JCSB are supported by NIAID grant 5T32AI060537-07 to the UCSF Microbial Pathogenesis Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blackstock R, Murphy JW. Secretion of the C3 component of complement by peritoneal cells cultured with encapsulated Cryptococcus neoformans. Infect Immun. 1997;65:4114–4121. doi: 10.1128/iai.65.10.4114-4121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts MR, Hull CM. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol. 2010;13:437–442. doi: 10.1016/j.mib.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow ED, Liu OW, O’Brien S, Madhani HD. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Curr Genet. 2007;52:137–148. doi: 10.1007/s00294-007-0147-9. [DOI] [PubMed] [Google Scholar]
- Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Madhani HD. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, et al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Poeta M. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot Cell. 2004;3:1067–1075. doi: 10.1128/EC.3.5.1067-1075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromer F, Aucouturier P, Clauvel JP, Saimot G, Yeni P. Cryptococcus neoformans antibody levels in patients with AIDS. Scand J Infect Dis. 1988;20:283–285. doi: 10.3109/00365548809032452. [DOI] [PubMed] [Google Scholar]
- Dromer F, Denning DW, Stevens DA, Noble SM, Hamilton JR. Anti-Cryptococcus neoformans antibodies during cryptococcoss in patients with acquired immunodeficiency syndrome. Serodiagnosis and Immunotherapy in Infectious Disease. 1995;7:181–188. [Google Scholar]
- Hejgaard J, Jacobsen S, Bjorn SE, Kragh KM. Antifungal activity of chitin-binding PR-4 type proteins from barley grain and stressed leaf. FEBS Lett. 1992;307:389–392. doi: 10.1016/0014-5793(92)80720-2. [DOI] [PubMed] [Google Scholar]
- Hull CM, Cox GM, Heitman J. The alpha-specific cell identity factor Sxi1alpha is not required for virulence of Cryptococcus neoformans. Infect Immun. 2004;72:3643–3645. doi: 10.1128/IAI.72.6.3643-3645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, DiBenedetto DJ. Paradoxical role of capsule in murine bronchoalveolar macrophage-mediated killing of Cryptococcus neoformans. J Immunol. 1989;142:659–665. [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luberto C, Martinez-Marino B, Taraskiewicz D, Bolanos B, Chitano P, Toffaletti DL, Cox GM, Perfect JR, Hannun YA, Balish E, et al. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J Clin Invest. 2003;112:1080–1094. doi: 10.1172/JCI18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland CM, Chang YC, Kwon-Chung KJ. High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens. Fungal Genetics and Biology. 2005;42:904–913. doi: 10.1016/j.fgb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Monari C, Bistoni F, Vecchiarelli A. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 2006a;6:537–542. doi: 10.1111/j.1567-1364.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- Monari C, Kozel TR, Paganelli F, Pericolini E, Perito S, Bistoni F, Casadevall A, Vecchiarelli A. Microbial immune suppression mediated by direct engagement of inhibitory Fc receptor. J Immunol. 2006b;177:6842–6851. doi: 10.4049/jimmunol.177.10.6842. [DOI] [PubMed] [Google Scholar]
- Monga DP. Role of macrophages in resistance of mice to experimental cryptococcosis. Infect Immun. 1981;32:975–978. doi: 10.1128/iai.32.3.975-978.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum G, Anandasabapathy S, Mukherjee J, Fan M, Casadevall A, Scharff MD. Molecular and idiotypic analyses of the antibody response to Cryptococcus neoformans glucuronoxylomannan-protein conjugate vaccine in autoimmune and nonautoimmune mice. Infect Immun. 1999;67:4469–4476. doi: 10.1128/iai.67.9.4469-4476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholzer JJ, Milam JE, Chen GH, Toews GB, Huffnagle GB, Olszewski MA. Role of dendritic cells and alveolar macrophages in regulating early host defense against pulmonary infection with Cryptococcus neoformans. Infect Immun. 2009;77:3749–3758. doi: 10.1128/IAI.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Rothman BL, Merrow M, Despins A, Kennedy T, Kreutzer DL. Effect of lipopolysaccharide on C3 and C5 production by human lung cells. J Immunol. 1989;143:196–202. [PubMed] [Google Scholar]
- Subramaniam K, Metzger B, Hanau LH, Guh A, Rucker L, Badri S, Pirofski LA. IgM(+) memory B cell expression predicts HIV-associated cryptococcosis status. J Infect Dis. 2009;200:244–251. doi: 10.1086/599318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teakle GR, Gilmartin PM. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem Sci. 1998;23:100–102. doi: 10.1016/s0968-0004(98)01174-8. [DOI] [PubMed] [Google Scholar]
- Tejle K, Magnusson KE, Rasmusson B. Phagocytosis and phagosome maturation are regulated by calcium in J774 macrophages interacting with unopsonized prey. Biosci Rep. 2002;22:529–540. doi: 10.1023/a:1022025903688. [DOI] [PubMed] [Google Scholar]
- Torres HA, Prieto VG, Raad II, Kontoyiannis DP. Proven pulmonary cryptococcosis due to capsule-deficient Cryptococcus neoformans does not differ clinically from proven pulmonary cryptococcosis due to capsule-intact Cr. neoformans. Mycoses. 2005;48:21–24. doi: 10.1111/j.1439-0507.2004.01068.x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz K, May RC. Cryptococcal Interactions with the Host Immune System. Eukaryot Cell. 2010;9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.