Abstract
The DENN domain is a common, evolutionarily ancient, and conserved protein module, yet it has gone largely unstudied; until recently, little was known regarding its functional roles. New studies reveal that various DENN domains interact directly with members of the Rab family of small GTPases and that DENN domains function enzymatically as Rab-specific guanine nucleotide exchange factors. Thus, DENN domain proteins appear to be generalized regulators of Rab function. Study of these proteins will provide new insights into Rab-mediated membrane trafficking pathways.
Keywords: Endocytosis, Inositol Phospholipid, Intracellular Trafficking, Membrane Trafficking, Vesicles, DENN Domain, DENND, GEF, GTPase, Rab
Introduction
Protein domains are modular cassettes with conserved folds that are often found in otherwise unrelated proteins. From an evolutionary point of view, modular domains are readily joined in new combinations, creating novel connections and cellular pathways (1). The DENN (differentially expressed in normal and neoplastic cells) domain is a poorly characterized protein module conserved throughout evolution, with DENN domain proteins found in species as diverse as humans, Caenorhabditis elegans, Arabidopsis thaliana, and Schizosaccharomyces pombe. There are 18 genes encoding DENN domain-containing proteins in humans. For some, the DENN domain is the only recognizable feature; for others, the DENN domain is found alongside other modular domains. The observation that several DENN domain proteins interact with Rab GTPases provided the first insight into the potential function of the domain (2). Rabs, with ∼70 members in humans, are the largest family of small GTPases. They cycle between an inactive GDP-bound state and an active GTP-bound state. In the active state, they recruit effectors that control multiple aspects of membrane trafficking (3, 4). A breakthrough in our understanding of the DENN domain came with the observation that the DENN domain from the connecdenn family of proteins interacts directly with Rab35 and functions as a guanine nucleotide exchange factor (GEF)3 for this GTPase (5–7). GEFs activate Rabs by mediating the exchange of GDP for GTP. As Rabs have diversified throughout evolution (9 Rabs/Ypts in S. pombe, 30 in C. elegans, and ∼70 in humans, for example), so have DENN domain proteins, with 1 in S. pombe, ∼5 in C. elegans, and 18 in humans. Thus, DENN domains may have evolved as generalized GEFs for Rabs, and in fact, all subfamilies of DENN domain proteins appear to possess Rab-directed GEF activity (8). Moreover, because at least some DENN domains interact with one Rab while mediating GEF activity toward a second, DENN domains may be at an interface between different Rab pathways. Here, we will provide an overview of all DENN domain proteins encoded in the human genome and will describe exciting new insights confirming that DENN domain-bearing proteins are an important class of membrane trafficking molecules and key regulators of Rab GTPases.
Identification of the DENN Domain
Chow and Lee (9) originally cloned an open reading frame that they named DENN based on its variable mRNA expression levels in tissues and cell lines. The DENN protein was independently identified as a binding partner of the cytoplasmic death domain of the TNF receptor (10). Here, the protein was named MADD (MAPK-activating protein containing a death domain) (10).
Databases such as Pfam and PROSITE were established in part to identify and annotate protein modules. These initiatives recognized that a portion of the N-terminal region of DENN/MADD was similar to regions in several otherwise unrelated proteins, resulting in the concept of a DENN domain. One such protein was Rab6IP1 (Rab6-interacting protein 1), identified in a two-hybrid screen with Rab6 (11). A subsequent bioinformatics analysis comparing Rab6IP1 and DENN/MADD with other potential DENN domain proteins led to the seminal observation that the domain is in fact tripartite, consisting of a central DENN module flanked by upstream (uDENN) and downstream (dDENN) modules (Fig. 1) (2). The modules are always found together but are separated by linkers of various lengths (Fig. 2). DENN domains are related to a series of conserved regions found in members of the Avl9 and Avl9-related protein families (12). Each member of these families is composed of five regions, called Avl9 homology (AH) 1–5, which are found in consecutive order. Alignment of the combined AH1–AH5 regions (AH domain) with DENN domains revealed weak but significant similarity (12). Interestingly, within the AH domain, in addition to homology to DENN domains, there is weak homology to TBC (Tre-2/Bub2/Cdc16) and RhoGEF domains, which are found in other regulators of GTPases (12). Thus, like DENN domains, AH domains may function in the regulation of GTPases.
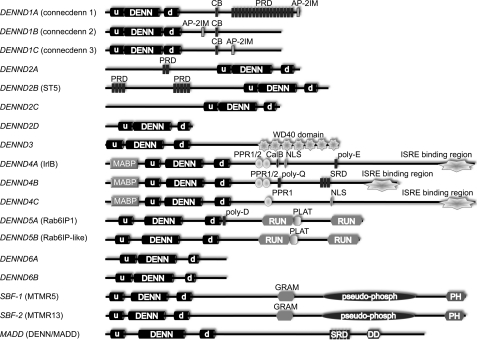
FIGURE 1.
Domain models of the 18 DENN domain-bearing proteins encoded in the human genome. The gene name is listed on the left, with the protein name in parentheses when appropriate. u, uDENN module; d, dDENN module; CB, clathrin-binding motif; PRD, proline-rich domain; AP-2IM, AP-2 interaction motif; MABP, MVB12-associated β-prism; PPR, pentatricopeptide repeat; CalB, calmodulin-binding domain; NLS, nuclear localization sequence; poly-E, polyglutamic acid domain; ISRE, interferon-stimulated response element; poly-Q, polyglutamine domain; SRD, serine-rich domain; poly-D, polyaspartic acid domain; PLAT, polycystin-1/lipoxygenase/α-toxin domain; GRAM, glucosyltransferase/Rab-like GTPase activator/myotubularin domain; pseudo-phosph, pseudo-phosphatase domain; DD, death domain.
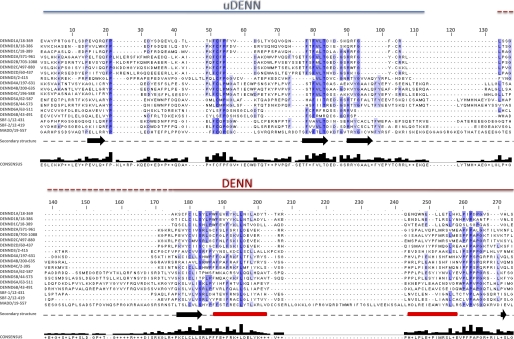
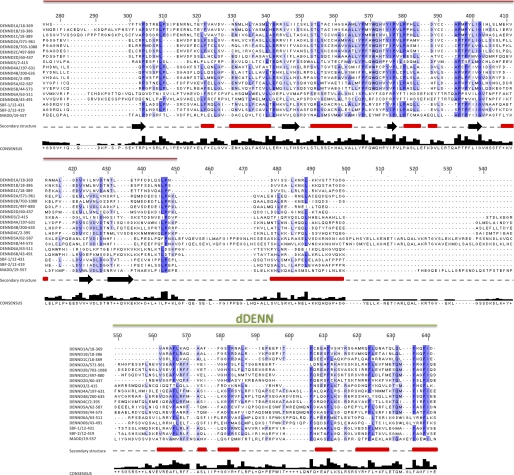
FIGURE 2.
Amino acid sequence alignment of the 18 human DENN domain proteins. The amino acid sequences of the DENN domains correspond to the sequences of the UniProt long isoforms listed in Table 1. Alignment was performed with ClustalW and manually modified using Jalview. Shaded boxes represent conserved residues, with darker shadings based on increasing percent identity. The dashed border for the N terminus of the DENN module is used to highlight the varying lengths of the linker regions between the uDENN and DENN modules, where some DENN modules begin at the start of the dashed line and others begin at the start of the solid line. Secondary structure predictions are also indicated along the gray dashed line below the alignment. β-Strands and α-helices are represented as black arrows and red rectangles, respectively.
DENN Domain Protein Families in Humans
Based on homology and domain organization, the 18 DENND (DENN domain) proteins in the human genome are grouped into eight families (Fig. 1 and Table 1). These are 1) DENND1A–1C, 2) DENND2A–2D, 3) DENND3, 4) DENND4A–4C, 5) DENND5A/5B, 6) DENND6A/6B, 7) MTMR5/13, and 8) DENN/MADD. In all cases, the DENN domain is located toward the N terminus, except for the DENND2 family, where it is located toward the C terminus. Outside the DENN domain, the protein families have no homology with other families (Fig. 1). The DENN domain families will be discussed in turn.
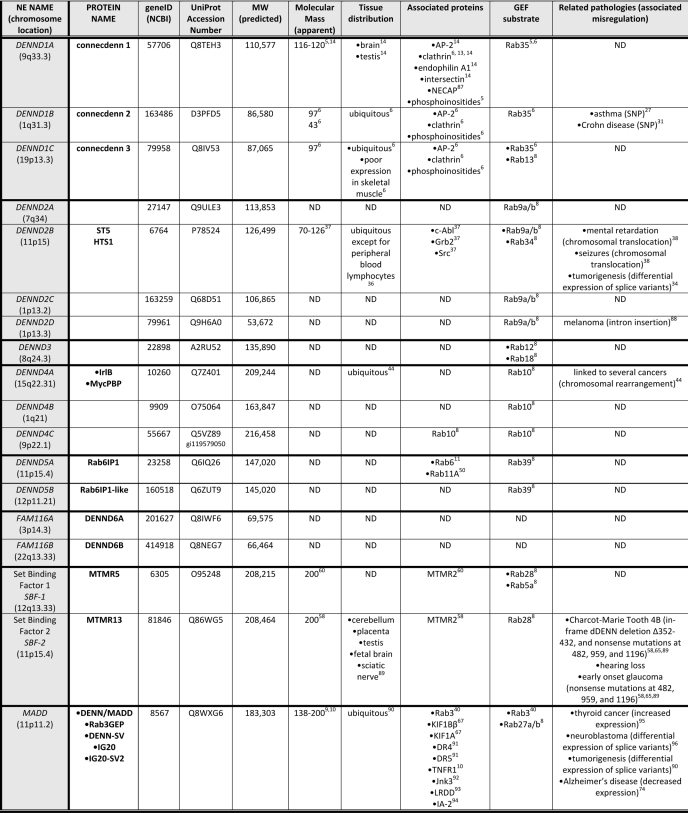
TABLE 1.
Human DENN domain proteins
All information regarding apparent molecular mass, tissue distribution, binding partners, and related pathologies has been determined experimentally. The predicted molecular weight is of the longest potential isoform of the protein. Some DENN domain proteins have defined or predicted splice variants accounting for the range in their apparent molecular masses. ND, not determined.
DENND1A–1C (Connecdenns 1–3)
One of the best characterized DENND families is DENND1A–1C, also known as connecdenns 1–3. The first family member, connecdenn 1, was detected in a proteomic analysis of clathrin-coated vesicles (13). In addition to the DENN domain, the protein contains consensus-binding motifs for clathrin, the clathrin adaptor AP-2, and Src homology 3 (SH3) domains (Fig. 1) (14). Connecdenn 1 binds directly to clathrin, AP-2, and the endocytic SH3 domain-bearing proteins endophilin and intersectin (5, 6, 14).
The first link between the connecdenns and Rab35 was demonstrated in an elegant mutagenesis screen in C. elegans aimed at identifying proteins involved in receptor-mediated endocytosis of yolk proteins. Two of the proteins identified were RME-4 (receptor-mediated endocytosis 4) and RME-5 (7). RME-4 is the C. elegans ortholog of the connecdenns, and RME-5 is Rab35 (7). Mutations in either protein disrupt endosomal recycling of the yolk receptor, leading to an indirect defect in yolk protein endocytosis. RME-4 binds Rab35 and directs it to clathrin-coated vesicles for transport to early endosomes. From there, Rab35 functions in a recycling route that traffics the yolk receptor back to the plasma membrane (7).
GEFs interact with the GDP-bound form of their substrate GTPases and catalyze the removal of GDP. This allows GTP, present at much higher levels in cells than GDP, to diffuse onto the GTPase. The GEF has low affinity for GTP-bound GTPase and dissociates. Thus, the observation that RME-4/connecdenn binds specifically to GDP-bound Rab35 suggested that it may function as a Rab35 GEF (7). This was investigated in mammalian systems, where it was discovered that all three connecdenns, via their DENN domains, serve as Rab35 GEFs (5, 6). These experiments were performed with highly purified DENN domains and Rab35. The best known Rab GEF module is the Vps9 domain (15). In direct comparison with the active GEF region from the well established Rab5 GEF Rabex-5 (residues 1–399), which contains a Vps9 domain, the connecdenn 1 DENN domain is dramatically more robust (5). Within 90 s at room temperature, >90% of the GDP on 19 pmol of preloaded Rab35 is exchanged for GTP by 1.5 pmol of connecdenn 1 DENN domain (5). In contrast, even after 3 min under identical conditions, Rabex-5 exchanges 5-fold less GDP preloaded on Rab5 (5). Interestingly, each of the connecdenn DENN domains has a distinct rate of GEF activity, with connecdenn 1 being the fastest and connecdenn 3 the slowest (6). However, even connecdenn 3 is more robust for Rab35 than is Rabex-5 for Rab5. Thus, unlike for Rabex-5, where the protein Rabaptin-5 is needed for maximum enzymatic activity (16), a cofactor is not strictly required for the connecdenn DENN domains.
A subsequent study confirmed the activity of connecdenns 1 and 2 toward Rab35 but failed to detect the GEF activity of connecdenn 3 toward Rab35, instead indicating activity toward Rab13 (8). This is surprising in that for every other DENND family, each member of the family shows GEF activity towards a common Rab (Table 1). Perhaps the GEF assays employed in this study (8), which utilized immunoprecipitated full-length proteins, lacked sufficient sensitivity to detect the activity of connecdenn 3 on Rab35. Also, given the broad scope of the study, in which the authors tested multiple DENN proteins against large panels of Rabs, kinetic studies were not performed, and instead, GEF activity was measured only as a function of GDP release or GTP binding after 20 min (8). It will be important to perform detailed kinetic analysis of each DENN domain family using purified DENN domains.
In addition to its role as a Rab35 GEF, the DENN domain of connecdenn 1 binds a broad spectrum of lipids with a slight preference for phosphatidylinositol 3-phosphate (PI(3)P) (5). The DENN domains of connecdenns 2 and 3 share this lipid-binding profile.4 However, the functional relevance of this property and the relationship to the GEF activity remain uncertain.
It is interesting that each member of each DENND family targets a common Rab (Table 1) (5, 6, 8). Perhaps different proteins within a family activate the Rab toward different cellular activities. For example, Rab35 has been found on the plasma membrane, clathrin-coated pits and vesicles, and endosomes, and it controls a cargo-specific fast recycling route from early endosomes (5, 7, 17–25). In addition, Rab35 co-localizes with actin at the leading edge of the cell and on stress fibers and has been implicated in regulation of actin-based cellular events (19–21, 26). Different connecdenns may therefore activate Rab35 at different cellular locations to control the diverse functions of this Rab.
Recently, a genome-wide association study linked DENND1B/connecdenn 2 to childhood asthma (Table 1) (27). However, this study must be interpreted with caution given that DENND1B/connecdenn 2 has not been linked to pro-inflammatory signaling (28) as implied by Sleiman et al. (27) and that two other studies have failed to replicate this linkage (29, 30). A more recent genome-wide association study suggests a link of DENND1B to Crohn disease (31).
DENND2A–2D
The DENND2 family is the only example wherein the DENN domain is found in the C-terminal region (Fig. 1). Each DENND2 protein acts as a GEF toward Rab9a/b, and as DENND2D is composed of a DENN domain alone, for this protein, GEF activity is via the DENN domain (8). Rab9 functions in retrograde trafficking of the mannose phosphate receptor from late endosomes to the trans-Golgi network (32), and consistently, depletion of DENND2A disrupts this trafficking process (8). However, depletion of other DENND2 family members did not influence mannose phosphate receptor trafficking (8), so perhaps these DENND2 proteins activate Rab9 toward other cellular activities such as biogenesis of lysosome-related organelles (33).
The best studied DENND2 family member is DENND2B, originally named ST5 (suppressor of tumorigenicity 5) (Table 1) (34). ST5 exists in three splice variants that all contain the C-terminal DENN domain (35). Interestingly, expression levels of the shortest isoform, p70, correlate positively with reduced tumorigenicity, and overexpression of p70 restores contact-regulated cell growth to tumor cells (36). Loss of the C terminus of p70 converts it from an inhibitor to an activator of transforming activity, demonstrating the importance of an intact DENN domain for tumor suppressor function (37). Disruption of ST5 is also associated with mental retardation, seizures, and congenital anomalies (38). The link of any of these activities of ST5 to its activation of Rab9 remains unclear.
DENND3
Full-length DENND3 (Fig. 1) has GEF activity toward Rab12 (8). Although it is likely that this activity is via the DENN domain, this has not been tested directly (8). This is an important consideration in that the GEF activity could be located outside the DENN domain, as is the case for the GEF activity of DENN/MADD toward Rab3 (39, 40). Rab12 localizes to small vesicles that accumulate on or near the Golgi, and the protein has been suggested to function in retrograde trafficking from the periphery to the perinuclear region (41, 42). The function of DENND3 has yet to be studied.
DENND4A–4C
The only studied member of the DENND4 family is DENND4A, originally named IrlB (43). IrlB contains a nuclear localization signal and binds the interferon-stimulated response element in the promoter of human Myc (43, 44). However, the functional role of DENND4A and other DENND4 family members in mammals is unknown, and it is unclear if they localize to the nucleus. In contrast, the Drosophila DENND4 ortholog Crag (calmodulin-binding protein related to a Rab3 GDP-GTP exchange protein) clearly regulates the polarized secretion of basement membrane components (45). Normally, these proteins are found on the basal side of the epithelium only, but in Crag mutants, they accumulate on both sides of the epithelium, resulting in a loss of epithelial integrity (45). Consistent with this function, DENND4A–4C proteins have GEF activity toward Rab10, a Rab involved in the regulation of basolateral trafficking in polarized cells as well as Glut4 recycling in adipocytes (8, 46–49). These data appear inconsistent with any role for DENND4 proteins in the nucleus.
DENND5A/5B (Rab6IP1/Rab6IP1-like Protein)
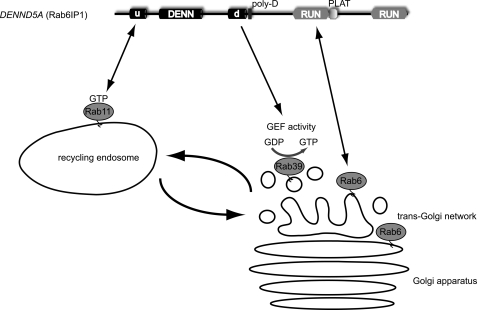
Rab6IP1 was identified as a Rab6-binding partner (11), and Rab6IP1-like protein is known only as its homolog. In addition to the DENN domain, both proteins contain two RUN (RPIP8/UNC-14/NESCA) domains (Fig. 1) (2, 11). RUN domains are found in numerous proteins associated with Rab GTPases, and Rab6IP1 binds Rab6 in a nucleotide-independent manner through the first RUN domain (Fig. 3) (50, 51).
FIGURE 3.
Coordination between endosome-derived carriers (Rab11) and Golgi acceptor compartments (Rab6 and Rab39) via Rab6IP1. Rab6IP1 binds GTP-bound Rab11, likely through its uDENN module (u), and binds Rab6 through its first RUN domain while functioning as a GEF toward Rab39. Rab6IP1 is targeted to the Golgi by Rab6 and to recycling endosomes by Rab11, providing a molecular link between Rab6, Rab11, and Rab39. d, dDENN domain; poly-D, polyaspartic acid domain; PLAT, polycystin-1/lipoxygenase/α-toxin domain.
Following from observations that Rab6 and Rab11 regulate sequential steps in retrograde transport from endosomes to the Golgi, Miserey-Lenkei et al. (50) set out to identify proteins that coordinate the function of these two GTPases and demonstrated that Rab6IP1 binds to GTP-bound Rab11 (Fig. 3). Rab6IP1 exists as two splice variants, A and B, due to a 24-amino acid insert within the uDENN module of isoform B. Only the A variant binds Rab11, suggesting that the uDENN module mediates the interaction (50). Rab6IP1 is targeted to the Golgi in a Rab6-dependent manner and also associates with Rab11 at recycling endosomes (50). Thus, Rab6IP1 provides a molecular link between Rab6 and Rab11 (Fig. 3), although the functional role of this process remains uncertain.
Both Rab6IP1 and Rab6IP1-like protein have GEF activity toward Rab39 (8), a poorly characterized Rab with two isoforms (52, 53). Rab39a regulates interleukin secretion (54). Rab39b is involved in trafficking of Golgi-derived vesicles required for normal growth cone function and synapse formation during neuronal development (55). Loss-of-function mutations in Rab39b result in X-linked mental retardation (55). Additional studies will be needed to determine the relationship between Rab39, Rab6, and Rab11.
DENND6A/B
The DENND6 family, including DENND6A and DENND6B, is composed of DENN domains exclusively (Fig. 1). This family remains completely uncharacterized.
MTMR5/13
Myotubular myopathy is a severe congenital skeletal muscle disease resulting from mutations in the lipid phosphatase myotubularin 1. Following the characterization of this protein, Blondeau et al. (56, 57) identified a large family of MTMR (myotubularin 1-related) proteins, including the DENN domain-bearing MTMR5 and MTMR13. Although MTMR5 and MTMR13 are homologous to other MTMR proteins, they are pseudo-phosphatases; however, they interact with MTMR2, an active lipid phosphatase (58–60). These interactions enhance the activity of MTMR2 toward its targets, PI(3)P and phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) (58–60). PI(3)P and PI(3,5)P2 are found on endosomes, and thus, the MTMR2-MTMR5 and MTMR2-MTMR13 complexes are thought to regulate endosomal function (58, 60).
MTMR5 and MTMR13 have GEF activity toward Rab28 (8). Rab28 is a poorly characterized and distant member of the Rab superfamily (8, 61). The association of MTMR5 and MTMR13 with endosomal function (62) now suggests that Rab28 could be involved in endosomal trafficking. In fact, Rab28 co-localizes with the ESCRT complex in the endosomal pathway, where it plays a role in late endocytic traffic (63).
Inactivating mutations in MTMR2 result in Charcot-Marie-Tooth type 4B1 neuropathy (64), and loss of the dDENN domain of MTMR13 also results in a form of Charcot-Marie-Tooth, type 4B2 (65). Although the mechanisms by which alterations in endosomal trafficking lead to Charcot-Marie-Tooth disease remain undefined, these results provide a direct link between a DENN domain mutation and human disease.
DENN/MADD/Rab3GEP
DENN/MADD was independently purified from brain extracts as a GEF for Rab3 and named Rab3GEP (Rab3 guanine nucleotide exchange protein) (40). In fact, DENN/MADD/Rab3GEP was the first protein containing a DENN domain shown to have GEF activity (40). However, the GEF activity maps to a region outside the DENN domain (39).
Rab3 is present on synaptic vesicles, and Rab3GEP knock-out mice have decreased numbers of synaptic vesicles and severely impaired neurotransmitter release (66). This could result from the inability of Rab3 to become activated in nerve terminals of these mice, which may disrupt synaptic vesicle docking. As an alternative mechanism, DENN/MADD/Rab3GEP was recently found to bind via its DENN domain to GTP-bound Rab3, a typical characteristic of a Rab effector (67). Through the death domain of the protein, located in the C-terminal region (Fig. 1), DENN/MADD binds KIF1Bβ and KIF1A, anterograde-directed microtubule motors (67). Therefore, DENN/MADD/Rab3GEP may function as an adaptor for motor-dependent transport of Rab3-positive vesicles to the presynaptic nerve terminal. Consistently, knockdown of the protein leads to decreased Rab3 in distal axons, which is rescued with full-length protein but not with a mutant lacking the death domain (67). Thus, Rab3 acts as a switch to regulate trafficking of Rab3-positive vesicles to nerve terminals. It will be important to resolve whether DENN/MADD/Rab3GEP is predominantly a Rab3 effector, a Rab3 GEF, or both.
DENN/MADD also has GEF activity toward Rab27a/b (8), an exocytic Rab closely related to Rab3 (68–70). This activity has not yet been mapped to any particular region of the DENN/MADD protein. Consistent with this dual GEF activity, Rab3a and Rab27b display overlapping roles in synaptic vesicle exocytosis (71). Moreover, the C. elegans ortholog of DENN/MADD, AEX-3, has GEF activity toward C. elegans Rab3 and Rab27 (72). AEX-3 is important for proper localization of Rab3 in nerve terminals to regulate synaptic vesicle release (73). Together, these data indicate that DENN/MADD/Rab3GEP is an important regulator of GTPase function. In addition, there is extensive literature on the role of DENN/MADD and its splice variant IG20 in TNF receptor signaling. The death domain of DENN/MADD binds to the death domain of the TNF receptor, inhibiting TNF signaling (10, 74). Knockdown of DENN/MADD makes cells overly susceptible to TNFα-induced apoptosis (75). It will be necessary to determine whether the membrane trafficking and signaling activities of DENN/MADD/Rab3GEP represent related or entirely distinct activities.
DENN Domains and DENN Domain Proteins as Integrators of Rab Pathways
An emerging concept in the field of GTPases is that of GEF cascades, whereby an upstream GTPase or its effectors or activators recruit a GEF, which activates a downstream GTPase. This process can couple distinct Rab pathways and provide directionality in trafficking. An example of a GEF cascade occurs during the maturation of Rab5-positive early endosomes into Rab7-positive late endosomes (76). Recruitment of Rab5 to endosomes by its GEF, Rabex-5, leads to the recruitment of Rab5 effectors such as PI3K that generates PI(3)P, characteristic of early endosome membranes. Accumulating levels of PI(3)P lead to the recruitment of SAND-1 (Mon1 in vertebrates), which binds to both PI(3)P and Rabex-5 (77). SAND-1/Mon1 plays multiple roles in endosome maturation. First, it displaces Rabex-5 from early endosomes, disrupting the positive feedback loop of Rab5 activation (77). Second, it simultaneously recruits Rab7 to endosomes (77). Finally, Mon1 in a complex with Ccz1 functions as a GEF for Rab7 (78). Activated Rab7 recruits a range of effectors that lead to maturation of late endosomes (76). Another example of a GEF cascade occurs between Rab11 and Rab8, whereby GTP-bound Rab11 interacts with Rabin8, a Rab8 GEF (79). This may allow for the recruitment of Rabin8 to recycling endosome-derived carriers that are targeted for fusion at the plasma membrane, a process that requires activated Rab8 (79). There are also countercurrent GAP (GTPase-activating protein) cascades, whereby a downstream Rab recruits a GAP that inactivates the upstream Rab (80).
It is intriguing to speculate that DENN domain proteins and perhaps DENN domains themselves function in certain forms of Rab integration such as GEF cascades. For example, Rab6IP1 binds to GTP-bound Rab11 on recycling endosomes, likely through its uDENN domain, and through its RUN domain, it binds Rab6 on the Golgi (Fig. 3) (11, 50, 51). Rab6IP1 also functions as a GEF for Rab39 (8), which is found on vesicles in the vicinity of the Golgi (Fig. 3). Thus, Rab6IP1 may function in a GEF cascade that coordinates the transition between endosome-derived carriers (Rab11) and Golgi acceptor compartments (Rab6 and Rab39) (Fig. 3). Another example is DENN/MADD, which binds GTP-bound Rab3 via its uDENN module (67) and functions as a GEF for Rab27. This would integrate these two Rabs, which are known to have an overlapping role in synaptic vesicle exocytosis (71).
Structure of the DENN Domain
There is currently no experimental structural information on DENN domains, although secondary structure predictions indicate all β, mixed α/β, and all α folds for the uDENN, DENN, and dDENN modules, respectively (Fig. 2). The DENN module is the largest at ∼180 residues, whereas the uDENN and dDENN modules are ∼70 residues. In the 18 DENN domain proteins from humans, the spacing between uDENN and DENN ranges from 0 to ∼70 amino acids, whereas DENN and dDENN are between ∼30 and ∼130 amino acids apart (Fig. 2). However, there is even greater variability in DENN domains from other species. For example, in the S. pombe DENN domain protein, there are 184 and 240 residues between uDENN/DENN and DENN/dDENN, respectively. Thus, the DENN domain appears to be a complex tripartite domain formed from three non-dissociable modules, each with a distinct fold. This hypothesis is supported by the observation that most protein domains are between 50 and 150 amino acids in length (1). The DENN modules have remained associated throughout evolution due to functional or structural constraints or both. Such an organization is most reminiscent of the FERM domain, which is composed of three modules that have intermodule contacts (81). There is considerable divergence in sequence among different FERM domains, which translates into structural differences between the individual modules as well as their relative orientation toward one another (82). This may also be the case with the DENN domain. This type of “clustering” of independent modules/domains is also observed with the Dbl homology (DH) domain, an extensively studied GEF that is almost always found N-terminal of a pleckstrin homology (PH) domain.
Given that DENN domains have multiple functions, including interactions with PIs, binding to Rabs in the GTP-bound and nucleotide-free/GDP-bound forms, and enzymatic GEF activity, it is likely that these different activities can be assigned to different modules. For example, the dDENN module has some sequence similarity to DH domains, and hydrophobic positions that participate in DH domain folds are conserved (2). Structure prediction programs weakly predict that the dDENN module of connecdenn 1 possesses a structure most similar to the DH domain of the Ras GEF Sos (son of sevenless). Alternatively, the various activities may require the direct cooperation of the modules. For example, in the Cdc42 GEF Dbs, the PH domain interacts directly with the Cdc42 switch 2 region, which is one of the regions that undergo a major conformational change upon GTP hydrolysis, indicating that both the DH and PH domains are required for GEF activity (83).
The DENN domain of the connecdenns bind lipids (5). Lipid binding to GEFs can be part of a mechanism that sequesters the enzyme to the membrane, thereby increasing the local concentration, or it can be used as a mechanism to regulate GEF activity by eliciting conformational changes that enhance activity (84). This type of modulation is seen with some DH-PH domains, where PH-lipid interaction leads to allosteric regulation of nucleotide exchange (83).
Although emphasizing the comparison of the dDENN module with DH domains, DH domains are not known Rab GEFs. The best known Rab GEF is the Vps9 domain, a 100-amino acid catalytic core composed of a helical bundle (15). It is important to note that several other Rab GEF complexes have been solved, and structural information has revealed that the catalytic cores of these GEFs use different structural mechanisms to perform nucleotide exchange (85). The DENN domains may in fact use a completely novel mechanism of nucleotide exchange. This underscores the biological importance of initiating structure-function studies to resolve these issues.
Conclusion and Perspectives
It is now clear that DENN domain proteins regulate Rab GTPases and represent a new class of membrane trafficking proteins. Moreover, for at least a subset of DENN domains, the domain itself interacts directly with Rabs and functions as a GEF to activate this important class of small GTPases. We now need to clarify that, for the many DENN domain proteins shown to have GEF activity, the activity is indeed mediated by the DENN domain. Another important issue pertains to how the GEF activity is regulated. Are there intramolecular interactions that control the GEF activity such as what is seen in the regulation of the GEF activity of intersectin-l (86)? Other potential modes of regulation could involve binding of PI or other GTPases to the DENN domain itself or phosphorylation of the module. Given the evidence that certain DENN domains and DENN domain proteins function as a GEF for one Rab while binding to another, it seems likely that DENN domain proteins mediate GEF cascades. The study of DENN domain proteins is thus highly likely to reveal new links between Rab pathways and provide invaluable new information with regard to vesicle trafficking. Fewer than half of the DENN domain proteins have been examined in terms of their cell biological properties, and this will be a priority moving forward. Finally, we need to understand the structure of the DENN domain to determine its mechanism of action and modes of regulation. With such studies, our knowledge of this intriguing protein domain and its regulation of membrane trafficking pathways will increase greatly and provide better understanding of several important human diseases.
This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
A. L. Marat and P. S. McPherson, unpublished data.
- GEF
- guanine nucleotide exchange factor
- uDENN
- upstream DENN
- dDENN
- downstream DENN
- AH
- Avl9 homology
- SH3
- Src homology 3
- PI(3)P
- phosphatidylinositol 3-phosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- DH
- Dbl homology
- PH
- pleckstrin homology.
REFERENCES
- 1. Pawson T., Nash P. (2003) Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 2. Levivier E., Goud B., Souchet M., Calmels T. P., Mornon J. P., Callebaut I. (2001) Biochem. Biophys. Res. Commun. 287, 688–695 [DOI] [PubMed] [Google Scholar]
- 3. Stenmark H. (2009) Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 4. Zerial M., McBride H. (2001) Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 5. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., Ritter B. (2010) Mol. Cell 37, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marat A. L., McPherson P. S. (2010) J. Biol. Chem. 285, 10627–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato M., Sato K., Liou W., Pant S., Harada A., Grant B. D. (2008) EMBO J. 27, 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., Barr F. A. (2010) J. Cell Biol. 191, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chow V. T., Lee S. S. (1996) DNA Seq. 6, 263–273 [DOI] [PubMed] [Google Scholar]
- 10. Schievella A. R., Chen J. H., Graham J. R., Lin L. L. (1997) J. Biol. Chem. 272, 12069–12075 [DOI] [PubMed] [Google Scholar]
- 11. Janoueix-Lerosey I., Jollivet F., Camonis J., Marche P. N., Goud B. (1995) J. Biol. Chem. 270, 14801–14808 [DOI] [PubMed] [Google Scholar]
- 12. Harsay E., Schekman R. (2007) Mol. Biol. Cell 18, 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Girard M., Allaire P. D., McPherson P. S., Blondeau F. (2005) Mol. Cell. Proteomics 4, 1145–1154 [DOI] [PubMed] [Google Scholar]
- 14. Allaire P. D., Ritter B., Thomas S., Burman J. L., Denisov A. Y., Legendre-Guillemin V., Harper S. Q., Davidson B. L., Gehring K., McPherson P. S. (2006) J. Neurosci. 26, 13202–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delprato A., Merithew E., Lambright D. G. (2004) Cell 118, 607–617 [DOI] [PubMed] [Google Scholar]
- 16. Lippé R., Miaczynska M., Rybin V., Runge A., Zerial M. (2001) Mol. Biol. Cell 12, 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouranti I., Sachse M., Arouche N., Goud B., Echard A. (2006) Curr. Biol. 16, 1719–1725 [DOI] [PubMed] [Google Scholar]
- 18. Walseng E., Bakke O., Roche P. A. (2008) J. Biol. Chem. 283, 14717–14727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chevallier J., Koop C., Srivastava A., Petrie R. J., Lamarche-Vane N., Presley J. F. (2009) FEBS Lett. 583, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 20. Zhang J., Fonovic M., Suyama K., Bogyo M., Scott M. P. (2009) Science 325, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 21. Shim J., Lee S. M., Lee M. S., Yoon J., Kweon H. S., Kim Y. J. (2010) Mol. Cell. Biol. 30, 1421–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patino-Lopez G., Dong X., Ben-Aissa K., Bernot K. M., Itoh T., Fukuda M., Kruhlak M. J., Samelson L. E., Shaw S. (2008) J. Biol. Chem. 283, 18323–18330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao Y., Balut C. M., Bailey M. A., Patino-Lopez G., Shaw S., Devor D. C. (2010) J. Biol. Chem. 285, 17938–17953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu C., Morohashi Y., Yoshimura S., Manrique-Hoyos N., Jung S., Lauterbach M. A., Bakhti M., Grønborg M., Möbius W., Rhee J., Barr F. A., Simons M. (2010) J. Cell Biol. 189, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanno E., Ishibashi K., Kobayashi H., Matsui T., Ohbayashi N., Fukuda M. (2010) Traffic 11, 491–507 [DOI] [PubMed] [Google Scholar]
- 27. Sleiman P. M., Flory J., Imielinski M., Bradfield J. P., Annaiah K., Willis-Owen S. A., Wang K., Rafaels N. M., Michel S., Bonnelykke K., Zhang H., Kim C. E., Frackelton E. C., Glessner J. T., Hou C., Otieno F. G., Santa E., Thomas K., Smith R. M., Glaberson W. R., Garris M., Chiavacci R. M., Beaty T. H., Ruczinski I., Orange J. M., Allen J., Spergel J. M., Grundmeier R., Mathias R. A., Christie J. D., von Mutius E., Cookson W. O., Kabesch M., Moffatt M. F., Grunstein M. M., Barnes K. C., Devoto M., Magnusson M., Li H., Grant S. F., Bisgaard H., Hakonarson H. (2010) N. Engl. J. Med. 362, 36–44 [DOI] [PubMed] [Google Scholar]
- 28. Marat A. L., McPherson P. S. (2010) N. Engl. J. Med. 363, 988–989 [DOI] [PubMed] [Google Scholar]
- 29. Ferreira M. A., McRae A. F., Medland S. E., Nyholt D. R., Gordon S. D., Wright M. J., Henders A. K., Madden P. A., Visscher P. M., Wray N. R., Heath A. C., Montgomery G. W., Duffy D. L., Martin N. G. (2010) Eur. J. Hum. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moffatt M. F., Gut I. G., Demenais F., Strachan D. P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W. O. (2010) N. Engl. J. Med. 363, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., Anderson C. A., Bis J. C., Bumpstead S., Ellinghaus D., Festen E. M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C. G., Montgomery G. W., Prescott N. J., Raychaudhuri S., Rotter J. I., Schumm P., Sharma Y., Simms L. A., Taylor K. D., Whiteman D., Wijmenga C., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Cohen A., Colombel J. F., Cottone M., Stronati L., Denson T., De V. M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van Gossum A., Guthery S. L., Halfvarson J., Verspaget H. W., Hugot J. P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panés J., Phillips A., Proctor D. D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A. H., Stokkers P. C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S. R., Brant S. R., Rioux J. D., D'Amato M., Weersma R. K., Kugathasan S., Griffiths A. M., Mansfield J. C., Vermeire S., Duerr R. H., Silverberg M. S., Satsangi J., Schreiber S., Cho J. H., Annese V., Hakonarson H., Daly M. J., Parkes M. (2010) Nat. Genet. 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carroll K. S., Hanna J., Simon I., Krise J., Barbero P., Pfeffer S. R. (2001) Science 292, 1373–1376 [DOI] [PubMed] [Google Scholar]
- 33. Kloer D. P., Rojas R., Ivan V., Moriyama K., van Vlijmen T., Murthy N., Ghirlando R., van der Sluijs P., Hurley J. H., Bonifacino J. S. (2010) J. Biol. Chem. 285, 7794–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lichy J. H., Modi W. S., Seuanez H. N., Howley P. M. (1992) Cell Growth Differ. 3, 541–548 [PubMed] [Google Scholar]
- 35. Majidi M., Hubbs A. E., Lichy J. H. (1998) J. Biol. Chem. 273, 16608–16614 [DOI] [PubMed] [Google Scholar]
- 36. Hubbs A. E., Majidi M., Lichy J. H. (1999) Oncogene 18, 2519–2525 [DOI] [PubMed] [Google Scholar]
- 37. Majidi M., Gutkind J. S., Lichy J. H. (2000) J. Biol. Chem. 275, 6560–6565 [DOI] [PubMed] [Google Scholar]
- 38. Göhring I., Tagariello A., Endele S., Stolt C. C., Ghassibé M., Fisher M., Thiel C. T., Trautmann U., Vikkula M., Winterpacht A., FitzPatrick D. R., Rauch A. (2010) J. Med. Genet. 47, 91–98 [DOI] [PubMed] [Google Scholar]
- 39. Coppola T., Perret-Menoud V., Gattesco S., Magnin S., Pombo I., Blank U., Regazzi R. (2002) Biochem. J. 362, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wada M., Nakanishi H., Satoh A., Hirano H., Obaishi H., Matsuura Y., Takai Y. (1997) J. Biol. Chem. 272, 3875–3878 [DOI] [PubMed] [Google Scholar]
- 41. Iida H., Noda M., Kaneko T., Doiguchi M., Mōri T. (2005) Mol. Reprod. Dev. 71, 178–185 [DOI] [PubMed] [Google Scholar]
- 42. Olkkonen V. M., Dupree P., Killisch I., Lütcke A., Zerial M., Simons K. (1993) J. Cell Sci. 106, 1249–1261 [DOI] [PubMed] [Google Scholar]
- 43. Postel E. H., Weiss V. H., Beneken J., Kirtane A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6892–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Semova N., Kapanadze B., Corcoran M., Kutsenko A., Baranova A., Semov A. (2003) Genomics 82, 343–354 [DOI] [PubMed] [Google Scholar]
- 45. Denef N., Chen Y., Weeks S. D., Barcelo G., Schüpbach T. (2008) Dev. Cell 14, 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuck S., Gerl M. J., Ang A., Manninen A., Keller P., Mellman I., Simons K. (2007) Traffic 8, 47–60 [DOI] [PubMed] [Google Scholar]
- 47. Babbey C. M., Ahktar N., Wang E., Chen C. C., Grant B. D., Dunn K. W. (2006) Mol. Biol. Cell 17, 3156–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi A., Chen C. C., Banerjee R., Glodowski D., Audhya A., Rongo C., Grant B. D. (2010) Mol. Biol. Cell 21, 2930–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. (2007) Cell Metab. 5, 293–303 [DOI] [PubMed] [Google Scholar]
- 50. Miserey-Lenkei S., Waharte F., Boulet A., Cuif M. H., Tenza D., El Marjou A., Raposo G., Salamero J., Héliot L., Goud B., Monier S. (2007) Traffic 8, 1385–1403 [DOI] [PubMed] [Google Scholar]
- 51. Recacha R., Boulet A., Jollivet F., Monier S., Houdusse A., Goud B., Khan A. R. (2009) Structure 17, 21–30 [DOI] [PubMed] [Google Scholar]
- 52. Stankovic T., Byrd P. J., Cooper P. R., McConville C. M., Munroe D. J., Riley J. H., Watts G. D., Ambrose H., McGuire G., Smith A. D., Sutcliffe A., Mills T., Taylor A. M. (1997) Genomics 40, 267–276 [DOI] [PubMed] [Google Scholar]
- 53. Cheng H., Ma Y., Ni X., Jiang M., Guo L., Ying K., Xie Y., Mao Y. (2002) Cytogenet. Genome Res. 97, 72–75 [DOI] [PubMed] [Google Scholar]
- 54. Becker C. E., Creagh E. M., O'Neill L. A. (2009) J. Biol. Chem. 284, 34531–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Giannandrea M., Bianchi V., Mignogna M. L., Sirri A., Carrabino S., D'Elia E., Vecellio M., Russo S., Cogliati F., Larizza L., Ropers H. H., Tzschach A., Kalscheuer V., Oehl-Jaschkowitz B., Skinner C., Schwartz C. E., Gecz J., Van Esch H., Raynaud M., Chelly J., de Brouwer A. P., Toniolo D., D'Adamo P. (2010) Am. J. Hum. Genet. 86, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Blondeau F., Laporte J., Bodin S., Superti-Furga G., Payrastre B., Mandel J. L. (2000) Hum. Mol. Genet. 9, 2223–2229 [DOI] [PubMed] [Google Scholar]
- 57. Laporte J., Blondeau F., Buj-Bello A., Mandel J. L. (2001) Trends Genet. 17, 221–228 [DOI] [PubMed] [Google Scholar]
- 58. Robinson F. L., Dixon J. E. (2005) J. Biol. Chem. 280, 31699–31707 [DOI] [PubMed] [Google Scholar]
- 59. Berger P., Berger I., Schaffitzel C., Tersar K., Volkmer B., Suter U. (2006) Hum. Mol. Genet. 15, 569–579 [DOI] [PubMed] [Google Scholar]
- 60. Kim S. A., Vacratsis P. O., Firestein R., Cleary M. L., Dixon J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee S. H., Baek K., Dominguez R. (2008) FEBS Lett. 582, 4107–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nicot A. S., Laporte J. (2008) Traffic 9, 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lumb J. H., Field M. C. (2010) Mol. Biol. Cell 21, (suppl.) Abstr. 1264 [Google Scholar]
- 64. Bolino A., Muglia M., Conforti F. L., LeGuern E., Salih M. A., Georgiou D. M., Christodoulou K., Hausmanowa-Petrusewicz I., Mandich P., Schenone A., Gambardella A., Bono F., Quattrone A., Devoto M., Monaco A. P. (2000) Nat. Genet. 25, 17–19 [DOI] [PubMed] [Google Scholar]
- 65. Senderek J., Bergmann C., Weber S., Ketelsen U. P., Schorle H., Rudnik-Schöneborn S., Büttner R., Buchheim E., Zerres K. (2003) Hum. Mol. Genet. 12, 349–356 [DOI] [PubMed] [Google Scholar]
- 66. Tanaka M., Miyoshi J., Ishizaki H., Togawa A., Ohnishi K., Endo K., Matsubara K., Mizoguchi A., Nagano T., Sato M., Sasaki T., Takai Y. (2001) Mol. Biol. Cell 12, 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Niwa S., Tanaka Y., Hirokawa N. (2008) Nat. Cell Biol. 10, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 68. Pereira-Leal J. B., Seabra M. C. (2001) J. Mol. Biol. 313, 889–901 [DOI] [PubMed] [Google Scholar]
- 69. Figueiredo A. C., Wasmeier C., Tarafder A. K., Ramalho J. S., Baron R. A., Seabra M. C. (2008) J. Biol. Chem. 283, 23209–23216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fukuda M. (2005) J. Biochem. 137, 9–16 [DOI] [PubMed] [Google Scholar]
- 71. Pavlos N. J., Grønborg M., Riedel D., Chua J. J., Boyken J., Kloepper T. H., Urlaub H., Rizzoli S. O., Jahn R. (2010) J. Neurosci. 30, 13441–13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mahoney T. R., Liu Q., Itoh T., Luo S., Hadwiger G., Vincent R., Wang Z. W., Fukuda M., Nonet M. L. (2006) Mol. Biol. Cell 17, 2617–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iwasaki K., Staunton J., Saifee O., Nonet M., Thomas J. H. (1997) Neuron 18, 613–622 [DOI] [PubMed] [Google Scholar]
- 74. Del Villar K., Miller C. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kurada B. R., Li L. C., Mulherkar N., Subramanian M., Prasad K. V., Prabhakar B. S. (2009) J. Biol. Chem. 284, 13533–13541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 77. Poteryaev D., Datta S., Ackema K., Zerial M., Spang A. (2010) Cell 141, 497–508 [DOI] [PubMed] [Google Scholar]
- 78. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 79. Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rivera-Molina F. E., Novick P. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14408–14413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. (2000) EMBO J. 19, 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ceccarelli D. F., Song H. K., Poy F., Schaller M. D., Eck M. J. (2006) J. Biol. Chem. 281, 252–259 [DOI] [PubMed] [Google Scholar]
- 83. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Groves J. T., Kuriyan J. (2010) Nat. Struct. Mol. Biol. 17, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee M. T., Mishra A., Lambright D. G. (2009) Traffic 10, 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Guipponi M., Antonarakis S. E., Kay B. K., Stossel T. P., Lamarche-Vane N., McPherson P. S. (2001) Nat. Cell Biol. 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 87. Ritter B., Denisov A. Y., Philie J., Allaire P. D., Legendre-Guillemin V., Zylbergold P., Gehring K., McPherson P. S. (2007) EMBO J. 26, 4066–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bloethner S., Mould A., Stark M., Hayward N. K. (2008) Genes Chromosomes Cancer 47, 1076–1085 [DOI] [PubMed] [Google Scholar]
- 89. Azzedine H., Bolino A., Taïeb T., Birouk N., Di Duca M., Bouhouche A., Benamou S., Mrabet A., Hammadouche T., Chkili T., Gouider R., Ravazzolo R., Brice A., Laporte J., LeGuern E. (2003) Am. J. Hum. Genet. 72, 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Efimova E. V., Al-Zoubi A. M., Martinez O., Kaithamana S., Lu S., Arima T., Prabhakar B. S. (2004) Oncogene 23, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 91. Ramaswamy M., Efimova E. V., Martinez O., Mulherkar N. U., Singh S. P., Prabhakar B. S. (2004) Oncogene 23, 6083–6094 [DOI] [PubMed] [Google Scholar]
- 92. Zhang Y., Zhou L., Miller C. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2586–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Telliez J. B., Bean K. M., Lin L. L. (2000) Biochim. Biophys. Acta 1478, 280–288 [DOI] [PubMed] [Google Scholar]
- 94. Hu Y. F., Zhang H. L., Cai T., Harashima S., Notkins A. L. (2005) Diabetologia 48, 2576–2581 [DOI] [PubMed] [Google Scholar]
- 95. Subramanian M., Pilli T., Bhattacharya P., Pacini F., Nikiforov Y. E., Kanteti P. V., Prabhakar B. S. (2009) J. Clin. Endocrinol. Metab. 94, 1467–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li L. C., Sheng J. R., Mulherkar N., Prabhakar B. S., Meriggioli M. N. (2008) Cancer Res. 68, 7352–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]