Abstract
RNA editing, catalyzed by the multiprotein editosome complex, is an essential step for the expression of most mitochondrial genes in trypanosomatid pathogens. It has been shown previously that Trypanosoma brucei RNA editing ligase 1 (TbREL1), a core catalytic component of the editosome, is essential in the mammalian life stage of these parasitic pathogens. Because of the availability of its crystal structure and absence from human, the adenylylation domain of TbREL1 has recently become the focus of several studies for designing inhibitors that target its adenylylation pocket. Here, we have studied new and existing inhibitors of TbREL1 to better understand their mechanism of action. We found that these compounds are moderate to weak inhibitors of adenylylation of TbREL1 and in fact enhance adenylylation at higher concentrations of protein. Nevertheless, they can efficiently block deadenylylation of TbREL1 in the editosome and, consequently, result in inhibition of the ligation step of RNA editing. Further experiments directly showed that the studied compounds inhibit the interaction of the editosome with substrate RNA. This was supported by the observation that not only the ligation activity of TbREL1 but also the activities of other editosome proteins such as endoribonuclease, terminal RNA uridylyltransferase, and uridylate-specific exoribonuclease, all of which require the interaction of the editosome with the substrate RNA, are efficiently inhibited by these compounds. In addition, we found that these compounds can interfere with the integrity and/or assembly of the editosome complex, opening the exciting possibility of using them to study the mechanism of assembly of the editosome components.
Keywords: Enzyme Inhibitors, Protein Assembly, Ribozyme, RNA Editing, RNA-Protein Interaction
Introduction
Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major are three major trypanosomatid pathogens that cause hundreds of thousands of deaths and infect millions of people in tropical and subtropical areas of the world (1). Current trypanocidal drugs have a number of limitations such as high rate of toxicity, low rate of efficacy, and drug resistance (2, 3). Therefore, it is important to look for a drug that is effective and does not produce harmful side effects. RNA editing is a unique post-transcriptional modification of mitochondrial mRNAs that is shared in all trypanosomatid pathogens (4, 5). Modification of specific editing sites, dictated by complementary guide RNAs (gRNAs),5 constitutes essential steps to ensure the production of translatable mRNAs that encode essential components of the mitochondrial respiratory system. Although gRNAs specify the number of uridylates (Us) to be added or deleted by base pairing at each editing block (6, 7), a 1.6-MDa multiprotein complex, the editosome, is responsible for catalysis of different steps of RNA editing. Although the complete composition of the editosome is being elucidated, most purified functional editosomes contain over 20 proteins (8, 9). The editosomes differ in their compositions, having at least three different complexes that sediment at ∼20 S on glycerol gradients, and have the essential RNA editing activities (10). These editosomes, however, contain a similar essential catalytic core whose proteins are functionally characterized, including two endoribonucleases (KREN1 and KREN2) (11, 12), two 3′-terminal uridylyltransferases (KRET1 and KRET2) (13, 14), two 3′-exoribonucleases (U-exoribonucleases) that are called KREX1 and KREX2 (15, 16), two RNA ligases (KREL1 and KREL2) (17, 18), and six proteins with predicted oligonucleotide binding (OB) folds (KREPA1–6) (8, 19–25). The letter K in the beginning of each protein name refers to kinetoplastid. To specify species, we have used species-specific abbreviations (e.g. TbREL1 for REL1 in T. brucei).
Although other ligases such as T7 DNA ligase have a catalytic domain and an OB fold domain, KREL1 and KREL2 contain only the catalytic domain (19, 26). The OB fold domain that is essential for interaction with the substrate RNA is provided in trans by KREPA2 and KREPA1, which interact via their zinc fingers with KREL1 and KREL2, respectively, and bring the substrate RNA and the catalytic domain into proximity (15, 27, 28). Although RNA editing ligases differ in their structure compared with DNA ligases (15), their overall mechanism is similar. Ligation requires three distinct and reversible steps: (i) the ligase adenylylation step in which the conserved catalytic lysine of the ligase attacks the α-phosphate of ATP and displaces pyrophosphate, forming an enzyme-AMP intermediate through the phosphoamide linkage; (ii) the ligase deadenylylation step in which the guide RNA/nicked mRNA duplex binds to the protein and AMP is transferred from the adenylylated ligase to the 5′-phosphate of the RNA molecule, forming an adenylylated RNA with a 5′,5′-phosphoanhydride bond; and (iii) the ligation step in which the free 3′-hydroxyl at the nick site attacks the phosphoanhydride bond of the adenylylated RNA fragment, forming a phosphodiester bond, ligating the double-stranded RNA, and releasing AMP.
TbREL1 is an essential enzyme for the editing process and parasite viability (18). The crystal structure of the TbREL1 N-terminal catalytic domain has been determined, making virtual screening of chemical compounds against TbREL1 possible (29, 30). The unique characteristics of the TbREL1 ATP-binding pocket and the absence of any close homolog in the human genome make TbREL1 an ideal target for design of selective inhibitors that block the essential RNA ligase function.
A recent study has found several inhibitors against TbREL1 using a combination of in silico analysis and in vitro adenylylation assays (29). Some of these inhibitors have been validated as inhibitors of full-round deletion RNA editing in the presence of the purified editosome (31). While we were preparing this manuscript, a second study (30) was published that reported a number of naphthalene-based inhibitors of TbREL1. In the present study, we have studied the effect of one of the naphthalene-based inhibitors provided by the National Cancer Institute, chemical number 162535 (previously referred to as V2 (30) and designated in this study as C35), on the editosome, and we report that upon addition of this new inhibitor all the different essential catalytic steps of RNA editing are inhibited, most likely as a result of losing the interaction of the core editosome with substrate RNA.
EXPERIMENTAL PROCEDURES
Structure Preparation
The starting structure for the virtual screen was taken from the Protein Data Bank (code 1XDN). The bound ATP, water molecules, and ions were removed from the complex. Missing terminal residues and hydrogen atoms were added. Protonation states were assigned using the H++ server (32). Visual inspection of all assigned protonation states was done in Sybyl 8.0 (Tripos Inc., St. Louis, MO), and adjustments were made as needed.
Virtual Screening
Virtual screening was performed using our in-house docking program.6 The program uses an empirical scoring function trained to reproduce the binding modes of known protein-ligand complexes. The program takes conformers generated by Omega (OpenEye Scientific Software, Santa Fe, NM) and carries out an exhaustive rigid docking of the ligand on a grid around the binding site with a grid spacing of 0.6 Å. The window and root mean square settings of Omega were set to 20 kcal/mol and 0.4 Å, respectively. We screened a 77,000-molecule druglike subset of National Cancer Institute compounds contained in the ZINC database (33). The 2000 top scoring compounds were then clustered based on structural similarity using Sybyl 8.0 (Tripos Inc.). These compounds were also rescored using the solvated interaction energy (SIE) binding free energy function (34, 35). Representatives from each cluster having a good docking score and SIE score were ordered for testing.
Solvated Interaction Energy
Binding free energies were estimated using the SIE method (34, 35). The SIE is the AMBER interaction energy augmented with the desolvation cost of binding consisting of a reaction field energy and cavity cost. The reaction field energy was obtained by solving the Poisson equation using the BRI BEM program (36, 37) and using a variable probe molecular surface (38) to define the dielectric boundary.
Preparation of Mitochondrial Extract and Tandem Affinity Purification of Ligase Complex
The wild-type T. brucei cell line 1.7A was used to extract the mitochondrial contents (39). The mitochondrial contents extracted from 11 × 109 cells were centrifuged on a linear 10–30% (v/v) glycerol gradient and fractionated into 21 fractions of 500 μl each as described before (40). Tagged TbREL1 complexes were purified from 4 liters of T. brucei cells as described before (9) with the following modifications. After TEV protease cleavage, the TEV eluates were obtained, loaded onto 10–30% (v/v) glycerol gradients, and fractionated into 500-μl fractions as above. The Western blot analysis was performed using four monoclonal antibodies against KREPA1, KREPA2, KREL1, and KREPA3 as described previously (31).
Preparation of RNAs
All RNAs used in full-round deletion RNA editing were prepared as described before (31). The fluorescently labeled 16-mer reporter substrate, 5′-FAM (6-carboxyfluorescein)-GAUCUAUUGUCUCACA-TAMRA (6-carboxytetramethylrhodamine)-3′, was synthesized and HPLC-purified by Eurogentec. The cytochrome b (Cyb) pre-edited RNA and its guide (gCyb) were synthesized as described before (41). RNAs for ligase and insertion assays (5′CL18, 3′CL13pp, gA6PC-0A, and gA6PC-1A) were synthesized as described previously (42). The substrates for the precleaved deletion assay (U4-5′CL, U4-3′CL, and gA6[14]PC-del) were prepared as described before (43). Radiolabeling of RNAs at the 3′ terminus was performed by 5′-[32P]pCp ligation (29). For 5′ terminus-labeled RNAs, γ-32P was used. All RNAs were purified by gel electrophoresis on a 9 or 15% denaturing polyacrylamide gel containing 7 m urea.
Adenylylation and Deadenylylation Assays
Compounds were dissolved in DMSO, and reactions with an equivalent concentration of DMSO served as controls. Adenylylation assays were performed with 0.5 or 5 μl of glycerol gradient fraction 11, which was incubated for 10 min on ice in the presence or absence of each compound followed by incubation at 28 °C in the reaction buffer containing 12.5 mm HEPES (pH 7.9), 25 mm KCl, 5 mm magnesium acetate, 0.25 mm DTT, 40 nm [α-32P]ATP, and 0.1% Triton X-100. Adenylylation of the tag-purified editosome (TEV eluate) was done for 10 min using the same protocol. The proteins were resolved by 10% SDS-PAGE, and radiolabeled proteins were detected by phosphorimaging. Deadenylylation assays were performed using 5 μl of fraction 11 from the glycerol gradient, which was preincubated with compounds as mentioned above followed by incubation with 40 nm [α-32P]ATP at 28 °C for 15, 90, and 150 min in reaction buffer containing 1× HHE (25 mm HEPES (pH 7.9), 10 mm Mg(OAc)2, 50 mm KCl, and 1 mm EDTA), 5 mm CaCl2, 0.1% Triton X-100, 83 ng/ml yeast torula RNA, and ligatable RNA fragments (used in the precleaved ligase assay). Deadenylylation of TEV eluate was carried out for 15 min using the same protocol. The reaction was stopped by SDS dye, and samples were resolved by electrophoresis on a 10% SDS gel and visualized by phosphorimaging.
In Vitro RNA Editing Assays
All in vitro RNA editing assays were performed using 5 μl of glycerol gradient fraction 11, which was preincubated for 10 min on ice in the presence or absence of each compound. 0.1% Triton X-100 was included in each reaction. Equivalent concentrations of DMSO and Triton X-100 were included in the controls that did not contain any compound. The full-round hammerhead ribozyme-based assay was performed as described previously (31). Real time measurement of the ribozyme activity was recorded at intervals of 1 min for a period of 2 h using Rotor Gene 3000. The emission spectra of FAM and TAMRA were 535 and 582 nm, respectively, and the excitation wavelength was 470 nm for FAM. The rate of increase of the signal from FAM was used to measure the RNA editing activity.
The endonuclease assay was performed as described previously (41) using 3′-end-labeled [32P]pCp pre-edited Cyb and Cyb guide RNA. Precleaved insertion, deletion, and ligation assays were performed using 5′CL18, 3′CL13pp, and gA6PC RNAs as described previously (42, 43). An equal volume of 10 m urea dye was added to all samples, which were run on a 9 or 15% polyacrylamide gel according to their sizes and visualized by phosphorimaging.
Gel Shift Assay
The gA6[14] guide RNA and A6 pre-edited mRNA used in this assay were prepared by T7 polymerase (Promega) transcription of PCR-generated templates as described previously (7). Gel shift assays were performed as described previously (21) with the following exception. Purified KREPA4 at 400 nm concentration or 5 μl of editosome (glycerol gradient fraction 11) was incubated in the presence or absence of a 20 μm concentration of each compound on ice for 10 min. This was followed by incubation with 9 nm 3′-end-labeled gA6[14] gRNA in the presence or absence of 4.5 nm A6 pre-mRNA. Alternatively, we incubated the editosome with 9 nm labeled gA6[14] for 30 min, then added 20 μm drug, and incubated for another 10 min. For shift-Western blotting, the gel shift assay was duplicated: one for autoradiography and one for Western blotting using a 4% acrylamide gel to better resolve heavy protein complexes. We used 15 μl of glycerol gradient fraction 11 and four monoclonal antibodies against KREPA1, KREPA2, KREL1, and KREPA3 for blotting. For gel shifts with TEV eluate, C35-treated, or untreated TEV eluate were run on a 10–30% glycerol gradient, and 10 μl of protein from every odd-numbered fraction was incubated with 9 nm 3′-end-labeled gA6[14] gRNA in the reaction buffer.
Guanylyltransferase Labeling
To visualize endogenous RNA associated with the editosome, the purified editosome from the tandem TEV/glycerol gradient (see above) was used, and fractions 1–6 (pool a), 7–12 (pool b), and 13–17 (pool c) were pooled together. RNA was extracted from each pooled fraction using phenol/chloroform. The RNA obtained from each pooled fraction was treated with guanylyltransferase (Epicenter Biotechnologies) in the presence of 50 μCi of [α-32P]GTP in a 20-μl reaction according to the manufacturer's instructions. An equal volume of 10 m urea dye was added to all samples, and samples were run on a 15% polyacrylamide gel and visualized by phosphorimaging.
RESULTS
Virtual Screening
We conducted a virtual screening of 77,000 compounds from the National Cancer Institute library for potential inhibitors of TbREL1 adenylylation. After clustering of the virtual screening hits, 12 top ranking representatives from various clusters were selected for experimental testing. Table 1 lists the chosen compounds along with their score and rank in the library. Compounds that are referenced explicitly in the text are also identified with a shorter identification label. The top two ranking compounds in the editosome assay were C35 and C10 in that order (see below). The compound that we call C35 was also found by a different method in a recent virtual screening that was published as we were preparing this manuscript (referred to as V2 in Ref. 30), indicating the robustness of this compound against the methodology used for virtual screening.
TABLE 1.
Virtual hits selected for experimental validation
| NCIa no. | IDb | VS scorec | SIEd | Rank |
|---|---|---|---|---|
| 614641 | −35.1 | −9.6 | 12 | |
| 45210 | C11 | −34.7 | −8.7 | 18 |
| 344553 | −34.6 | −8.7 | 20 | |
| 162535 | C35 | −33.9 | −8.3 | 38 |
| 79710 | C10 | −33.9 | −8.8 | 41 |
| 641601 | −33.4 | −10.1 | 58 | |
| 89166 | C66 | −33.0 | −7.7 | 79 |
| 641753 | −32.8 | −8.2 | 92 | |
| 7809 | −32.8 | −8.1 | 96 | |
| 37204 | C04 | −32.7 | −8.0 | 102 |
| 674000 | −31.0 | −8.8 | 257 | |
| 623766 | −29.4 | −8.1 | 562 |
a National Cancer Institute.
b Short identification label.
c Values are in arbitrary units. These are the empirical scores from the docking step of the high throughput virtual screening (VS). More negative numbers suggest better affinity.
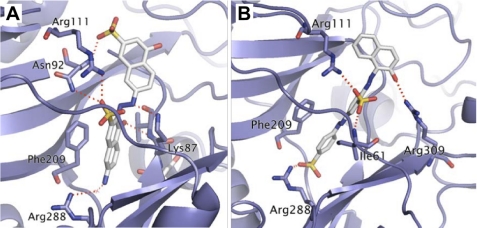
Fig. 1 shows the predicted binding modes of C35 and C10 in TbREL1. The interactions of C35 with the protein mimic certain aspects of ATP binding. Similar to the adenine moiety of ATP, one of the naphthalene rings forms π-π stacking interactions with Phe209. Also, similar to the polyphosphate tail of ATP, a sulfonate group forms an ion pair with the guanidine group of Arg111. In addition, the aminonaphthyl group forms a water-mediated interaction with Arg288 as is seen with the adenine N1 atom in ATP. However, our predicted binding mode is rotated almost 180° compared with that described by Durrant et al. (30) in which the aminonaphthyl group is exposed rather than buried. Durrant et al. (30) docked their compounds on an ensemble of about 30 protein conformations obtained from molecular dynamics simulations. The favored binding mode that they found for C35 required a protein conformation in which the Glu60 side chain is somewhat separated from Arg111 with the aminonaphthyl packing between Glu60 and Arg111. That pose is incompatible with the crystal structure of the protein in which there is little room between Glu60 and Arg111. In our case, all our docking calculations were performed on a single protein conformation based on the crystal structure. Even for this single protein conformation, our exhaustive docker was able to find a favorable pose with good interactions with the protein. Hence, we present a binding mode that does not require the extra step of invoking a conformational change in the protein. However, it is not possible without further experimental work to decide which is the correct binding mode for C35.
FIGURE 1.
Predicted binding modes of TbREL1 inhibitors. The inhibitors C35 (A) and C10 (B) in TbREL1 are represented in stick form with hydrogen atoms not shown for clarity. The protein is represented in ribbon form with key interacting residues represented as sticks. Red dotted lines represent hydrogen bonding interactions.
Inhibition of RNA Editing by Selected Compounds
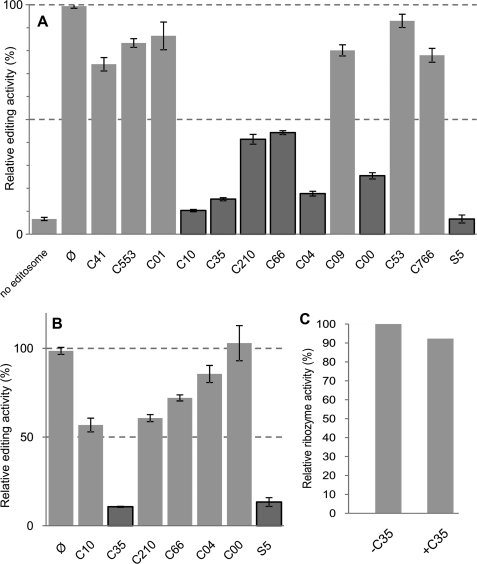
Adenylylation is the first essential step of RNA editing ligation. Therefore, inhibition of this step results in inhibition of ligase activity and consequently inhibition of full-round RNA editing. Using our recently developed fluorescence-based hammerhead ribozyme assay (31), we tested our initial 12 compounds for their ability to inhibit the full-round RNA editing along with S5, a previously reported inhibitor of TbREL1 adenylylation and full-round RNA editing (29, 31), as the positive control. In the reaction mixture, a 20 μm concentration of each compound was incubated with 5 μl of the editosome purified from whole cell mitochondrial extract by glycerol gradient fractionation. The complete RNA editing reaction mixture, which was used for all the experiments throughout this study, contained pre-edited mRNA, gRNA, and fraction 11, which is the most active fraction of the glycerol gradient. We found that three compounds, C10, C35, and C04, led to an ∼7 times decrease in RNA editing activity (Fig. 2A). To test whether the inhibition observed was due to promiscuous aggregation, we repeated the assay by including Triton X-100 (0.1%, w/v) for the compounds that had resulted in a 50% or greater reduction in full-round RNA editing activity. Triton X-100 prevents compounds from aggregation or nonspecific inhibition as described previously (29). In the presence of this detergent, five of the six examined compounds did not retain their inhibitory effect and therefore were considered as promiscuous aggregators (Fig. 2B). However, we found that C35 has an even more pronounced inhibitory effect on full-round RNA editing compared with the previously reported S5. To exclude the possibility of an inhibitory effect of C35 on the function of the reporter ribozyme system, the interaction of this compound with the active ribozyme in the presence of gRNA and fluorescent substrate was examined (Fig. 2C). The ribozyme activity was not significantly altered by C35 at a concentration of 20 μm. Therefore, the inhibition observed in full-round RNA editing assay was not due to nonspecific binding of C35 to the reporter ribozyme in the assay. Our results prompted us to test whether this compound can be used to selectively block the ligase activity in vitro.
FIGURE 2.
Effect of selected compounds that inhibit editosome activity. A, results of the first round of screening using a FRET-based RNA editing assay are shown here. The reactions were performed in the absence (Ø) or presence of various compounds in a 30-μl reaction volume under multiple turnover conditions. Compounds resulting in more than 50% inhibition are indicated by dark gray bars. B, effect of Triton X-100 on the compounds prone to aggregation is shown. Triton X-100 (0.1%, w/v) was used to monitor any nonspecific inhibition due to aggregation. The compounds C35 and S5, which still showed more than 50% inhibition, are indicated by dark gray bars. The error bars represent the experimental variation (S.D.) from three independent repetitions. C, the effect of the best inhibitor compound, C35, on ribozyme activity was evaluated. Cleavage activity of active ribozyme was measured in the absence of the compound (−C35) and presence of the compound (+C35). In all graphs, the y axis represents relative percentages of the cleavage activity for edited ribozyme in each experiment.
Inhibition of Ligase Adenylylation at Low Protein Concentrations by C35 and S5
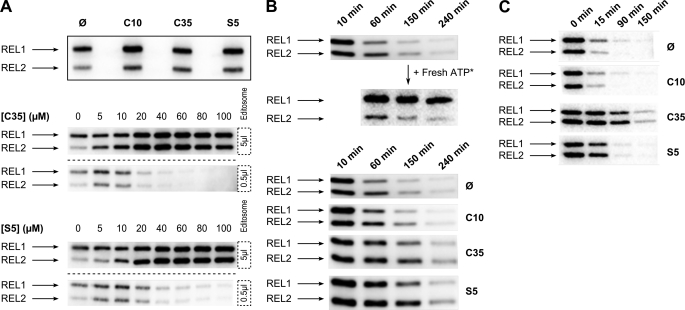
To determine the specificity of inhibition, we analyzed adenylylation activities of editosome ligases in the absence and presence of 20 μm C35 and S5 as well as the less inhibitory C10 as a negative control. Adenylylation of ligase is an RNA-independent activity in which the enzyme reacts with ATP, becomes covalently linked to the adenylyl moiety via a lysine, and releases a pyrophosphate. Upon incubation of the editosome in the presence of Mg2+ and [α-32P]ATP, the RNA editing ligases TbREL1 and TbREL2 are adenylylated. Unexpectedly, however, when we incubated 5 μl of the editosome with [α-32P]ATP in the presence of 20 μm C35, S5, or C10, we did not observe inhibition of adenylylation of either TbREL1 or TbREL2 (Fig. 3A, upper panel). It should be noted that the amount of editosome and the concentration of the compounds in this experiment were the same as in the full-round RNA editing assay in which we saw the inhibition of editing by C35 and S5 (see the previous section). Next, we tested various concentrations of the compounds to determine whether adenylylation inhibition could occur at higher compound concentrations. Surprisingly, not only did we not observe inhibition of adenylylation in the presence of C35 and S5, but we also saw an increase in the adenylylation levels of both TbREL1 and TbREL2 with increasing concentrations of these compounds (Fig. 3A, middle and lower panels, 5 μl of editosome). This is not in agreement with the previous reports (29, 30) in which both S5 and C35 were suggested to compete with ATP for binding to the recombinant TbREL1 adenylylation pocket. We noticed that the estimated concentration of TbREL1 in our reaction mixture (with 5 μl of editosome) was about 10 times higher than the concentration of recombinant TbREL1 used in the previous studies (29, 30). To test whether the difference in concentration of TbREL1 could explain our observations, the editosome was diluted 10 times and subjected to various compound concentrations (Fig. 3A, middle and lower panels, 0.5 μl of editosome). Inhibition in adenylylation was observed by both C35 and S5 as their concentrations were increased, suggesting that the inhibitory compounds can block adenylylation activity but only at lower concentrations of the editosome. It has been reported that the adenylylation activities of TbREL1 and TbREL2 are significantly increased in the presence of their interacting partners, KREPA2 and KREPA1, respectively (15, 28). 10-Fold dilution of the editosome might result in depletion and dissociation of interacting partners and consequently lower the adenylylation efficiency, thus leading to effective inhibition of the basal levels of adenylylation activities by the compounds. The effect of these compounds on TbREL2 is not surprising as TbREL1 and TbREL2 are highly similar in sequence and structure (19, 44), and it has been reported that S5 can efficiently inhibit the function of the more distantly related T4 RNA ligase (29). Nevertheless, the inhibition observed in full-round RNA editing cannot be explained by inhibition of adenylylation activity alone because in contrast to adenylylation activity full-round RNA editing can be inhibited at high concentrations of the editosome in the presence of C35 and S5.
FIGURE 3.
Effect of inhibitory compounds on adenylylation and deadenylylation steps of RNA editing ligases. A, adenylylation and the effect of inhibitory compounds are shown. The upper panel is an autoradiograph of 5 μl of fraction 11 from the glycerol gradient labeled with [α-32P]ATP in the absence (Ø) and presence of 20 μm C10, C35, and S5. In both the middle and lower panels, either 5 μl of editosome protein (above the dashed line) or 0.5 μl of editosome protein (below the dashed line) was subjected to various concentrations of C35 and S5 as indicated. The concentrations of compounds are indicated above each gel (0–100 μm). B, time course experiments to monitor changes in the state of adenylylation are shown. The upper panel shows the adenylylation of 5 μl of editosome in the absence of any compounds at different time points (10, 60, 150, and 240 min). The middle panel indicates that the ligases maintained their adenylylation activity during the 3-h time course as they were able to bind the freshly added radiolabeled ATP (Fresh ATP*). The lower panel shows the effect of C10, C35, and S5 on spontaneous deadenylylation of ligases as indicated. C, time course experiments to monitor changes in the state of deadenylylation at different time points are shown. 5 μl of fraction 11 was labeled with [α-32P]ATP for 10min. Then, ligatable RNA substrate, 5′CL18, CL13pp, and gA6PC-0A were added along with 20 μm C10, C35, or S5 and incubated for 0, 15, 90, or 150 min. A control lacking any chemicals was also included. The results indicate that deadenylylation is blocked as soon as 15 min after addition of C35 or S5 in the presence of ligatable substrate. The adenylylated TbREL1 and TbREL2 bands are indicated by arrows.
Inhibition of Deadenylylation by C35 and S5
To better understand the mechanism responsible for the selective action of C35 and S5 on the editosome function, we monitored editosome adenylylation for the same duration of time required for the full-round RNA editing reaction (3 h) in the absence or presence of a 20 μm concentration of the compounds. Interestingly, the results showed that in the absence of the inhibitory C35 and S5 compounds adenylylated TbREL1 and TbREL2 gradually become deadenylylated during the course of 3 h (Fig. 3B, upper panel). We hypothesized that the reduction in adenylylated enzymes might have occurred as a result of deadenylylation of the enzymes by endogenous RNA substrates that are present in the editosome fraction and/or as a result of instability and inactivation of the ligases after the 3-h incubation at 28 °C. To test these two possibilities, we replenished the reactions with additional radiolabeled ATP at different time points and observed that the adenylylation of the ligases was restored by binding to the freshly added ATP during and at the end of the 3-h incubation period (Fig. 3B, upper panel, + Fresh ATP*). These data suggest that the loss of adenylylation signal during the incubation is most likely due to deadenylylation of ligases by the endogenous RNAs, highlighting a dynamic interaction of the editosomes with the endogenous RNA substrates. Interestingly, in the presence of C35 and S5, the level of deadenylylation was lower, suggesting that the compounds directly or indirectly inhibit ligase deadenylylation (Fig. 3B, lower panel).
To directly test for deadenylylation inhibition by C35 and S5, we monitored the deadenylylation activity in the presence of exogenously added ligatable double-stranded RNAs. In the presence of ligatable double-stranded RNAs, the AMP is transferred from the adenylylated enzyme (E-AMP) to the 5′-phosphate group of the 3′-RNA fragment, thereby adenylylating the RNA substrate and deadenylylating the enzyme. In the presence of ligatable RNA substrates and the absence of inhibitors, the deadenylylation occurred quickly in the first 15 min as evident by the reduction in the adenylylated enzymes. In the presence of S5 and particularly C35, the deadenylylation rate was significantly reduced (Fig. 3C). Inhibition of the ligase deadenylylation step supports the observation that adenylylated RNA editing ligases are accumulated in the presence of C35 and S5 compounds (see the previous section).
Inhibition of Different Steps of RNA Editing by C35 and S5
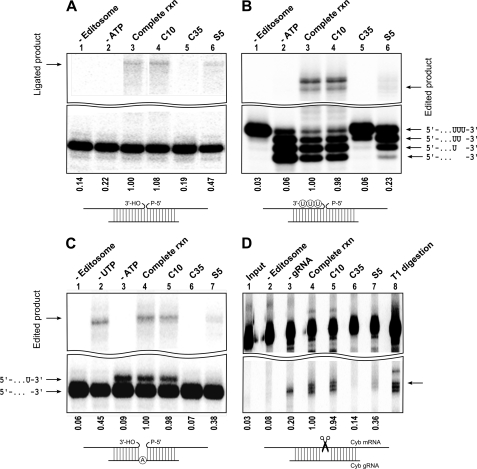
We further characterized the effect of the compounds on different steps of RNA editing by carrying out in vitro precleaved editing assays as described previously (42, 43, 45) in the absence and presence of the compounds. First, we studied precleaved ligation by performing the editing reaction in the absence or presence of inhibitory compounds. As expected, we detected a significant inhibition of the ligation when the reaction contained C35 (5 times inhibition) and to a lesser extent S5 (2 times inhibition) compared with the reaction containing C10 or controls (Fig. 4A). The difference in the efficiency of inhibition by C35 and S5 is consistent with the observation that C35 is also a more potent inhibitor of the deadenylylation step (Fig. 3C).
FIGURE 4.
Effect of inhibitory compounds on different steps of RNA editing. A, precleaved ligation reactions were performed using 5′-labeled 5′CL18, 3′CL13pp, and gPCA6-0A gRNA in the absence of any compounds (lane 3) or presence of 20 μm C10, C35, or S5 (lanes 4–6, respectively). Lane 1 does not contain the editosome, and lane 2 does not have ATP as indicated above the gel. The ligated product is indicated by an arrow. B, precleaved deletion editing reactions were performed with 5′-labeled U4-5′CL, U4-3′CL, and gA6[14]PC-del, which specifies the deletion of three Us in the presence of the editosome (lane 3). Lane 1 does not contain the editosome, and lane 2 does not contain ATP. Lanes 4–6 represent the complete reaction in the presence of 20 μm C10, C35, or S5, respectively. Input RNA and added Us are represented schematically. The edited product is also shown. The top band represents unedited ligated product, which represents the ligation of the 5′- and 3′-fragments without deletion of Us. C, precleaved insertion editing reactions were performed using 5′-labeled 5′CL18, 3′CL13pp, and gPCA6-1A gRNA that specifies the insertion of one U. Lanes 1–3 represent the reactions with no editosome, no UTP, and no ATP, respectively. Lane 4 contains all the components for insertion RNA editing (complete reaction), and lanes 5–7 contain 20 μm C10, C35, or S5, respectively. The edited product is indicated by an arrow. The pre-edited 5′-fragment and U-inserted 5′-fragment are represented schematically. D, the endonuclease assay was performed by gRNA-dependent cleavage of 3′-labeled Cyb mRNA at editing site 1 in the absence (lane 4) and presence of 20 μm C10, C35, or S5 (lanes 5–7), respectively. Lane 1 contains only the input RNA, lane 2 contains no editosome, and lane 3 contains no gRNA. Lane 8 represents the T1 digestion of input RNA. Numbers below the panels indicate percentage of edited/ligated product in the presence of compounds normalized to the edited/ligated product in the absence of any compound (complete reaction (rxn)).
To examine the effect of compounds on U removal as well as RNA ligation steps of deletion editing, we used a precleaved in vitro deletion assay that directs the deletion of three Us from the substrate RNA (Fig. 4B). Incubation of the substrate RNAs, gRNA, and the editosome in the absence of ATP resulted in removal of all three Us as expected. Addition of ATP resulted in ligation of the 3′-fragment with the 5′-fragment from which all three Us had been removed. In the presence of C35, a significant inhibition of both U removal (U-exoribonuclease activity) and subsequent ligation activity was observed (Fig. 4B). The addition of compound S5 to the editing reaction also affected U-exoribonuclease and ligase activities but to a lesser extent, whereas the no-compound control and the C10 control showed no inhibition on U removal or RNA ligation.
We also used the precleaved RNA editing insertion assay to test the effect of C35 and S5 on the terminal uridylyltransferase activity of the editosome. Incubation of the 5′- and 3′-fragments with gRNA (designed to direct insertion of one U into the substrate RNA) in the presence of UTP, ATP, and the editosome resulted in an RNA product with a single added U and the ligated edited RNA (Fig. 4C). As expected, in the absence of UTP or the editosome, neither the U addition product nor the edited RNA was detected, and in the absence of UTP, only the 5′- and 3′-input RNA fragments were ligated. Also, omission of ATP resulted in a major product with a single U added, which is an expected intermediate before the ATP-requiring ligation step. Interestingly, again both C35 and, to a lesser extent, S5 were able to inhibit terminal uridylyltransferase activity (Fig. 4C).
We next tested the effect of compounds on the endonuclease activity of the editosome, which is the required initial step for cleaving pre-edited mRNA, directed by complementary gRNA at the editing site. We used the Cyb pre-mRNA and its gRNA to monitor endonuclease activity. As expected, in the presence of the editosome, we detected both the gRNA-independent and gRNA-dependent cleavage activities (Fig. 4D). In the presence of C35 and, to a lesser extent, S5, the endonuclease activity of the editosome was completely lost (Fig. 4D).
Inhibitory Compounds Affect Editosome RNA Binding Activity
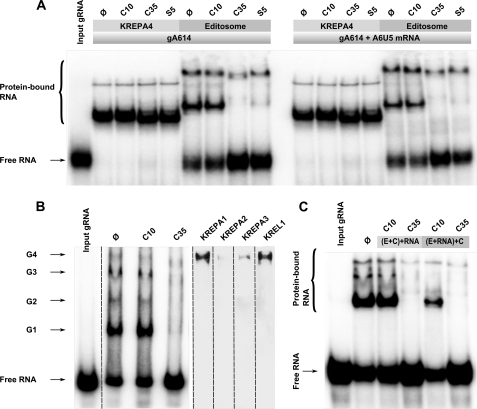
The RNA editing model implies the requirement for substrate RNA and gRNA binding by various components of the editosome for positioning of the catalytic core (20, 21, 46, 47). We noticed that one of the simplest explanations for the inhibitory effect of C35 and S5 on all editing-related activities is that the editosome cannot interact with its substrate RNA in the presence of these compounds. We used a native gel mobility shift assay, which allows for detection of ribonucleoprotein complexes (20), to test this hypothesis. The 32P-labeled gA6[14] gRNA, which specifies the first editing site of the ATPase subunit 6 (A6) pre-mRNA, was incubated with the editosome or recombinant KREPA4 in the absence or presence of pre-mRNA either with or without the inhibitory compounds. KREPA4 is a known gRNA-binding protein and a core component of the editosome. The assembled complexes were analyzed by native gel electrophoresis. As expected, the untreated controls showed the characteristic pattern of RNA-protein interaction for both recombinant KREPA4 and the editosome (Fig. 5A). We saw that although the KREPA4-RNA interaction was not affected by C35 and S5 these compounds completely inhibited the formation of a major gRNA-protein complex that was seen in the editosome preparation (Fig. 5A). Also, a significant reduction in the amount of a heavier gRNA-protein complex was evident. Further separation on a 4% gel showed four distinct RNA-protein complexes, as also reported before (46, 48), of which the lightest complex, previously designated as G1, showed the most dramatic inhibition by C35. The other three (G2–G4) also showed significant inhibitions (Fig. 5B). To further investigate the composition of each of these gRNA-containing ribonucleoprotein complexes, we used shift-Western blotting using antibodies against KREPA1, KREPA2, KREPA3, and KREL1. Interestingly, we observed that only the heaviest complex (G4), which was least affected by C35, contained these four proteins. These results suggest that RNA-protein interaction is one of the earliest steps affected by C35; however, the major target of C35 is none of the four proteins that we analyzed by shift-Western.
FIGURE 5.
Effect of inhibitory compounds on RNA binding activity of editosome complex. A, 32P-labeled gA6[14] gRNA alone (left panel) or 32P-labeled gA6[14] gRNA in the presence of A6 pre-mRNA (right panel) was incubated with the recombinant KREPA4 protein or the editosome (5 μl of glycerol gradient fraction 11) in the absence (Ø) or presence of C10, C35, and S5 as indicated. The protein-bound RNA and unbound input RNAs are indicated. The reactions were resolved on 10% denaturing polyacrylamide gels. The positions of free RNA and protein-bound RNA are indicated. B, treated and untreated editosome proteins (similar to A, left) were resolved on a 4% polyacrylamide gel for better separation of high molecular weight ribonucleoprotein complexes. Complexes G1–G4 are shown by the arrowheads (left). Also, the presence of KREPA1, KREPA2, KREPA3, and KREL1 in the untreated sample was investigated by shift-Western (right). It can be seen that these proteins primarily exist in the G4 complex. C, the editosome preparation was treated first with C35/C10, and then RNA was added ((E+C)+RNA), or the RNA was added first, and then the RNA-protein complex was subjected to C35/C10 treatment ((E+RNA)+C). It can be seen that in both cases C35 inhibited RNA-protein interaction, whereas C10 had no effect.
To understand the dynamics of inhibition of RNA-protein interaction by C35, we used alternative orders of addition of RNA and C35 to the editosome. We observed that C35 can inhibit RNA-protein interaction even after the RNA-protein complex is formed, suggesting that it can disengage the RNA that is already bound to the protein (Fig. 5C).
It has been shown previously that treatment of the editosome complex with nucleases results in disassembly of the complex, suggesting a role for RNA to maintain 20 S editosome complex integrity (41). Furthermore, our data on deadenylylation of TbREL1 and TbREL2 in the absence of exogenous substrate RNA indicate that the purified editosome complex already contains non-negligible amounts of endogenous RNA substrate (Fig. 3B, upper panel), which may be responsible for the integrity of the purified editosome. Effective inhibition of editosome-substrate interaction by C35 prompted us to examine whether this drug can promote dissociation of editosome components as discussed in the next section.
20 S Editosome Complex Integrity Is Affected by C35 Treatment
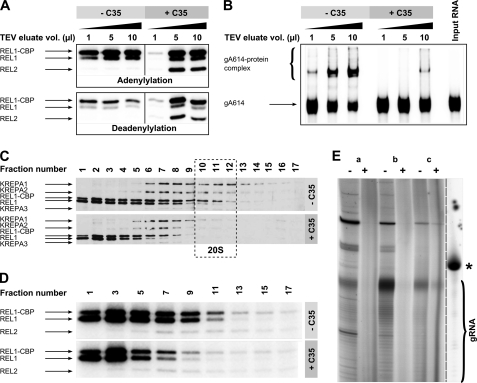
To examine the editosome complex integrity in the presence and absence of the more potent inhibitory compound C35, we prepared editosome by affinity purification of tagged TbREL1, and after elution of the purified editosome complex using TEV protease, we incubated different amounts of the TEV eluate with 20 μm C35. Consistent with our previous experiments on the glycerol gradient-purified editosome, adenylylation of TbREL1 and TbREL2 could be inhibited by C35 only when low amounts of affinity-purified editosome were present; at higher editosome concentrations, although the adenylylation was not inhibited, the drug could effectively reduce the rate of deadenylylation at 15 min (Fig. 6A). Furthermore, again we saw a very significant inhibition of gRNA-protein interaction in the presence of C35 (Fig. 6B). These data prompted us to test the effect of C35 treatment on editosome complex integrity. We treated the TEV eluate with C35, and then we fractionated the TEV eluate on a 10–30% glycerol gradient. As shown in Fig. 6C, a nearly complete loss of the 20 S editosome complex can be seen upon treatment with C35, and the profiles of proteins are shifted toward low density fractions. The adenylylation assay on glycerol gradient fractions of C35-treated sample confirmed that TbREL1 and TbREL2 are mostly present in complexes that are smaller than 20 S, indicating disintegration of the 20 S editosome after treatment (Fig. 6D). To investigate the effect of C35 on endogenous RNA that accompanies the editosome, we extracted RNA from the tandem TEV/glycerol gradient-purified editosome that was or was not treated with C35. As shown in Fig. 6E, although an abundance of gRNA molecules is present in complex with the editosome, especially in fractions 7–12, treatment with C35 almost completely abolishes these RNA molecules. These data suggest that upon C35 treatment the endogenous protein-bound RNA disengages from the protein complex and thus becomes susceptible to degradation.
FIGURE 6.
Analysis of sedimentation profile and activity of ligase-associated complexes in the presence of C35. A, adenylylation and deadenylylation of REL1-TAP TEV eluate were studied in the absence and presence of C35. The upper gel represents adenylylation of various amounts of REL1-TAP TEV eluate (1, 5, and 10 μl) with [α-32P]ATP in the absence or presence of 20 μm C35. The lower gel represents the deadenylylation of the same volumes of REL1-TAP TEV eluate as indicated above with [α-32P]ATP and ligatable RNA substrate, 5′CL18, CL13pp, and gA6PC-0A in the absence or presence of 20 μm C35. B, a gel shift assay was performed with 32P-labeled gA6[14] gRNA using different amounts of REL1-TAP TEV eluate (1, 5, and 10 μl) in the absence or presence of 20 μm C35. The rightmost lane represents unbound gA6[14] in the absence of any protein. The protein-bound RNA and unbound input RNAs are indicated by arrows. C, REL1-TAP TEV eluates in the absence of C35 (upper gel) or presence of 20 μm C35 (lower gel) were fractionated on 10–30% glycerol gradients. Fractions were collected from the top of the gradients, resolved by SDS-PAGE, blotted, and probed with mAbs against MP81, MP63, REL1, and MP42. D, 10 μl of odd-numbered glycerol gradient fractions from both untreated (upper gel) and treated (lower gel) REL1-TAP TEV eluate was adenylylated with [α-32P]ATP and run on an SDS gel. E, a guanylyltransferase assay showed that C35 treatment effectively abolishes most RNA species in the tandem TEV/glycerol gradient-purified editosome fractions. Letters a, b, and c correspond to pooled fractions 1–6, 7–12, and 13–17, respectively. − and + signs indicate non-treated control and C35-treated sample, respectively. The rightmost lane shows in vitro transcribed [α-32P]GTP-labeled gA6[14], which corresponds to 70 nucleotides (indicated by the asterisk). The ladder-like pattern below the major gRNA band in the non-treated samples corresponds to gRNA molecules with different poly(U) tails. CBP, calmodulin-binding peptide.
DISCUSSION
In the course of this work, we have examined the mechanism of action of two naphthalene-based inhibitors of the RNA editing process, C35 and S5, and have shown that these compounds affect virtually all editosome activities that require editosome-RNA interaction, suggesting that they block the interaction of the editosome with its substrate RNA. This hypothesis is further supported by direct experiments showing that the editosome cannot interact with substrate RNA in the presence of C35 and S5. Therefore, although the exact molecular mechanism of action of naphthalene-based compounds remains to be determined, our data present C35 and S5 as the first known druglike compounds that can interfere with editosome-RNA interaction. In addition, we examined the effect of the more potent compound C35 on complex integrity and showed that this compound could interfere with the integrity and/or assembly of the editosome complex, shifting the 20 S editosome sedimentation to the lower 5–10 S region. This resembles the results obtained from treatment of core editosome proteins by RNases (47), suggesting that the editosome requires its substrate RNA for initiating assembly and/or maintaining its integrity.
It has been reported that C35 and S5 inhibit the adenylylation step of ligation with the inhibitory compounds competing with ATP for binding to the adenylylation pocket (29, 30). However, in our partially purified editosome fraction, as estimated, the minimum concentration of TbREL1 that is able to perform full-round editing is about 50 nm, which is about 10 times higher than the concentration of recombinant TbREL1 used in previous reports. With this amount of editosome, we could not observe any inhibition at the adenylylation step. However, we were able to see inhibition of adenylylation after we diluted our editosome preparation 10 times, indicating that adenylylation inhibition only occurs at very low protein concentrations. On the other hand, we observed efficient inhibition of editing using the non-diluted amount of editosome that is required for successful completion of full-round in vitro RNA editing, suggesting that adenylylation inhibition is most likely not the mechanism through which C35 and S5 inhibit the editing process at higher protein concentrations.
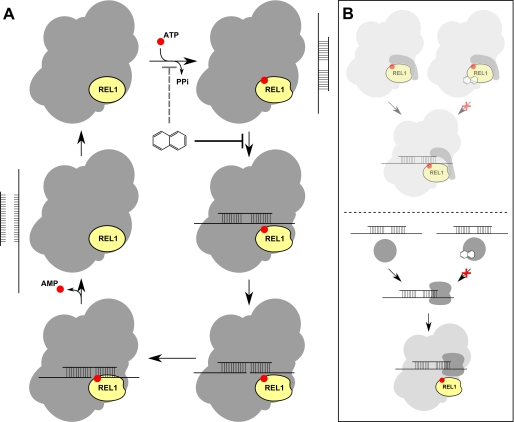
Our results suggest that the main mechanism of action of C35 is through inhibition of a protein-RNA interaction that is integral to the process of editing (Fig. 7A). Alternative molecular mechanisms can lead to such observation. For example, freezing the OB fold-containing interacting partner of ligases in a closed conformational state can lead to their inability to interact with the RNA substrate. During the deadenylylation step in capping enzymes and DNA ligases, it has been shown that the OB fold rotates and positions itself at a distance from the AMP-binding pocket that allows the ligase to have an open conformation to bind to the nicked double-stranded DNA (49). Thus, one possible model is that in the presence of compound when the OB fold provides the ATP for the adenylylation pocket the OB fold gets locked and induces the closed ligase-interacting partner conformation, which does not allow the interaction of protein with RNA (Fig. 7B, top panel). In other words, due to the compound-induced locked position of the OB fold on the adenylylated catalytic pocket, the OB fold cannot rotate and is not capable of positioning its nucleic acid binding surface toward the active site awaiting nick binding. Although this model corresponds to in silico modeling studies that suggest the interaction of C35 and S5 with TbREL1 (this work as well as Refs. 29 and 30), it is not fully supported by our experimental data especially in that the main target of C35 does not seem to be in association with TbREL1 (Fig. 5B). Alternatively, C35 may inhibit a yet unidentified protein(s) that is responsible for bringing the substrate RNA into the editosome complex (Fig. 7B, bottom panel). This may result from similarity of physiochemical properties of RNA-binding regions of proteins to nucleoside triphosphate-binding clefts; C35 was designed to be positioned in the adenylylation pocket of RNA editing ligases, which can structurally be similar to other nucleotide- and nucleic acid-binding pockets. Furthermore, we have shown that in the presence of C35 there is a shift in the 20 S editosome toward 5–10 S. This result is very similar to the reports showing that upon treatment of the editosome complex with nucleases the editosome is disintegrated (47), suggesting that the loss of integrity after treatment of the editosome with C35 is because the editosome loses its RNA-interacting capacity. Treatment with the compounds does not result in loss of RNA binding activity of KREPA4, suggesting that these compounds do not directly affect KREPA4 and that they do not block protein-RNA interactions in a nonspecific manner but rather target a particular protein.
FIGURE 7.
Alternative models for mechanism of action of C35 and S5. Although C35 and S5 are designed to inhibit adenylylation, our data suggest that they are only weekly able to block this step of editing (A, dashed line). Instead, C35 and S5 act primarily by blocking the interaction of RNA with the editosome, therefore inhibiting all subsequent steps. The exact molecular mechanism for this protein-RNA interaction inhibition is yet to be determined. Two likely possibilities are depicted in B. Top, C35 and S5 bind to the ligases of the editosome and inhibit the induced conformational change of their interacting partners. Because this conformational change is necessary for RNA interaction, binding of these drugs blocks the interaction of the editosome with RNA. This model is supported by the presence of a possible drug-binding cleft in a conformationally variable region of TbREL1 (Glu60–Arg111) (30) that may be responsible for inducing the conformational change of the interacting partner. However, it is not supported by our experimental data and thus is not highlighted in this figure. Bottom, alternatively, C35 and S5 may bind directly to an RNA-binding protein that, independently of TbREL1 and TbREL2, is responsible for recruiting the substrate RNA into the protein. In this figure, for simplicity, we have only shown TbREL1 and not TbREL2, which might be targeted by the compounds as well. PPi, inorganic pyrophosphate.
The RNA-protein complex that we found to be most affected by C35 has been previously designated as G1 (46). The most abundant gRNA-binding protein of G1 has been shown to be a 25-kDa protein. G1 has been proposed to be an intermediate during the formation of the complete editosome (48). This supports our observation that C35 inhibits the formation of high molecular weight RNA-containing editosome complexes. Another candidate target of C35 can be RBP38, a previously identified essential protein that has been shown to be able to bind to both single-stranded and double-stranded RNA and is important for the stability of mitochondrial RNA (50). The properties of this protein match the observation that C35 abolishes the interaction of its target with both single-stranded gRNA and double-stranded gRNA-mRNA complex (Fig. 5A), which leads to instability of RNA after drug treatment as shown by the guanylyltransferase assay (Fig. 6E).
Although our data support a role for RNA to maintain editosome complex integrity, there are contradicting reports showing that mutants that lack mitochondrial DNA and hence lack mitochondrial mRNA and gRNA contain catalytically active editosomes (51). Resolving the exact mechanism of inhibitory compounds such as C35 would provide a more detailed understanding for assembly of functional editosomes.
Acknowledgment
We thank Dr. Kenneth Stuart (Seattle Biomedical Research Institute) for kindly providing monoclonal antibodies against KREPA1, -A2, -A3, and KREL1.
This work was supported in part by Natural Sciences and Engineering Research Council of Canada Grant 328186 (to R. S.).
H. Hogues and E. Purisima, manuscript in preparation.
- gRNA
- guide RNA
- U
- uridylate
- Cyb
- cytochrome b
- TbREL1
- T. brucei RNA editing ligase 1
- OB
- oligonucleotide binding
- SIE
- solvated interaction energy
- TEV
- tobacco etch virus
- FAM
- 6-carboxyfluorescein
- TAMRA
- 6-carboxytetramethylrhodamine
- TAP
- tandem affinity purification.
REFERENCES
- 1. Stuart K., Brun R., Croft S., Fairlamb A., Gürtler R. E., McKerrow J., Reed S., Tarleton R. (2008) J. Clin. Investig. 118, 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Denise H., Barrett M. P. (2001) Biochem. Pharmacol. 61, 1–5 [DOI] [PubMed] [Google Scholar]
- 3. Delespaux V., de Koning H. P. (2007) Drug Resist. Updat. 10, 30–50 [DOI] [PubMed] [Google Scholar]
- 4. Simpson L., Sbicego S., Aphasizhev R. (2003) RNA 9, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. (2005) Trends Biochem. Sci. 30, 97–105 [DOI] [PubMed] [Google Scholar]
- 6. Kable M. L., Heidmann S., Stuart K. D. (1997) Trends Biochem. Sci. 22, 162–166 [DOI] [PubMed] [Google Scholar]
- 7. Seiwert S. D., Heidmann S., Stuart K. (1996) Cell 84, 831–841 [DOI] [PubMed] [Google Scholar]
- 8. Panigrahi A. K., Gygi S. P., Ernst N. L., Igo R. P., Jr., Palazzo S. S., Schnaufer A., Weston D. S., Carmean N., Salavati R., Aebersold R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panigrahi A. K., Schnaufer A., Ernst N. L., Wang B., Carmean N., Salavati R., Stuart K. (2003) RNA 9, 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carnes J., Trotter J. R., Peltan A., Fleck M., Stuart K. (2008) Mol. Cell. Biol. 28, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carnes J., Trotter J. R., Ernst N. L., Steinberg A., Stuart K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trotter J. R., Ernst N. L., Carnes J., Panicucci B., Stuart K. (2005) Mol. Cell 20, 403–412 [DOI] [PubMed] [Google Scholar]
- 13. Aphasizhev R., Sbicego S., Peris M., Jang S. H., Aphasizheva I., Simpson A. M., Rivlin A., Simpson L. (2002) Cell 108, 637–648 [DOI] [PubMed] [Google Scholar]
- 14. Ernst N. L., Panicucci B., Igo R. P., Jr., Panigrahi A. K., Salavati R., Stuart K. (2003) Mol. Cell 11, 1525–1536 [DOI] [PubMed] [Google Scholar]
- 15. Schnaufer A., Ernst N. L., Palazzo S. S., O'Rear J., Salavati R., Stuart K. (2003) Mol. Cell 12, 307–319 [DOI] [PubMed] [Google Scholar]
- 16. Kang X., Rogers K., Gao G., Falick A. M., Zhou S., Simpson L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cruz-Reyes J., Zhelonkina A. G., Huang C. E., Sollner-Webb B. (2002) Mol. Cell. Biol. 22, 4652–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schnaufer A., Panigrahi A. K., Panicucci B., Igo R. P., Jr., Wirtz E., Salavati R., Stuart K. (2001) Science 291, 2159–2162 [DOI] [PubMed] [Google Scholar]
- 19. Worthey E. A., Schnaufer A., Mian I. S., Stuart K., Salavati R. (2003) Nucleic Acids Res. 31, 6392–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kala S., Salavati R. (2010) RNA 16, 1951–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salavati R., Ernst N. L., O'Rear J., Gilliam T., Tarun S., Jr., Stuart K. (2006) RNA 12, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo X., Ernst N. L., Carnes J., Stuart K. D. (2010) PLoS One 5, e8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarun S. Z., Jr., Schnaufer A., Ernst N. L., Proff R., Deng J., Hol W., Stuart K. (2008) RNA 14, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niemann M., Brecht M., Schlüter E., Weitzel K., Zacharias M., Göringer H. U. (2008) Nucleic Acids Res. 36, 4465–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Law J. A., O'Hearn S. F., Sollner-Webb B. (2008) RNA 14, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McManus M. T., Shimamura M., Grams J., Hajduk S. L. (2001) RNA 7, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Swift R. V., Durrant J., Amaro R. E., McCammon J. A. (2009) Biochemistry 48, 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao G., Rogers K., Li F., Guo Q., Osato D., Zhou S. X., Falick A. M., Simpson L. (2010) Protist 161, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amaro R. E., Schnaufer A., Interthal H., Hol W., Stuart K. D., McCammon J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17278–17283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Durrant J. D., Hall L., Swift R. V., Landon M., Schnaufer A., Amaro R. E. (2010) PLoS Negl. Trop. Dis. 4, e803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moshiri H., Salavati R. (2010) Nucleic Acids Res. 38, e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., Onufriev A. (2005) Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Irwin J. J., Shoichet B. K. (2005) J. Chem. Inf. Model. 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Naïm M., Bhat S., Rankin K. N., Dennis S., Chowdhury S. F., Siddiqi I., Drabik P., Sulea T., Bayly C. I., Jakalian A., Purisima E. O. (2007) J. Chem. Inf. Model. 47, 122–133 [DOI] [PubMed] [Google Scholar]
- 35. Cui Q., Sulea T., Schrag J. D., Munger C., Hung M. N., Naïm M., Cygler M., Purisima E. O. (2008) J. Mol. Biol. 379, 787–802 [DOI] [PubMed] [Google Scholar]
- 36. Purisima E. O. (1998) J. Comput. Chem. 19, 1494–1504 [Google Scholar]
- 37. Purisima E. O., Nilar S. H. (1995) J. Comput. Chem. 16, 681–689 [Google Scholar]
- 38. Bhat S., Purisima E. O. (2006) Proteins 62, 244–261 [DOI] [PubMed] [Google Scholar]
- 39. Allison J., Rothwell V., Newport G., Agabian N., Stuart K. (1984) Nucleic Acids Res. 12, 9051–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneider A., Charrière F., Pusnik M., Horn E. K. (2007) Methods Mol. Biol. 372, 67–80 [DOI] [PubMed] [Google Scholar]
- 41. Salavati R., Panigrahi A. K., Morach B. A., Palazzo S. S., Igo R. P., Stuart K. (2002) Mol. Biochem. Parasitol. 120, 23–31 [DOI] [PubMed] [Google Scholar]
- 42. Igo R. P., Jr., Palazzo S. S., Burgess M. L., Panigrahi A. K., Stuart K. (2000) Mol. Cell. Biol. 20, 8447–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Igo R. P., Jr., Weston D. S., Ernst N. L., Panigrahi A. K., Salavati R., Stuart K. (2002) Eukaryot. Cell 1, 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shaneh A., Salavati R. (2009) J. Mol. Model. 16, 61–76 [DOI] [PubMed] [Google Scholar]
- 45. Sabatini R., Hajduk S. L. (1995) J. Biol. Chem. 270, 7233–7240 [DOI] [PubMed] [Google Scholar]
- 46. Read L. K., Göringer H. U., Stuart K. (1994) Mol. Cell. Biol. 14, 2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aphasizhev R., Aphasizheva I., Nelson R. E., Gao G., Simpson A. M., Kang X., Falick A. M., Sbicego S., Simpson L. (2003) EMBO J. 22, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Göringer H. U., Koslowsky D. J., Morales T. H., Stuart K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1776–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doherty A. J., Suh S. W. (2000) Nucleic Acids Res. 28, 4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sbicego S., Alfonzo J. D., Estévez A. M., Rubio M. A., Kang X., Turck C. W., Peris M., Simpson L. (2003) Eukaryot. Cell 2, 560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Domingo G. J., Palazzo S. S., Wang B., Pannicucci B., Salavati R., Stuart K. D. (2003) Eukaryot. Cell 2, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]