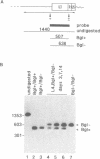
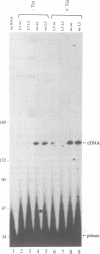
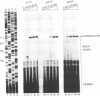
Abstract
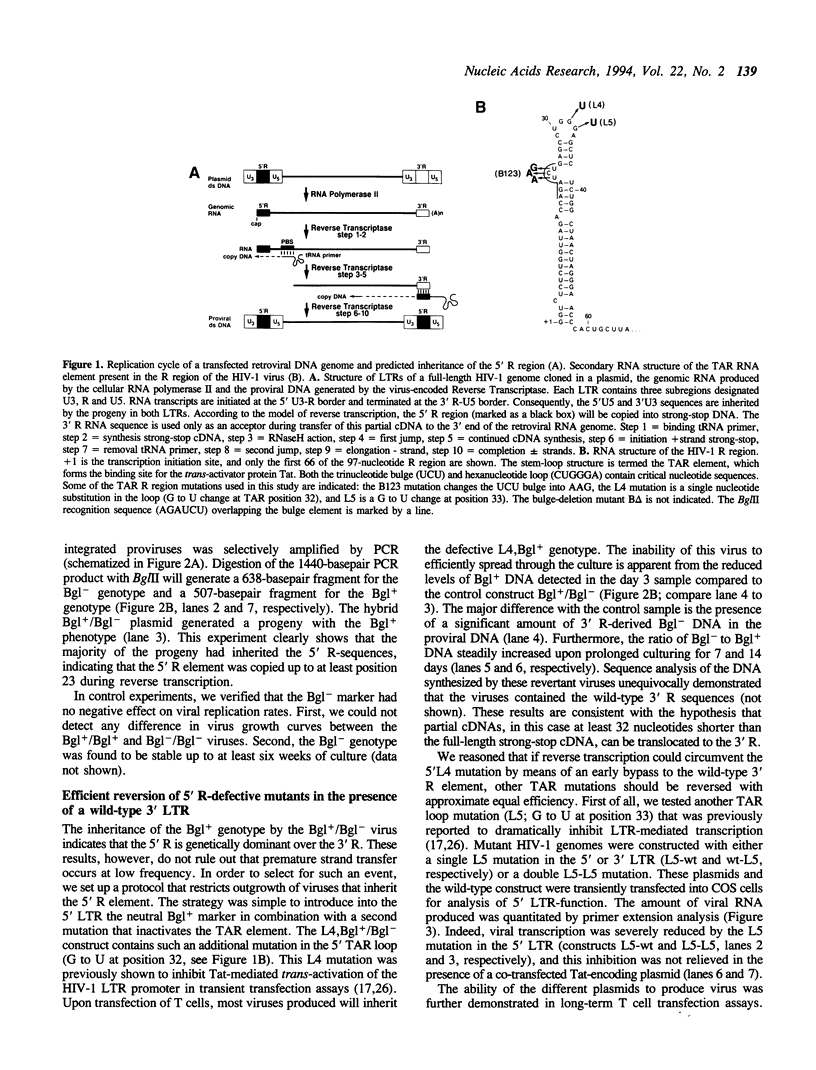
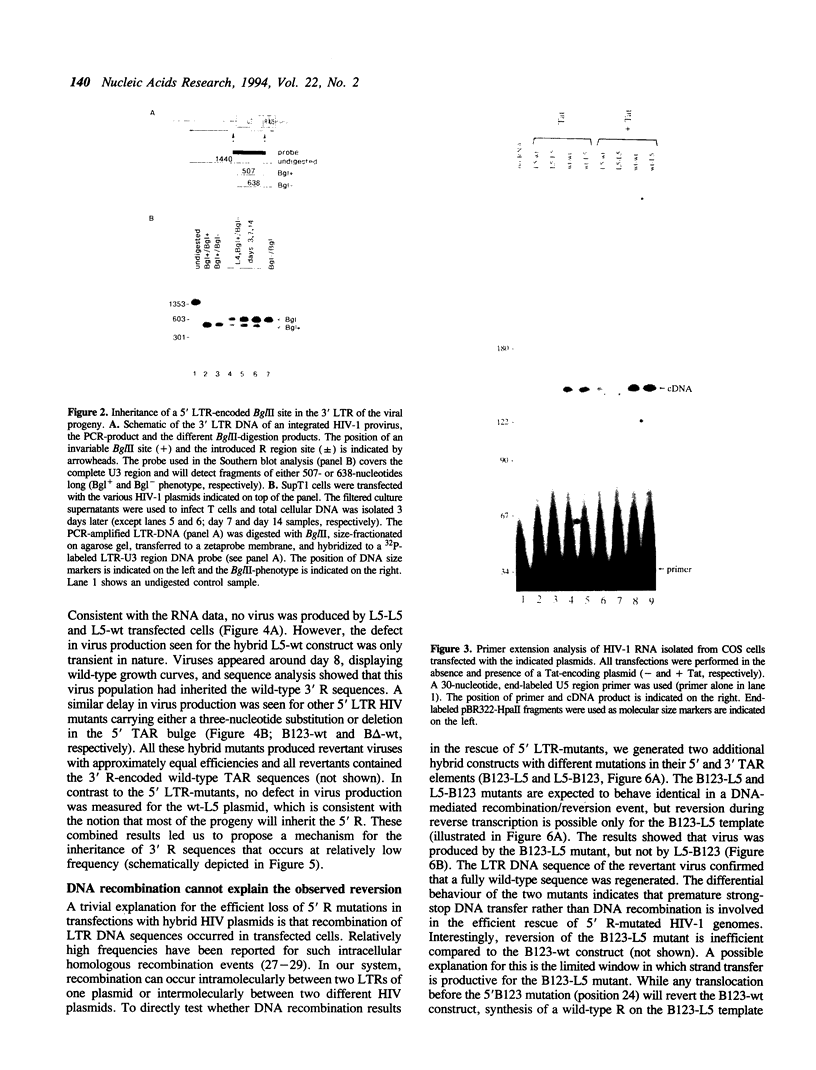
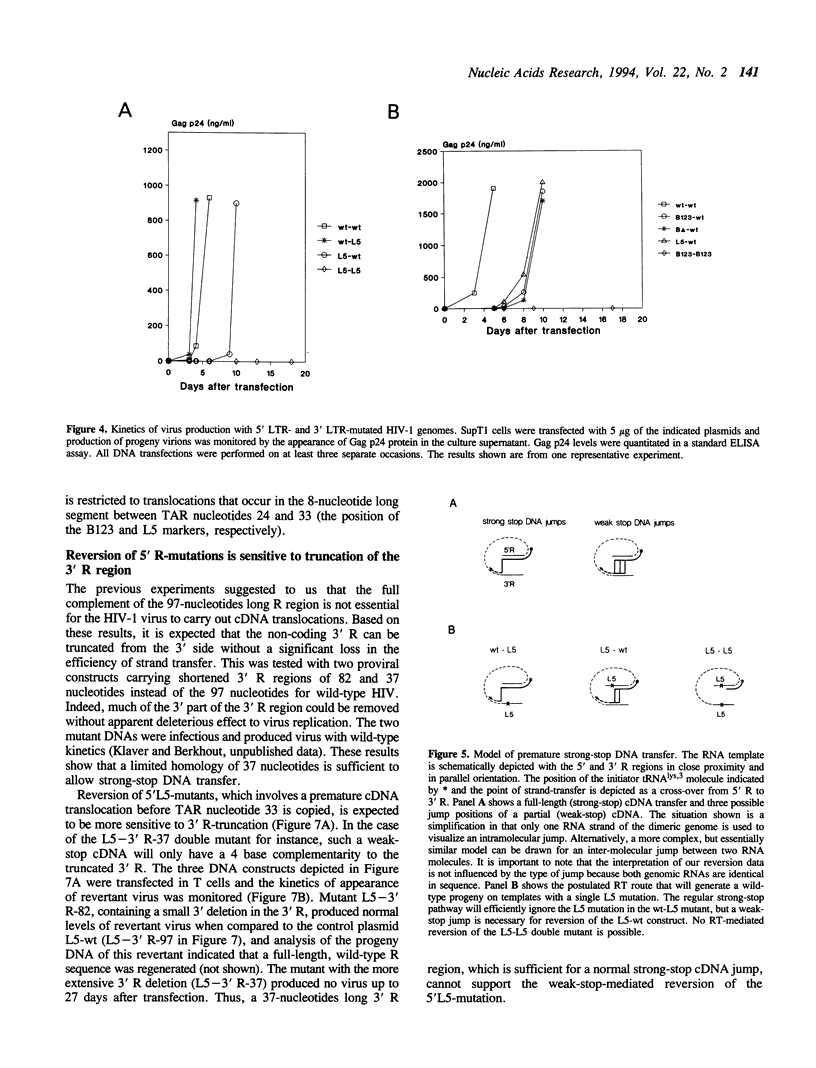
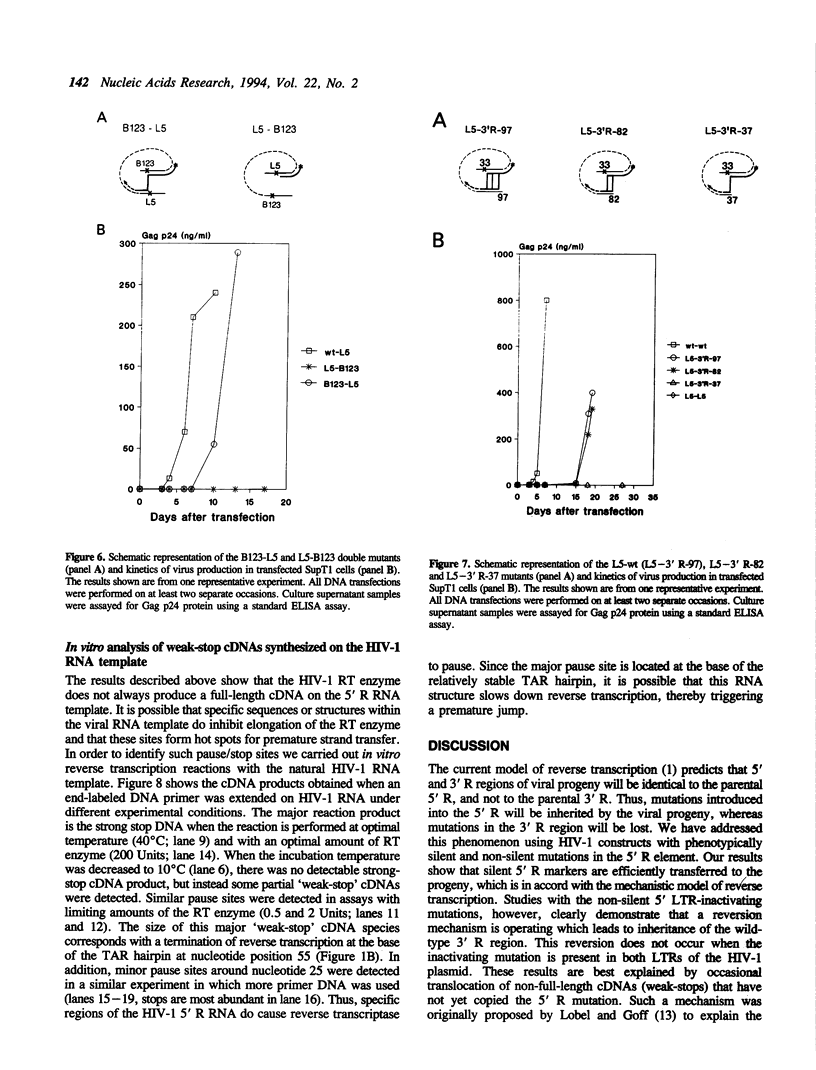
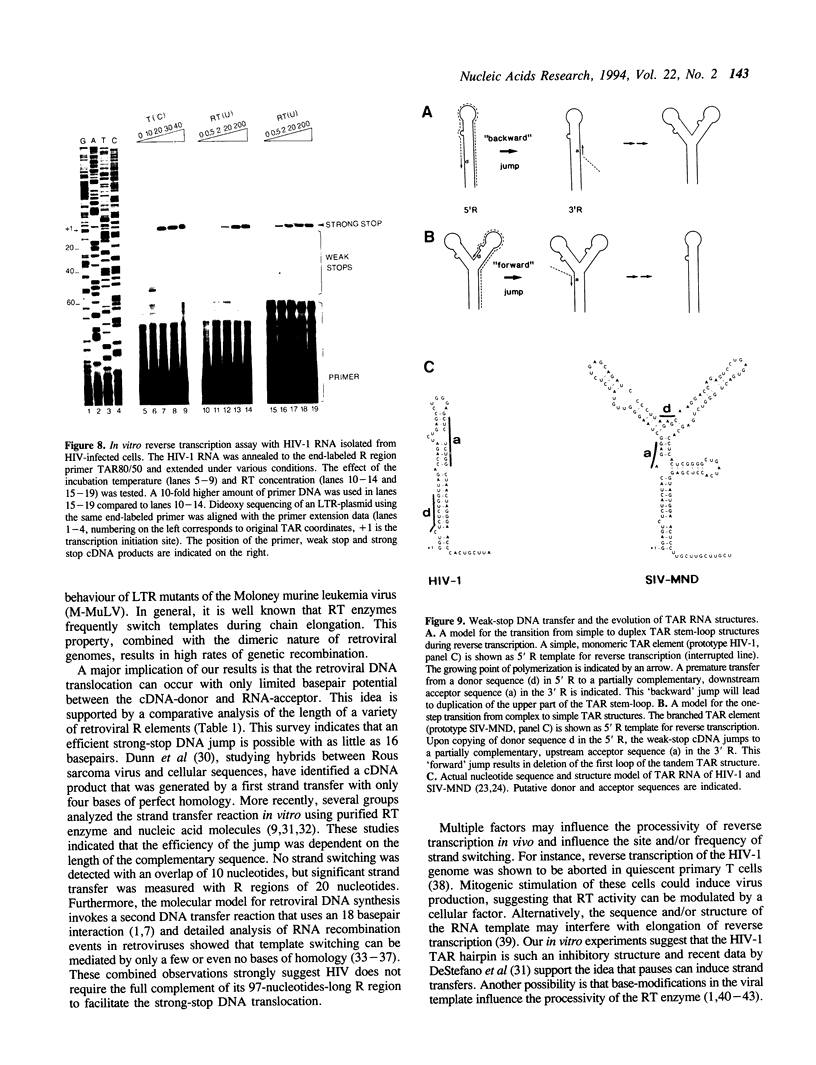
Reverse transcription of retroviral genomes starts near the 5' end of the viral RNA by use of an associated tRNA primer. According to the current model of reverse transcription, the initial cDNA product, termed minus-strand strong-stop DNA, 'jumps' to a repeated sequence (R region) at the 3' end of the RNA template. The human retroviruses have relatively long R regions (97-247 nucleotides) when compared to murine and avian viruses (16-68 nucleotides). This suggests that the full complement of the R region is not required for strand transfer and that partial cDNA copies of the 5' R can prematurely jump to the 3' R. To test this hypothesis, we generated mutants of the human immunodeficiency virus with R region changes and analyzed whether 5' or 3' R sequences were inherited by the progeny. We found that in most cases, 5' R-encoded sequences are dominant, which is consistent with the model of reverse transcription. Using a selection protocol, however, we were also able to identify progeny viruses with R sequences derived from the original 3' R element. These results suggest that partial strong stop cDNAs can be transferred with R region homologies much shorter than 97 nucleotides.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkhout B., Jeang K. T. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991 Nov 25;19(22):6169–6176. doi: 10.1093/nar/19.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989 Dec;63(12):5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992 Jan 11;20(1):27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard A. D., Powell R., Braddock M., Kingsman A. J., Kingsman S. M. An adenosine at position 27 in the human immunodeficiency virus type 1 trans-activation response element is not critical for transcriptional or translational activation by Tat. J Virol. 1992 Nov;66(11):6769–6772. doi: 10.1128/jvi.66.11.6769-6772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R., Sol C. J., Salimans M. M., Jansen C. L., Wertheim-van Dillen P. M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R. A model for reverse transcription by a dimeric enzyme. J Gen Virol. 1993 Apr;74(Pt 4):691–697. doi: 10.1099/0022-1317-74-4-691. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992 Sep;56(3):375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J. J., Mallaber L. M., Rodriguez-Rodriguez L., Fay P. J., Bambara R. A. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J Virol. 1992 Nov;66(11):6370–6378. doi: 10.1128/jvi.66.11.6370-6378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. M., Olsen J. C., Swanstrom R. Characterization of unintegrated retroviral DNA with long terminal repeat-associated cell-derived inserts. J Virol. 1992 Oct;66(10):5735–5743. doi: 10.1128/jvi.66.10.5735-5743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A. J., Stavnezer E., Bishop J. M. Blocked, methylated 5'-terminal sequence in avian sarcoma virus RNA. Nature. 1975 Oct 16;257(5527):618–620. doi: 10.1038/257618a0. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Hajjar A. M., Linial M. L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993 Jul;67(7):3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrich D., Garcia J., Mitsuyasu R., Gaynor R. TAR independent activation of the human immunodeficiency virus in phorbol ester stimulated T lymphocytes. EMBO J. 1990 Dec;9(13):4417–4423. doi: 10.1002/j.1460-2075.1990.tb07892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Klarmann G. J., Schauber C. A., Preston B. D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993 May 5;268(13):9793–9802. [PubMed] [Google Scholar]
- Lania L., Di Cristofano A., Strazzullo M., Pengue G., Majello B., La Mantia G. Structural and functional organization of the human endogenous retroviral ERV9 sequences. Virology. 1992 Nov;191(1):464–468. doi: 10.1016/0042-6822(92)90211-7. [DOI] [PubMed] [Google Scholar]
- Leonard J., Parrott C., Buckler-White A. J., Turner W., Ross E. K., Martin M. A., Rabson A. B. The NF-kappa B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989 Nov;63(11):4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Stephenson A. J., Kuiper L. J., Burrell C. J. Double-stranded strong-stop DNA and the second template switch in human immunodeficiency virus (HIV) DNA synthesis. Virology. 1993 May;194(1):82–88. doi: 10.1006/viro.1993.1237. [DOI] [PubMed] [Google Scholar]
- Lobel L. I., Goff S. P. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J Virol. 1985 Feb;53(2):447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987 Feb;61(2):480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988 Aug 26;241(4869):1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K., Emerman M., Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991 Dec;185(2):661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991 Sep;65(9):4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey C. A., Panganiban A. T. Replication of the retroviral terminal repeat sequence during in vivo reverse transcription. J Virol. 1993 Jul;67(7):4114–4121. doi: 10.1128/jvi.67.7.4114-4121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen L., Bass B., Sonenberg N., Weintraub H., Groudine M. Tat-dependent adenosine-to-inosine modification of wild-type transactivation response RNA. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8096–8100. doi: 10.1073/pnas.88.18.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A., York D., Jannoun-Nasr R., Kalyanaraman S., Swan D., Benson J., Bohan C., Luciw P. A., Schnoll S., Robinson R. A. Generation of hybrid human immunodeficiency virus by homologous recombination. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6388–6392. doi: 10.1073/pnas.86.16.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dimock K. Evidence of methylation of B77 avian sarcoma virus genome RNA subunits. J Virol. 1976 May;18(2):586–595. doi: 10.1128/jvi.18.2.586-595.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Mechanism of transduction by retroviruses. Science. 1992 Feb 14;255(5046):841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Simian virus 40 recombinants are produced at high frequency during infection with genetically mixed oligomeric DNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2876–2880. doi: 10.1073/pnas.76.6.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York D. F., Vigne R., Verwoerd D. W., Querat G. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992 Aug;66(8):4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. Reverse transcriptase pauses at N2-methylguanine during in vitro transcription of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3751–3754. doi: 10.1073/pnas.76.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack J. A., Haislip A. M., Krogstad P., Chen I. S. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992 Mar;66(3):1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Temin H. M. 3' junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993 Apr;67(4):1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]