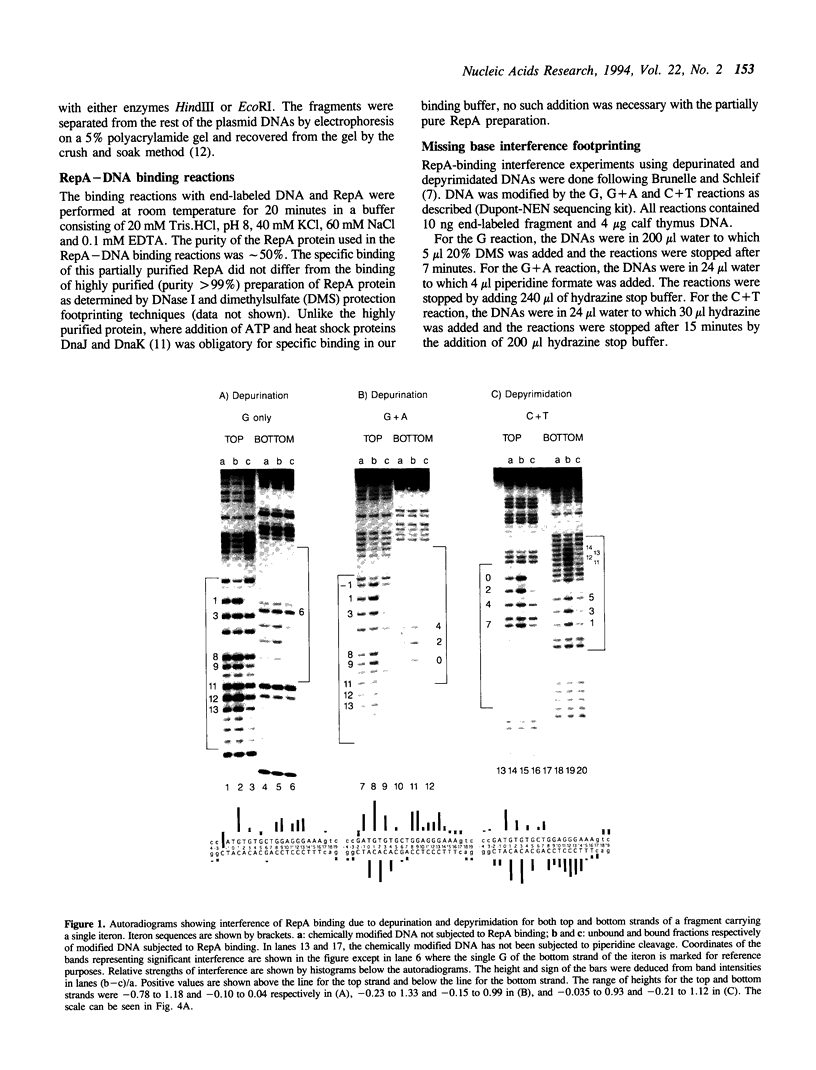
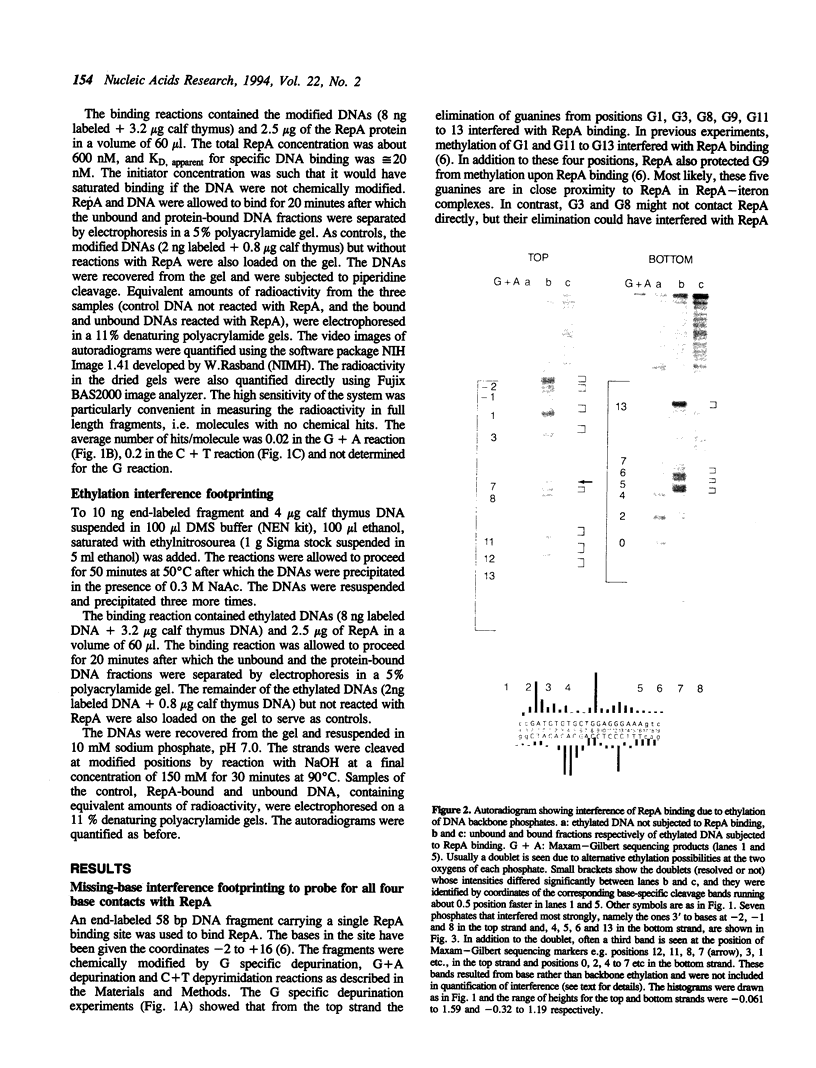
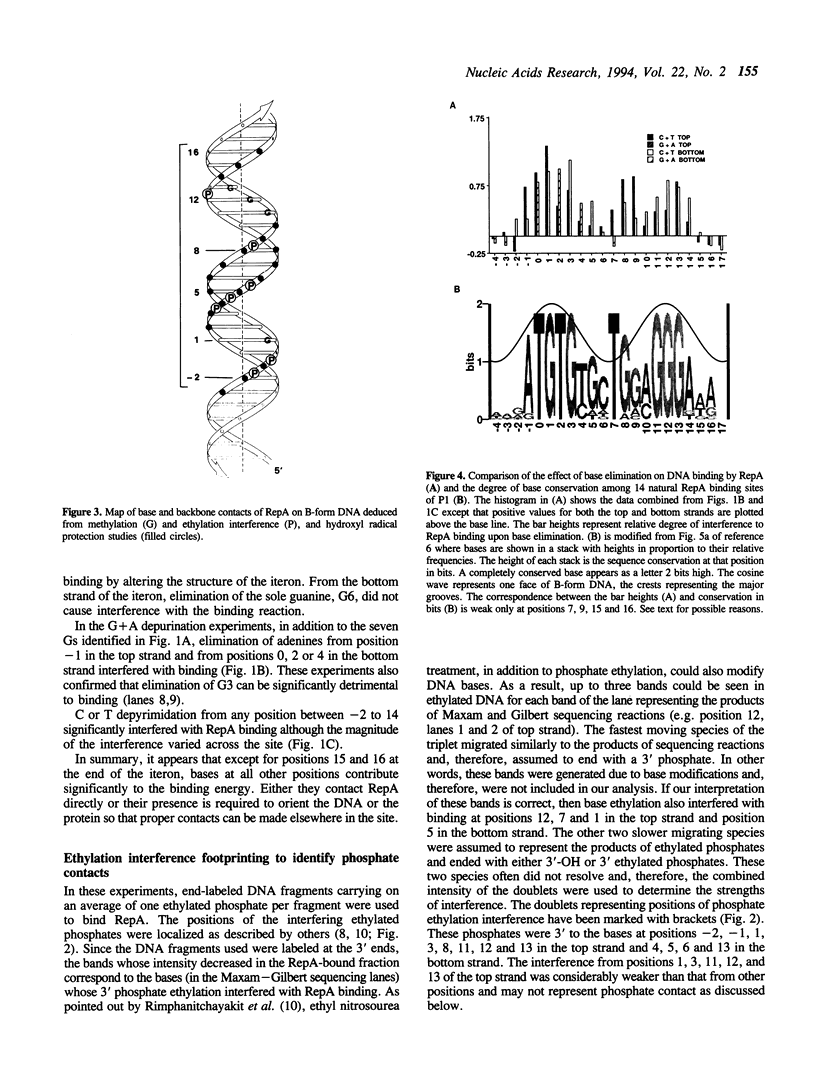
Abstract
RepA, the replication initiator protein of plasmid P1, binds to specific 19 bp sequences on the plasmid DNA. Earlier footprinting studies with dimethylsulfate identified the guanines that contact RepA through the major groove of DNA. In this study, base elimination was used to identify the contribution of all four bases to the binding reaction. Depurination and depyrimidation of any base in the neighborhood of the contacting guanines was found to decrease RepA binding. These results are consistent with the notion that RepA contacts bases of two consecutive major grooves on the same face of DNA. We also observed that depurination but not methylation of three guanines (G3, G8 and G9) affected binding. We identified the DNA phosphate groups (3 in the top strand, one of which mapped between G8 and G9, and 4 in the bottom strand, one of which was adjacent to C3) that strongly interfered with RepA binding upon ethylation. These results indicate that certain bases (e.g. G3, G8 and G9) may not contact RepA directly but contribute to base and backbone contacts by maintaining proper structure of the binding site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta S., Mukhopadhyay G., Papp P. P., Lewis M. S., Chattoraj D. K. Activation of DNA binding by the monomeric form of the P1 replication initiator RepA by heat shock proteins DnaJ and DnaK. J Mol Biol. 1993 Jul 5;232(1):23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Koudelka G. B., Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992 Jan 2;355(6355):89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Adhya S. Effect of ethylation of operator-phosphates on Gal repressor binding. DNA contortion by repressor. J Mol Biol. 1989 Jul 20;208(2):217–223. doi: 10.1016/0022-2836(89)90383-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay G., Chattoraj D. K. Conformation of the origin of P1 plasmid replication. Initiator protein induced wrapping and intrinsic unstacking. J Mol Biol. 1993 May 5;231(1):19–28. doi: 10.1006/jmbi.1993.1253. [DOI] [PubMed] [Google Scholar]
- Pal S. K., Mason R. J., Chattoraj D. K. P1 plasmid replication. Role of initiator titration in copy number control. J Mol Biol. 1986 Nov 20;192(2):275–285. doi: 10.1016/0022-2836(86)90364-5. [DOI] [PubMed] [Google Scholar]
- Papp P. P., Chattoraj D. K., Schneider T. D. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993 Sep 20;233(2):219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- Rimphanitchayakit V., Hatfull G. F., Grindley N. D. The 43 residue DNA binding domain of gamma delta resolvase binds adjacent major and minor grooves of DNA. Nucleic Acids Res. 1989 Feb 11;17(3):1035–1050. doi: 10.1093/nar/17.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Sozhamannan S., Chattoraj D. K. Heat shock proteins DnaJ, DnaK, and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. J Bacteriol. 1993 Jun;175(11):3546–3555. doi: 10.1128/jb.175.11.3546-3555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Chattoraj D. K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]