Summary
Here we report that Cyclooxygenase-2 (COX-2) mediates the formation of electrophilic fatty acid oxo-derivatives (EFOXs) from the omega-3 fatty acids (ω-3 FA) docosahexaenoic, docosapentaenoic and docosatetraenoic acid. EFOXs produced by activated macrophages were discovered by a mass spectrometry-based “fishing” method, using the nucleophile β-mercaptoethanol (BME) as bait. Intracellular EFOX concentrations ranged from 65 to 350 nM, with acetylsalicylic acid (ASA, aspirin) increasing both rate of production and intracellular concentrations. Due to an electrophilic nature, EFOXs adducted protein cysteine and histidine residues and the cysteine of glutathione (GSH) in activated macrophages. A role for EFOXs as signalling mediators was confirmed by the observations that 17-EFOX-D6 and 17-EFOX-D5 are PPARγ agonists and activate Nrf2-dependent antioxidant responses. They also inhibited cytokine production and inducible nitric oxide synthase (NOS-2) expression in activated macrophages within a biological concentration range. Thus, EFOXs are signalling mediators that can transduce the clinically beneficial effects of ω-3 FA, COX-2 and ASA.
The major ω-3 FAs (5Z,8Z,11Z,14Z,17Z)-eicosapentaenoic acid (EPA) and (4Z,7Z,10Z,13Z,16Z,19Z)-docosahexaenoic acid (DHA) have been associated with beneficial health effects in humans. For example, brain and retina are enriched with DHA in healthy individuals, with DHA being necessary for the normal development and function of these tissues1, 2. Moreover, the consumption of DHA in the diet has been implicated in reducing neurocognitive decline3, improving insulin resistance in diabetics4, decreasing incidence of myocardial infarction5 and reducing inflammation6. Despite these salutary effects, the molecular mechanisms of ω-3 FA actions remain largely uncharacterized.
Multiple anti-inflammatory lipid mediators have recently been reported that promote resolution of inflammation by suppressing NF-κB activation, modulating cytokine expression, activating G-protein coupled receptors7 and promoting cyto-protective responses8. Oxygenases, including COX-2 and lipoxygenases (LOXs) are involved in both the initiation of inflammatory events and in an orchestrated resolution9. Other bioactive lipids generated by non-enzymatic redox reactions include reactive electrophilic species (RES), such as nitro-fatty acids (NO2-FAs) and cyclopentenone isoprostanes and neuroprostanes10, 11.
Biological RES are characterized by an electron-withdrawing functional group that renders the β-carbon electron-poor, promoting Michael addition reactions with cellular nucleophiles (e.g. cysteine and histidine residues of protein). By adducting nucleophilic protein residues, RES modulate protein activity, subcellular localization, and electrophile-sensitive gene expression. The reactivity of the electrophile influences downstream signaling actions12, with irreversible adducts conveying toxic effects13. RES can also modulate cellular redox status by changing GSH/GSSG levels and ratios, further impacting cellular metabolism. Recently, there has been a move towards employing RES in the prevention or treatment of various diseases such as neurodegeneration, cancer, and other pathologies presenting a significant inflammatory component. For example, electrophilic neurite outgrowth-promoting prostaglandin compounds display protective effects during cerebral ischemia/reperfusion, an event attributed to their accumulation in neurons and subsequent activation of the Keap1/Nrf2 pathway14. Other RES (e.g. avicins15 and Bis(2-hydroxybenzylidene)acetone16, isothiocyanates17) are potential chemopreventative agents, due to their abilities to induce apoptosis of precancerous cells and tumor cells. Additionally, the electrophile 15-deoxy-Δ (12,14)-prostaglandin J2 (15d-PGJ2) is protective in animal models of acute lung injury18.
EFOX are produced by activated macrophages
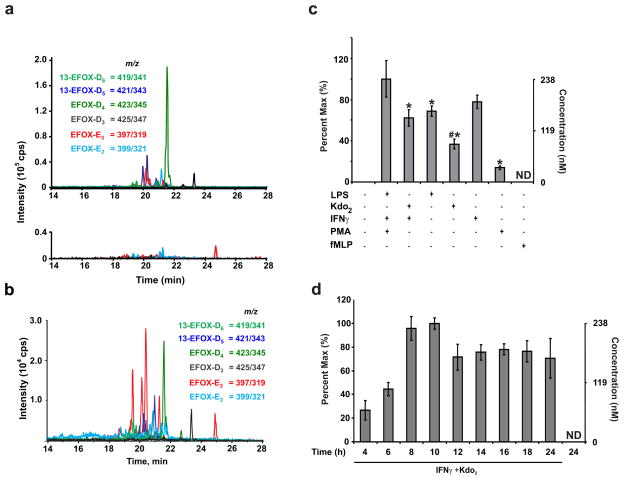
To search for electrophilic products formed during inflammation a screening method was applied that is based on BME reaction with electrophiles, followed by resolution with reverse phase-high pressure liquid chromatography and detection by tandem mass spectrometry (RP-HPLC-MS/MS)19. Alkylation with BME standardizes the MS/MS conditions for adducted RES, conferring similar ionization and fragmentation properties on a range of RES, each with their own particular MS/MS characteristics. This permits the sensitive detection of free RES and RES that a) are reversibly adducted to nucleophiles, b) fragment poorly during MS/MS, c) were previously at or below limits of detection, d) have not been previously predicted. Using this method to screen fatty acid-derived RES, the neutral loss of BME (78 amu) by RP-HPLC-MS/MS revealed six new and abundant RES following activation of RAW264.7 cells (a murine monocyte/macrophage cell line) by phorbol myristic acetate (PMA), LPS, and IFNγ (Fig. 1a). Identical RES were also detected in PMA, Kdo2-lipid A (Kdo2) and IFNγ-activated THP-1 cells (a human monocyte cell line) and in primary murine bone marrow-derived macrophages (BMDMs, Fig. 1b and Supplementary Fig. 2–3). Although relative abundance of these RES differed between the two cell lines, MS/MS spectra showed similar characteristic neutral losses and fragment ion intensity ratios (not shown). The BME reaction was further tested for the detection and quantification of reversibly reactive RES by comparing the mass spectrometric characteristics of several RES containing α,β-unsaturated carbonyls using three different approaches: the BME method, selected ion monitoring (SIM) and multiple ion monitoring (MRM) for loss of CO2 (Supplementary Fig. 4). In all cases, the BME method was unique in specificity for RES and superior in terms of signal intensity, low background levels and linearity of signal response versus concentration. For these reasons and ease of identification, electrophile-BME adducts were studied for identifying unknown RES in biological samples.
Figure 1. EFOXs are produced during macrophage activation.
a, MRM scans following the neutral loss of 78 reveal EFOXs adducted with BME in cell extracts from activated (upper) and non-activated (N/A, lower) RAW 264.7 cells. b, EFOX levels in PMA-differentiated THP-1 cells 8 h post activation. c, EFOX-D5 levels in RAW264.7 cells activated with the indicated compounds at the following concentrations: LPS (0.5 μg/ml), Kdo2 (0.5 μg/ml), IFNγ (200 U/ml), PMA (3.24 μM), fMLP (1 μM). Data are expressed as mean ± S.D. (n=4), where * = significantly different (p<0.01) from “PMA + IFNγ + LPS,” and # = significant difference (p<0.01) between “LPS” and “Kdo2 + IFNγ” (one-way ANOVA, post-hoc Tukey’s test). d, EFOX-D5 levels in RAW264.7 cells were quantified at the indicated time points post activation. ND indicates not detectable.
Macrophages were activated with various combinations of LPS, IFNγ, PMA, fMLP, and Kdo2 (Fig. 1c and Supplementary Fig. 5). Kdo2, a synthetic endotoxin, was used to avoid the contribution of potential LPS contaminants. Since the combination of Kdo2 and IFNγ induced responses similar to LPS, it was used for subsequent experiments. EFOX formation under different inflammatory conditions was confirmed by treating the cells with a variety of stimuli. Intracellular concentrations obtained for different species ranged from 65 to 350 nM (Table 1). EFOX formation became detectable 4–6 h post-activation and reached maximum levels approximately 10 h after activation (Fig. 1d and Supplementary Fig. 6).
Table 1.
Summary of EFOX concentrations and characteristics.
| Name | EFOX-D6 | EFOX-D5 | EFOX-D4 | EFAD-D3 | EFAD-E3 |
|---|---|---|---|---|---|
| Cellular concentration nM | 65 ± 5 | 238 ± 16 | 348 ± 26 | 106 ± 6 | 326 ± 15 |
| Mass (m/z) | 341.2 | 343.2 | 345.2 | 347.2 | 319.2 |
| FA precursors | 22:6 | 18:3n-6, 20:5 | 20:4, 18:3n-6, 20:5, 18:3n-3 | 18:3n-6, 20:4 | 18:3n-6 |
| Series | ω-3 | ω-3 | ω-3 and ω-6 | ω-6 | ω-6 |
| Identity | 13-oxoDHA 17-oxoDHA |
13-oxoDPA 17-oxoDPA |
oxoDTA | oxoDTrA | oxoETrA |
EFOXs are oxo-derivatives of ω-3 FA
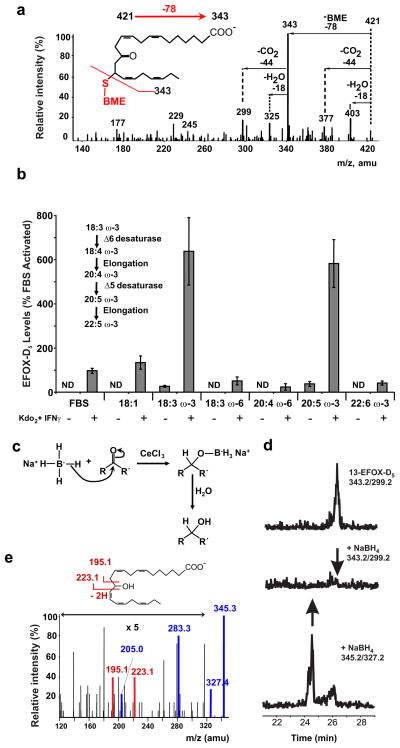
Accurate mass TOF data (mass accuracy ≤10 ppm), elution profile and loss of CO2 upon fragmentation indicated EFOX-D5 was a mono-oxygenated derivative of a 22-carbon FA with five double bonds. The MS/MS spectrum for BME-adducted EFOX-D5 (m/z 421 [M-H]−) displayed characteristic fragment ions at m/z 403 ([M-H-H2O]−), 377 ([M-H-CO2]−), 343 ([M-H-BME]−), 325 ([M-H-BME-H2O]−), and 299 ([M-H-BME- CO2]−) (Fig. 2a). EFOX-D6 and EFOX-D4 were identified as mono-oxygenated derivatives of 22-carbon FAs with a total of 6 and 4 double bonds, respectively. To elucidate metabolic precursors in vivo, cells were supplemented with different FAs. EFOX-D5 was significantly increased in activated RAW264.7 cells supplemented with 18:3 ω-3 (α-linolenic acid) and 20:5 ω-3 (EPA), and slightly decreased when ω-6 FA were provided (Fig. 2b), indicating that EFOX-D5 was derived from ω-3 FA. Addition of 22:6 ω-3 (DHA) did not increase EFOX-D5 levels, consistent with the precept that mammalian cells can desaturate and elongate shorter chain FAs and generally do not resaturate DHA. The formation of EFOX-D6 in activated RAW264.7 cells was increased only by supplementation of 22:6 ω-3, while EFOX-D4 was increased by both ω-3 and ω-6 FA supplementation, indicating precursors could be either ω-3 or ω-6 DTA (Supplementary Fig. 7a–b). Overall, EFOX-D6, EFOX-D5 and a percentage of EFOX-D4 were derivatives of the ω-3 FA DHA, (7Z,10Z,13Z,16Z,19Z)-docosapentaenoic acid (DPA) and docosatetraenoic (DTA) respectively, while EFOX-D3, EFOX-E3 and EFOX-E2 were synthesized from ω-6 and ω-9 FAs (Table 1).
Figure 2. EFOX-D5 is an α,β-unsaturated oxo-derivative of DPA.
a, Characteristic BME-EFOX fragmentation pattern derived from the EPI of BME-EFOX-D5. b, EFOX-D5 levels in activated RAW264.7 cells grown for 3 days in medium supplemented with the indicated FA. ND indicates not detectable. c, Diagram of NaBH4 reaction. d, MRM scans monitoring for the m/z transitions 343.2/299.2 (13-EFOX-D5/loss of CO2; upper and middle panel) and 345.2/327.2 (hydroxy-DPA/loss of H2O; lower panel) in RAW264.7 cell lysates purified for EFOX-D5 ± NaBH4. e, MS/MS fragmentation of EFOX-D5 purified from activated RAW 264.7 cells and reduced with NaBH4.
The Luche reaction, where NaBH4 selectively reduces conjugated carbonyls to allylic alcohols without loss of regioselectivity20 (Fig. 2c), confirmed that the EFOX electrophilic functional group was an α,β-unsaturated carbonyl. Lipid extracts from activated RAW264.7 cells were first fractionated by HPLC and then fractions containing EFOX-D5 were purified and reduced with NaBH4. This significantly decreased the signal corresponding to EFOX-D5 and induced the presence of a previously absent peak at the transition 345/327 (reduced EFOX-D5, hydroxy-DPA; Fig. 2d). The enhanced MS/MS fragmentation of hydroxy groups also revealed the location of the carbonyl group in precursor EFOXs. In addition to the commonly observed ion fragments m/z 327 ([M-H]-H2O) and 283 ([M-H]-H2O-CO2), the following diagnostic ions for 13-hydroxy-DPA (13-OH-DPA) were observed in the EFOX-D5 enriched fraction reduced with NaBH4: m/z 223, 205 (223-H2O) and 195 (Fig. 2e). These findings revealed that EFOX-D5 corresponded to 13-oxoDPA and EFOX-D4 was an oxo-derivative of DTA (Table 1).
COX-2 mediates EFOX production
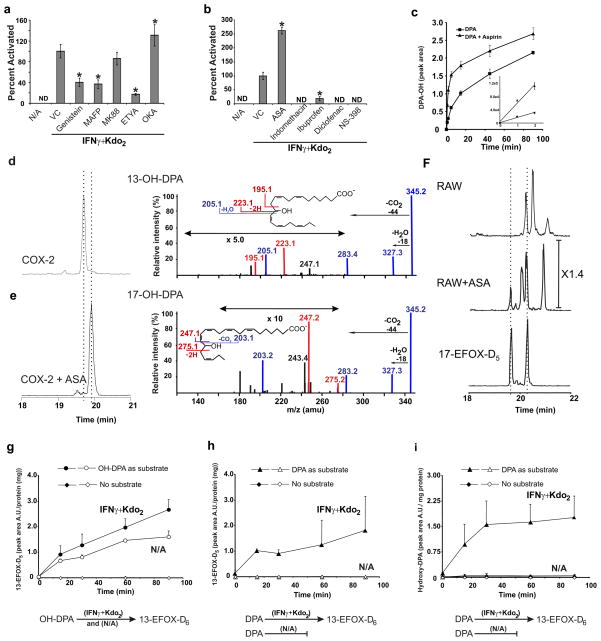
EFOX levels were quantified in activated RAW264.7 cells treated with a variety of inhibitors (Fig. 3a and Supplementary Fig. 8). Both genistein, a general tyrosine kinase inhibitor, and methyl arachidonyl fluorophosphonate (MAFP) inhibited EFOX production by over 50%. Inhibition by MAFP, a selective irreversible inhibitor of both calcium-dependent (cPLA2) and calcium-independent cytosolic phospholipase A2 (iPLA2), indicated that a significant proportion of FA substrates for EFOX generation came from the hydrolysis of complex lipids. MK886, an inhibitor of 5-LOX activating protein (FLAP)-dependent activation of 5-LOX and had no significant effect on EFOX formation. Eicosatetraynoic acid (ETYA), a nonspecific inhibitor of COX and LOX enzymes, strongly inhibited EFOX formation. The general phosphatase inhibitor, okadaic acid (OKA), only slightly increased EFOX formation. To further characterize EFOX formation by COX, EFOX levels were quantified in activated RAW264.7 cells treated with COX inhibitors at concentrations ≥ 5-fold the IC5021 (Fig. 3b and Supplementary Fig. 9). Indomethacin and diclofenac completely inhibited EFOX formation, while ibuprofen inhibition was 80%. The selective COX-2 inhibitor NS-398, fully inhibited EFOX formation, and in contrast ASA significantly increased production of all EFOX derivatives, with the exception of EFOX-D3 and EFOX-D2. This effect of ASA is consistent with ASA acetylation of COX-2 Ser530 favoring the mono-oxygenation of long chain FAs22.
Figure 3. EFOX-D5 formation is dependent on COX-2 activity.
a, b, EFOX-D5 levels in RAW264.7 activated and treated with the indicated inhibitors at the following concentrations: genistein (25 μM), MAFP (25 μM), MK886 (500 nM), ETYA (25 μM), OKA (50 nM), ASA (200 μM), indomethacin (25 μM), ibuprofen (100 μM), diclofenac (1 μM) and NS-398 (4 μM). ND indicates not detectable. Data are expressed as mean ± S.D. (n=4), where * = significantly different (p<0.01) from “Kdo2 + IFNγ” (one-way ANOVA, post-hoc Tukey’s test). c, Temporal formation of hydroxy-precursors (MRM 345/327) of EFOX-D5 synthesized in vitro by purified ovine COX-2 + DPA ± ASA. d, e, Chromatographic profiles (left panels) and spectra (right panels) of the two isomers formed by COX-2 ± ASA. f, BME-adducted 17-EFOX-D5 standard (MRM 421.2/343.2) and EFOX-D5 BME-adducts from activated RAW264.7 cells ± ASA. g–i, Production of EFOX-D5 and OH-DPA by RAW264.7 cell lysates supplemented with OH-DPA, DPA, or vehicle alone. Full and empty symbols indicate activated and non-activated cell lysates respectively.
The role of COX-2 in EFOX formation was affirmed by observing that purified ovine COX-2 generated the EFOX-D5 precursor, hydroxy-DPA (OH-DPA), from DPA (Fig. 3c–e) and the EFOX-D6 precursor, hydroxy-DHA (OH-DHA), from DHA (Supplementary Fig. 10a–b). ASA increased the rate and extent of formation of OH-DPA (Fig. 3c) and shifted the distribution of hydroxy regioisomers from C-13 to C-17 (Fig. 3d–e, and Supplementary Fig. 10a–b). The fragmentation pattern of COX-derived 13-OH-DPA showed the characteristic m/z 195 and 223 ions, corresponding to the hydroxyl-induced fragmentation observed in NaBH4-reduced RAW264.7 cell-derived products (Fig. 3d and 2e). In contrast, when COX-2 was treated with ASA, characteristic fragment ions corresponding to a 17-OH group were detected (Fig 3e). In activated RAW264.7 cells ASA addition similarly promoted the production of 17-EFOX-D5 (Fig. 3f) and 17- EFOX-D6 (Supplementary Fig. 10c).
Since purified COX-2 produced hydroxy rather than carbonyl derivatives of FA, a hydroxy-dehydrogenase reaction was viewed as necessary. When non-activated RAW264.7 cell lysates were incubated with OH-DPA in presence of NAD+, there was a time-dependent production of EFOX-D5. This indicates that the dehydrogenase activity responsible for the conversion of OH-DPA to its oxo-derivative was constitutively expressed (Fig. 3g–i). Only activated cells converted DPA into oxo-DPA, due to the requirement of COX-2 for the first oxidation step.
EFOXs adduct to proteins and GSH
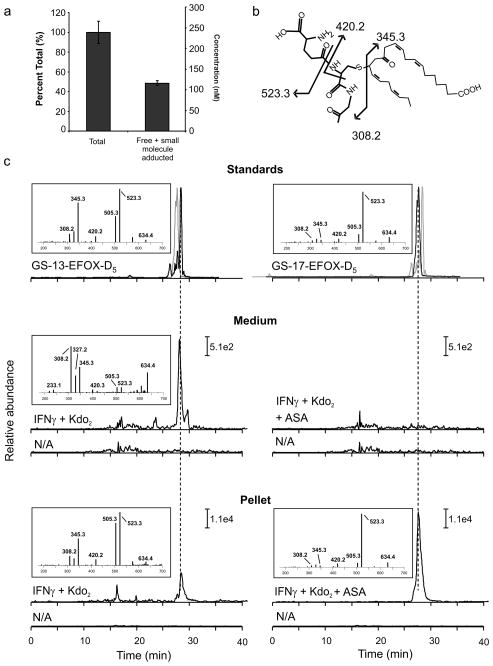
Typically, electrophiles form adducts with nucleophilic amino acid residues in the cell, including the cysteine of GSH23–26. Total EFOX content was quantified and compared with the pool of free EFOXs (including small molecule EFOX adducts, such as GSH). The difference between these two EFOX pools gave the percentage of EFOXs adducted to proteins (~50%) (Fig. 4a). The difference in the kinetics of BME reaction with free and adducted EFOXs further confirmed the distribution of intracellular EFOXs. Reaction rates of BME with free electrophiles were relatively fast, with a pseudo first order reaction rate constant between 3×10−3 and 5 ×10−3 sec−1 for different α,β-unsaturated carbonyl derivatives (15d-PGJ2, 17-EFOX-D5, 17-EFOX-D6 and 15-oxoeicosatetraenoic acid, 15-oxoETE)(Supplementary Fig 11a). In contrast, BME exchange reactions with adducted electrophiles were slower, a consequence of the koff reaction rate of Cys-EFOX and His-EFOX adducts. The time-dependence of these reactions further confirmed that biomolecule-adducted populations constituted ~50% of the total EFOX pool present in cell lysates (Supplementary Fig. 11b). To more specifically test the capability of EFOXs to react with nucleophilic residues of proteins, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized. GAPDH is a well-characterized target for electrophiles that becomes inactivated by s-nitrosation, oxidation or nucleophilic addition. Proteomic analysis revealed adducts between 17-EFOX-D5 and the Cys244, Cys149, His163 and His328 residues of GAPDH in vitro (Supplementary Fig. 12).
Figure 4. EFOXs form adducts with proteins and GSH following activation of RAW264.7.
a, Cell lysates from activated RAW264.7 cells were split into two groups: treatment with BME followed by protein precipitation with acetonitrile (“Total”) and protein precipitation followed by BME treatment (“Free + small molecule adducted”). b, Chemical structure and fragmentation pattern of GS-13-EFOX-D5. c, Chromatographic profiles and positive ion mass spectra of GSH adducts of 13-EFOX-D5 and 17-EFOX-D5 derived from standards (upper panels), cell medium (middle panel) and cell pellet (lower panel), N/A profiles correspond to non-activated cell samples. Grey chromatograms represent GS-17-EFOX-D5 (upper panel left) and GS-13-EFOX-D5 (upper panel right) standards. Fragments 345.3 and 523.3 were monitored in cell media and cell pellet samples respectively.
Since RES react with GSH, EFOXs were tested as substrates for glutathione S-transferase (GST) and the cellular formation of GS-EFOX products were evaluated. Incubation of EFOXs without or with GST resulted in EFOX-GSH adduction rates dependent on the amount of GST added, confirming that EFOXs are GST substrates (Supplementary Fig. 13). GS-EFOX adducts were measured in activated RAW264.7 cell lysates and media, and compared with synthetic standards of GS-17-EFOX-D5 and GS-17-EFOX-D6 (Fig. 4b–c and Supplementary Fig. 14). The fragmentation patterns and retention times observed for cellular GSH adducts of EFOX-D5 and EFOX-D6 corresponded with their synthetic homologues, confirming cellular GS-EFOX adduction. The addition of ASA enhanced the formation of GS-EFOX adducts to extents consistent with the concomitant increase in EFOX synthesis. GS-EFOX adducts were also found extracellularly, in the absence of cell lysis, suggesting multi-drug resistance protein transport.
Anti-inflammatory signaling actions of EFOXs
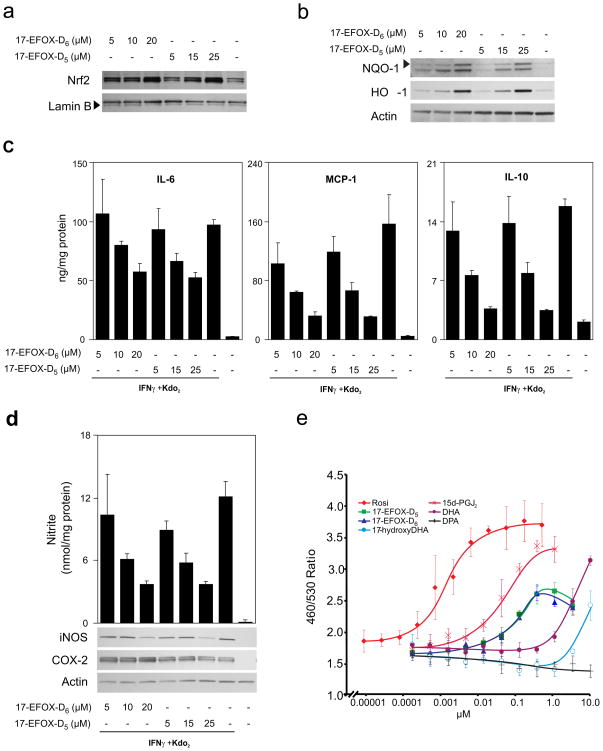
Many RES can promote expression of the Nrf2-dependent phase II genes24 by thiol-dependent modification of the Nrf2 inhibitor Keap1. 17-EFOX-D5 and 17- EFOX-D6 induced dose-dependent cellular Nrf2 nuclear accumulation and expression of the cytoprotective enzymes heme oxygenase 1 (HO-1) and NAD(P)H:quinone oxidoreductase 1 (NQO1) (Fig. 5a–b). Besides activating the Nrf2-dependent anti-oxidant response, 17-EFOX-D5 and 17- EFOX-D6 modulated the inflammatory response generated by Kdo2 and IFNγ treatment. 17-EFOX-D5 and 17-EFOX-D6 dose-dependently repressed IL-6, MCP-1 and IL-10 expression (Fig. 5c), inducible nitric oxide synthase (NOS-2) expression (Fig. 5d) and accumulation of the nitric oxide metabolites, nitrate (NO3−) and nitrite (NO2−), in the media of both RAW264.7 cells and BMDMs (Supplementary Fig. 15). COX-2 expression levels were not affected by EFOXs.
Figure 5. 17-EFOX-D6 and 17-EFOX-D5 modulate anti-oxidant and inflammatory responses.
a, b, RAW264.7 cells treated with 17-EFOX-D6 and 17-EFOX-D5 were harvested at 1 h for nuclear Nrf2 (a) and 18 h for of HO-1 and NQO-1 (b). c, d, RAW264.7 cells pretreated for 6 h with 17-EFOX-D6 and 17-EFOX-D5 were harvested 12 h post activation and IL-6, MCP-1, IL-10 levels, and nitrite levels were measured in the cell media and normalized to protein. d, iNOS and COX-2 levels were detected in cell lysates. e, PPARγ beta-lactamase reporter assays for Rosiglitazone, 17-EFOX-D6, 17-EFOX-D5, 15d-PGJ2, 17-hydroxyDHA, DPA, and DHA.
Oxo-FAs, including 15d-PGJ227, 5-oxoEPA, and the synthetic 4-oxoDHA28 covalently bind and activate the peroxisome proliferator-activated receptor γ (PPARγ). PPARγ beta-lactamase reporter assays revealed that 17-EFOX-D5 and 17-EFOX-D6 activated PPARγ within a physiological concentration range (Fig. 5e), with a slightly higher EC50 (~40 nM) than 15d-PGJ2 (~25 nM) and orders of magnitude lower than the EFOX precursor, 17-OH-DPA, DHA and DPA.
A BME “trapping” strategy revealed six COX-2-derived EFOXs, (as well as oxoETE, not shown) produced by activated macrophages. These ω-3 FA-derived species have not been described before as mediators of inflammation or as metabolic products of mammalian cells, although EFOX-E3 may correspond to the oxoETrE produced by plants29. Interestingly, concomitant monitoring of 15d-PGJ2 generation showed this species was below the limit of detection, implying that EFOXs may in some cases be responsible for effects often attributed to endogenous 15d-PGJ2.
EFOXs were discovered by a new chemical profiling strategy as negatively-charged hydrophobic molecules with reversible electrophilic activity. This BME-based method increased MS/MS sensitivity for diverse RES and standardized their behavior during MS/MS analysis. For example, oxo-FA derivatives do not fragment as well as their corresponding hydroxy-derivatives, preventing structural identification. In particular, EFOX-D4, EFOX-D5 and EFOX-D6 correspond to oxoDHA, oxoDPA and oxoDTA (with different isomers being favored by addition of ASA). EFOX-D3, EFOX-E3 and EFOX-E2 are derived from ω-6 and ω-9 FAs, with detailed structural characterization of these latter species not yet performed.
While COX-2 accounts for EFOX formation, additional mechanisms cannot be excluded. Concerning initial oxygenation, LOX and cytochrome p450 (CYP) monooxygenase activities can also catalyze FA hydroxylation9, 30. While the formation of monohydroxy-derivatives of FAs has already been described22, 31, further oxidation to the corresponding oxo-species has only been observed for hydroxy-ETA32. Moreover, despite an appreciation of oxoETE and oxo-octadecadienoic acid generation, similar 22-carbon species have not been described.
The enzymatic oxidation of hydroxyls on bioactive lipids has been generally viewed as a step in metabolic inactivation, but present data indicate that this reaction instead confers new biologic activity. This report also reveals an alternative reaction for the hydroxy-derivatives of ω-3 FAs that are proposed to be further oxidized by LOXs to resolvins and lipoxins. Alternatively, we observe that monohydroxy-FA derivatives can be readily converted to EFOXs by cellular dehydrogenases or by purified dehydrogenases in vitro. For example, 3a-hydroxysteroid dehydrogenases (3α-HSDs) converted 13- and 17-OH-DPA and 13- and 17-OH-DHA into corresponding EFOXs in the presence of NAD(P)+ in vitro (Supplementary Fig. 10). Members of the aldo/keto reductase superfamily, 3α-HSDs, regulate steroid hormone levels in mammals33. Other candidate dehydrogenases leading to EFOX generation could be the 5- and 15-hydroxyeicosanoid dehydrogenases (5- and 15-HEDH), which convert LOX products to 5-and 15-oxoETE32. The formation of 5-oxoETE is favored by a high NADP+:NADPH ratio, a condition symptomatic of cells under oxidative stress. Notably, while HEDH activity is present in myeloid cells, it is further induced following differentiation to macrophages using PMA32. Thus, 15-HEDH can confer electrophilic reactivity to EFOX precursors, since it catalyzes the oxidation of hydroxyl groups to carbonyl products for a wide range of substrates, including AA-derived prostaglandins, 15-hydroxyeicosatetraenoic acid, EPA-derived Resolvin E1 and DHA-derived Resolvin D1 at position 1734–36.
Approximately 50% of the EFOXs extracted from activated RAW264.7 cells were adducted to protein, but this value did not include EFOXs that were bound to small molecules such as GSH. These covalent adducts of EFOXs to protein and GSH revealed a capacity to modulate protein function and mediate electrophilic signaling by covalently binding to transcriptional regulators. The modulation of several signaling mechanisms by EFOXs confirmed a role as endogenously produced anti-inflammatory signaling mediators. 17- EFOX-D5 and 17- EFOX-D6 induced Nrf2-dependent translocation to the nucleus and the expression of two key Nrf2 target genes, HO-1 and NQO-1. Biologic concentrations of the 17-EFOX standards also activated PPARγ, implying potential anti-inflammatory actions through PPARγ-regulated genes37. Anti-inflammatory signaling properties of EFOXs were also observed when 17- EFOX-D5 and 17- EFOX-D6 dose-dependently inhibited IFNγ and Kdo2-induced cytokine production and iNOS expression. While recognized electrophile-sensitive signaling pathways were tested, it is probable that EFOXs display additional unique signaling profiles and receptors.
The present discovery gains physiological relevance, considering that EFOXs derived from ω-3 FA are produced by aspirin-enhanced COX-2 oxidation reactions. The beneficial roles of COX-2 and ω-3 FA in the resolution of inflammation and the demonstrably significant role of ω-3 FA in cardiovascular protection38 suggest that COX-2-derived EFOXs can contribute to a component of these actions.
Methods Summary
Cell Culture and treatment
RAW264.7 and THP-1 cell lines were obtained from ATCC (USA) and maintained according to ATCC guidelines. For treatment with EFOXs coupled with pro-inflammatory stimulation, 17- EFOX-D5 and 17- EFOX-D6 was followed by addition of Kdo2 and IFNγ at 6h.
Trans-alkylation reaction of electrophiles with BME
Upon thawing, lysates were exposed to BME (500 mM + internal standard, 5-OxoETE-d7 (1.25 ng/ml)) and incubated at 37 °C for 1 h in 50 mM phosphate buffer (pH= 7.4) as previously described19. Proteins were precipitated with cold acetonitrile and the supernatant was analyzed by HPLC-ESI-MS/MS.
HPLC-ESI-MS/MS
For experimental details see Methods. Samples were separated by reverse-phase HPLC. The analysis and quantification of BME adducts were performed using a hybrid triple quadrupole-linear ion trap mass spectrometer (4000 Q trap, Applied Biosystems/MDS Sciex) in the neutral loss (NL) scan mode, MRM scan mode, and the enhanced product ion analysis (EPI) mode. EFOXs were quantified using external synthetic standards, when available, and by comparing peak area ratios between analytes and a 5-OxoETE-d7 internal standard. EFOX values represented as percentages were normalized to internal standard and cell lysates protein concentration.
COX-2 reactions
COX-2 reactions were performed as described previously21, 22. The reactions were initiated by addition of FA (10 μM) and terminated by addition of ice-cold acetonitrile (9 x volume) and protein removed by centrifugation. Product formation was monitored by RP-HPLC-MS/MS in MRM mode following the loss of CO2 (m/z 345/301 and m/z 343/299 for OH-DPA and OH-DHA, respectively).
Nitrate/Nitrite measurement
Total nitrite and nitrate concentration was measured in cell culture media by Griess reaction using the Nitrate/Nitrite Colorimetric Assay Kit (Cayman Chemical).
Measurement of glutathione adducts
For experimental details see Methods. GS-adducts were analyzed by nano-LC-MS/MS using nanoACQUITY UltraPerformance LC coupled with Thermo-Fisher LTQ.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants, American Diabetes Association (FJS), and Ri MED Foundation, Italy (CC).
Footnotes
Conflict of Interest: BAF acknowledges financial interest in Complexa, Inc.
Nomenclature: To be consistent with prostaglandin and isoprostane nomenclature EFOX are designated according to the length of the carbon chain (eicosanoids and docosanoids) and “x” number of double bonds (EFOX-Dx and EFOX-Ex). In the case of regoisomers the number preceding the EFOX species indicates the location of the carbonyl group.
References
- 1.Connor WE. Importance of n-3 fatty acids in health and disease. Am J Clin Nutr. 2000;71:171S–175S. doi: 10.1093/ajcn/71.1.171S. [DOI] [PubMed] [Google Scholar]
- 2.Neuringer M, Anderson GJ, Connor WE. The essentiality of n-3 fatty acids for the development and function of the retina and brain. Annu Rev Nutr. 1988;8:517–541. doi: 10.1146/annurev.nu.08.070188.002505. [DOI] [PubMed] [Google Scholar]
- 3.Morris MC, Evans DA, Tangney CC, Bienias JL, Wilson RS. Fish consumption and cognitive decline with age in a large community study. Arch Neurol. 2005;62:1849–1853. doi: 10.1001/archneur.62.12.noc50161. [DOI] [PubMed] [Google Scholar]
- 4.Fedor D, Kelley DS. Prevention of insulin resistance by n-3 polyunsaturated fatty acids. Curr Opin Clin Nutr Metab Care. 2009;12:138–146. doi: 10.1097/MCO.0b013e3283218299. [DOI] [PubMed] [Google Scholar]
- 5.Dietary supplementation with n-3 polyunsaturated fatty acids and vitamin E after myocardial infarction: results of the GISSI-Prevenzione trial. Gruppo Italiano per lo Studio della Sopravvivenza nell’Infarto miocardico. Lancet. 1999;354:447–455. [PubMed] [Google Scholar]
- 6.Duda MK, et al. Fish oil, but not flaxseed oil, decreases inflammation and prevents pressure overload-induced cardiac dysfunction. Cardiovasc Res. 2009;81:319–327. doi: 10.1093/cvr/cvn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arita M, et al. Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J Exp Med. 2005;201:713–722. doi: 10.1084/jem.20042031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim EH, Surh YJ. 15-deoxy-Delta12,14-prostaglandin J2 as a potential endogenous regulator of redox-sensitive transcription factors. Biochem Pharmacol. 2006;72:1516–1528. doi: 10.1016/j.bcp.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui T, et al. Nitrated fatty acids: Endogenous anti-inflammatory signaling mediators. J Biol Chem. 2006;281:35686–35698. doi: 10.1074/jbc.M603357200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musiek ES, et al. Electrophilic cyclopentenone neuroprostanes are anti-inflammatory mediators formed from the peroxidation of the omega-3 polyunsaturated fatty acid docosahexaenoic acid. J Biol Chem. 2008;283:19927–19935. doi: 10.1074/jbc.M803625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin D, Saleh S, Liebler DC. Reversibility of covalent electrophile-protein adducts and chemical toxicity. Chem Res Toxicol. 2008;21:2361–2369. doi: 10.1021/tx800248x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satoh T, et al. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophillic phase II inducers. Proc Natl Acad Sci U S A. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haridas V, et al. Avicins: triterpenoid saponins from Acacia victoriae (Bentham) induce apoptosis by mitochondrial perturbation. Proc Natl Acad Sci U S A. 2001;98:5821–5826. doi: 10.1073/pnas.101619098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinkova-Kostova AT, Cory AH, Bozak RE, Hicks RJ, Cory JG. Bis(2-hydroxybenzylidene)acetone, a potent inducer of the phase 2 response, causes apoptosis in mouse leukemia cells through a p53-independent, caspase-mediated pathway. Cancer Lett. 2007;245:341–349. doi: 10.1016/j.canlet.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Thornalley PJ. Isothiocyanates: mechanism of cancer chemopreventive action. Anticancer Drugs. 2002;13:331–338. doi: 10.1097/00001813-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki M, et al. Role of 15-deoxy delta(12,14) prostaglandin J2 and Nrf2 pathways in protection against acute lung injury. Am J Respir Crit Care Med. 2005;171:1260–1266. doi: 10.1164/rccm.200406-755OC. [DOI] [PubMed] [Google Scholar]
- 19.Schopfer FJ, et al. Detection and quantification of protein adduction by electrophilic fatty acids: mitochondrial generation of fatty acid nitroalkene derivatives. Free Radic Biol Med. 2009;46:1250–1259. doi: 10.1016/j.freeradbiomed.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean-Louis Luche LR-HaPC. Reduction of natural enones in the presence of cerium trichloride. J Chem Soc, Chem Commun. 1978:601–602. [Google Scholar]
- 21.Gierse JK, Koboldt CM. In: Cyclooxygenase Assays. ESJ, editor. John Wiley and Sons, Inc; 1998. [Google Scholar]
- 22.Serhan CN, et al. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med. 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa T, Esterbauer H, Sies H. Role of cardiac glutathione transferase and of the glutathione S-conjugate export system in biotransformation of 4-hydroxynonenal in the heart. J Biol Chem. 1986;261:1576–1581. [PubMed] [Google Scholar]
- 24.Levonen AL, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batthyany C, et al. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J Biol Chem. 2006;281:20450–20463. doi: 10.1074/jbc.M602814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy RC, Zarini S. Glutathione adducts of oxyeicosanoids. Prostaglandins Other Lipid Mediat. 2002;68–69:471–482. doi: 10.1016/s0090-6980(02)00049-7. [DOI] [PubMed] [Google Scholar]
- 27.Waku T, Shiraki T, Oyama T, Morikawa K. Atomic structure of mutant PPARgamma LBD complexed with 15d-PGJ2: novel modulation mechanism of PPARgamma/RXRalpha function by covalently bound ligands. FEBS Lett. 2009;583:320–324. doi: 10.1016/j.febslet.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Itoh T, et al. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat Struct Mol Biol. 2008:924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch T, Hoskovec M, Boland W. Efficient syntheses of (10E,12Z,15Z)-9-oxo- and (9Z,11E,15E)-13-oxo-octadecatrienoic acids; two stress metabolites of wounded plants. Tetrahedron. 2002;58:3271–3274. [Google Scholar]
- 30.Stark K, Wongsud B, Burman R, Oliw EH. Oxygenation of polyunsaturated long chain fatty acids by recombinant CYP4F8 and CYP4F12 and catalytic importance of Tyr-125 and Gly-328 of CYP4F8. Arch Biochem Biophys. 2005;441:174–181. doi: 10.1016/j.abb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Schwartzman ML, Falck JR, Yadagiri P, Escalante B. Metabolism of 20-hydroxyeicosatetraenoic acid by cyclooxygenase. Formation and identification of novel endothelium-dependent vasoconstrictor metabolites. J Biol Chem. 1989;264:11658–11662. [PubMed] [Google Scholar]
- 32.Erlemann KR, et al. Regulation of 5-hydroxyeicosanoid dehydrogenase activity in monocytic cells. Biochem J. 2007;403:157–165. doi: 10.1042/BJ20061617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penning TM, et al. Mammalian 3 alpha-hydroxysteroid dehydrogenases. Steroids. 1997;62:455–456. doi: 10.1016/s0039-128x(97)00025-1. [DOI] [PubMed] [Google Scholar]
- 34.Wei C, Zhu P, Shah SJ, Blair IA. 15-Oxo-Eicosatetraenoic Acid, a Metabolite of Macrophage 15-Hydroxyprostaglandin Dehydrogenase that Inhibits Endothelial Cell Proliferation. Mol Pharmacol. 2009 doi: 10.1124/mol.109.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arita M, et al. Metabolic inactivation of resolvin E1 and stabilization of its anti-inflammatory actions. J Biol Chem. 2006;281:22847–22854. doi: 10.1074/jbc.M603766200. [DOI] [PubMed] [Google Scholar]
- 36.Sun YP, et al. Resolvin D1 and its aspirin-triggered 17R epimer. Stereochemical assignments, anti-inflammatory properties, and enzymatic inactivation. J Biol Chem. 2007;282:9323–9334. doi: 10.1074/jbc.M609212200. [DOI] [PubMed] [Google Scholar]
- 37.Malur A, et al. Deletion of PPAR gamma in alveolar macrophages is associated with a Th-1 pulmonary inflammatory response. J Immunol. 2009;182:5816–5822. doi: 10.4049/jimmunol.0803504. [DOI] [PubMed] [Google Scholar]
- 38.Lavie CJ, Milani RV, Mehra MR, Ventura HO. Omega-3 polyunsaturated fatty acids and cardiovascular diseases. J Am Coll Cardiol. 2009;54:585–594. doi: 10.1016/j.jacc.2009.02.084. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez MN, Trujillo M, Radi R. Peroxynitrite formation from biochemical and cellular fluxes of nitric oxide and superoxide. Methods in Enzymology. 2002;359:353–366. doi: 10.1016/s0076-6879(02)59198-9. [DOI] [PubMed] [Google Scholar]
- 40.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 41.Davies JQ, Gordon S. Isolation and culture of murine macrophages. Methods Mol Biol. 2005;290:91–103. doi: 10.1385/1-59259-838-2:091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.