Abstract
Anhaptoglobinemic patients run the risk of severe anaphylactic transfusion reaction because they produce serum haptoglobin antibodies. Being homozygous for the haptoglobin gene deletion allele (HPdel) is the only known cause of congenital anhaptoglobinemia, and detection of HPdel before transfusion is important to prevent anaphylactic shock. In this study, we developed a loop-mediated isothermal amplification (LAMP)-based screening for HPdel. Optimal primer sets and temperature for LAMP were selected for HPdel and the 5′ region of the HP using genomic DNA as a template. Then, the effects of diluent and boiling on LAMP amplification were examined using whole blood as a template. Blood samples diluted 1:100 with 50 mmol/L NaOH without boiling gave optimal results as well as those diluted 1:2 with water followed by boiling. The results from 100 blood samples were fully concordant with those obtained by real-time PCR methods. Detection of the HPdel allele by LAMP using alkaline-denatured blood samples is rapid, simple, accurate, and cost effective, and is readily applicable in various clinical settings because this method requires only basic instruments. In addition, the simple preparation of blood samples using NaOH saves time and effort for various genetic tests.
The absence of a serum protein such as IgA or haptoglobin (Hp) is one of the factors that can lead to anaphylactic transfusion reactions due to production of serum antibodies against the absent protein after a transfusion.1 At present, a homozygous deletion of the haptoglobin gene (HPdel) is the only known cause of anhaptoglobinemia.
Human Hp has a genetic polymorphism of two codominant alleles, HP1 and HP2, that give rise to the three common phenotypes, Hp1, Hp2-1, and Hp2.2,3 The HP2 allele appears to have occurred by a 1.7-kb intragenic duplication of exons 3 and 4 of the HP1 allele (Figure 1A). Anomalous inheritance of the Hp phenotypes was encountered during determinations of parentage, and HPdel was identified by genetic analyses of one such family in Japan.4 The HPdel allele lacks an approximately 28-kb segment of chromosome 16 extending from the promoter region of the HP gene to intron 4 of the haptoglobin-related gene (HPR) (Figure 1A).4,5
Figure 1.
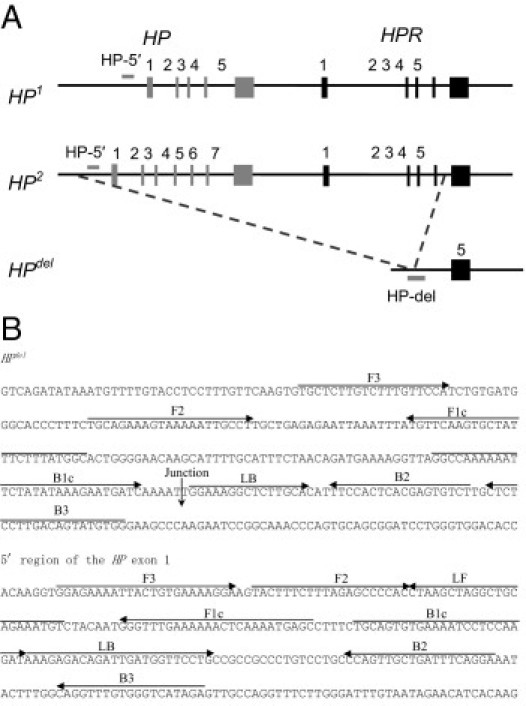
Gene structure of HP1, HP2, and HPdel, and positions of the target regions (A) and locations of two primer sets used in the LAMP reaction (B). The position and direction of each primer are shown by arrows. A downward arrow indicates the junction of gene deletion of the HPdel.
The HPdel allele has been found only in East and Southeast Asian populations (Chinese, Korean, Japanese, Mongols, Thais, and Indonesians), not in African, West and South Asian, and European populations so far.5–9 Detection of homozygosity for HPdel before blood transfusion or blood component infusion is important to prevent severe side effects of transfusion because washed red blood cells and platelet concentrate do not cause transfusion-related anaphylactic reactions.10 Recently, we established two real-time PCR methods for detection of HPdel by use of a 5′-nuclease assay using dual-labeled (TaqMan; Applied Biosystems, Foster City, CA) probes and SYBR Green I (Invitrogen, Carlsbad, CA).11,12 These methods are rapid, robust, and suitable for high-throughput analysis but require a real-time PCR apparatus.
A technique called loop-mediated isothermal amplification (LAMP) of DNA has recently been developed.13 LAMP employs a DNA polymerase with strand-displacement activity; the reaction proceeds when the forward inner primer anneals to the target DNA and the first strand is synthesized, and then the outer forward primer hybridizes and displaces the first strand, forming a loop structure at one end. The single-stranded DNA serves as a template for backward inner primer–initiated DNA synthesis and subsequent outer backward–primed strand-displacement DNA synthesis. The stem loop formed acts as a template, and the final products are stem-loop DNAs with several inverted repeats of the target DNA. Loop primers (forward and backward), which are additional primers designed to anneal at the loop structure (between F1c and F2, B1c and B2) in LAMP amplicons, can accelerate and enhance the sensitivity of the LAMP reaction. In DNA polymerization by DNA polymerase, pyrophosphate ions are released from dNTP as a by-product, which react with magnesium ions in the LAMP reaction buffer, yielding an insoluble white precipitate.13,14 This method amplifies DNA with high specificity, efficiency, and rapidity under isothermal conditions, and the advantages are i) easy identification of positive reaction by visual inspection of turbidity, and ii) requirement of only a heating block or water bath.13 In this study, we developed a LAMP reaction for detection of HPdel with the aim of establishing a feasible detection method for HPdel in clinical diagnostic laboratories.
Materials and Methods
The study protocol was approved by the ethics committee of Kurume University School of Medicine.
LAMP Amplification
LAMP primers, which specifically amplify two regions, the region encompassing the HPdel breakpoint (HP-del) and the 5′ region of HP exon 1 (HP-5′), were designed with the aid of PrimerExplorer V4 (http://primerexplorer.jp/elamp4.0.0, last accessed December 2, 2010) and synthesized by Operon Biotechnologies (Tokyo, Japan). The locations and sequences of primers that brought the optimal results are presented in Figure 1 and Table 1. The LAMP reaction was performed in a volume of 25 μL containing 12.5 μL of 2× Reaction Mix, 1 μL of BstDNA Polymerase (Eiken, Tokyo, Japan), 40 pmol each of the forward inner and the backward inner primers, 20 pmol each of the backward loop primer and/or the forward loop primer, 5 pmol each of the outer forward and the outer backward primers, and 2 μL of the template (diluted and/or heated venous whole blood or genomic DNA) in a Loopamp Reaction tube (Eiken). Wells without a template were included as negative controls. Reactions for HP-del and HP-5′ were conducted in discrete tubes. The reaction mixture was incubated at 60°C to 65°C for 60 minutes, and turbidity was measured every 6 seconds using a real-time turbidity meter (Loopamp EXIA; Eiken).15 The reaction was followed by incubation at 80°C for 5 minutes for inactivation of the enzyme. At least three assays using different samples were performed for every group for assay validation. The turbidities were also assessed by visual inspection. Fluorescent Detection Reagent (Eiken) was added to the reaction mix before the reaction to inspect the amplification visually by the color.
Table 1.
Sequences of Primers Used in the LAMP Assay
| Primer sequence | |
|---|---|
| Set for HPdel (HP-del) | |
| F3 | 5′-TGCTCTTGTCTTTGTTCCA-3′ |
| B3 | 5′-CCACATACTGTCAAGGAGAG-3′ |
| FIP (F1c + F2) | 5′-GCCATAAAGAAATAGCACTTGAACATGCAGAAAGTAAAAATTGCCT-3′ |
| BIP (B1c + B2) | 5′-GGCCAAAAAATTCTATATAAAGAATGATCAGACACTCGTGAGTGGAA-3′ |
| LB | 5′-GGAAAGGCTCTTGCA-3′ |
| Set for 5′ region of HP (HP-5′) | |
| F3 | 5′-GGAGAAAATTACTGTGAAAAGGA-3′ |
| B3 | 5′-TCTATGACCCACAAACCTG-3′ |
| FIP (F1c + F2) | 5′-GCTCATTTTGAGTTTTTTCAAACCCTACTTTCTTTAGAGCCCCAC-3′ |
| BIP (B1c + B2) | 5′-TGCAGTGTGAAAATCCTCCAAGATTCCTGAAATCAGCAACTGG-3′ |
| LF | 5′-ACATTTCTGCAGCCTAGCTTAG-3′ |
| LB | 5′-AAAGAGACAGATTGATGGTTCCTG-3′ |
Preparation of Templates
To determine the optimal primer sets and temperature for detection of HPdel, LAMP reactions were performed using 10 ng of genomic DNAs from known genotypes lacking HPdel (HP/HP), heterozygous for HPdel (HP/HPdel), and homozygous for HPdel (HPdel/HPdel) as templates. For determination of the detection limit of the method, a dilution series of HP/HPdel DNA, 100, 10, 1, 0.1, 0.01, and 0.001 ng/reaction, was used. To determine analytical sensitivity, a dilution series of HP/HPdel DNA, 0.2, 0.1, 0.05, 0.025, and 0.0125 for Hp-del and 0.02, 0.01, 0.005, 0.0025, and 0.00125 for Hp-5′ (in nanograms/reaction), was also examined. The detection limit was estimated by using probit analysis (SAS Statistical Software Package, SAS, Cary, NC). In addition, 1:10, 1:30, 1:100, 1:300, 1:1000, and 1:3000 dilutions of whole-blood samples of HP/HPdel individuals with 50 mmol/L NaOH were prepared for evaluation of NaOH dilution. For investigation of the effect of diluents (water or NaOH) and heat treatment (98°C for 5 minutes), we prepared blood samples diluted 1:2 with water and heated, 1:10 with water and heated, 1:100 with NaOH and heated, and 1:100 with NaOH. Denaturation (boiling) of diluted blood samples was performed in a block incubator and subsequent centrifugation at 12,000 rpm for 3 minutes. We also determined the optimal proportion of NaOH in the mixture by using 1, 2, 4, and 8 μL of 1:100 diluted samples with 50 mmol/L NaOH in a 25-μL mixture.
Confirmation of Specific Amplification of LAMP Products
After amplification, 2 μL of reaction mixtures were electrophoresed on 1.5% agarose gel and stained by ethidium bromide. Some fragments were gel purified and inserted into a pUC118 plasmid using a Mighty Cloning Kit (TaKaRa, Shiga, Japan). Sequence analyses of three clones for each of band were performed using BigDye Terminators v1.1 Cycle Sequencing Kit and ABI PRISM 310 Genetic Analyzer (Life Technologies Japan, Tokyo, Japan).
Results
To determine the optimal primer sets and temperature for detection of HPdel, LAMP reactions were performed using genomic DNA of HP/HP, HP/HPdel, and HPdel/HPdel as templates. The optimal results were obtained by incubation at 61.5°C for both amplifications of HP-del and HP-5′. An amplification curve of HP-del was obtained with HP/HPdel and HPdel/HPdel, whereas that of HP-5′ was obtained with HP/HP and HP/HPdel (Figure 2A). No increase in turbidity was detected in the negative controls of HP-del and HP-5′. As expected, the HP-5′ results obtained using genomic DNA of HP1/HP1, HP1/HP2, and HP2/HP2 were indistinguishable from each other (n = 3, data not shown), and no amplifications were observed using HP-del in these genomic DNAs. In addition, only HP-5′ tubes were positive when using DNA of three Ghanaians with acquired anhaptoglobinemia (without any causal mutations in the promoter and coding regions) as templates.6 The threshold time required for the turbidity of the solution to develop was about 25 minutes for HP-del and about 23 minutes for HP-5′. After amplification, all of the positive reactions produced white precipitates and a characteristic ladder of multiple bands on an agarose gel, whereas no bands were detected in negative controls. In addition, sequence analyses revealed the target HP sequences in the plasmids inserted the LAMP products (data not shown). These results confirmed the specific amplifications of the HPdel and 5′ region of HP exon 1.
Figure 2.
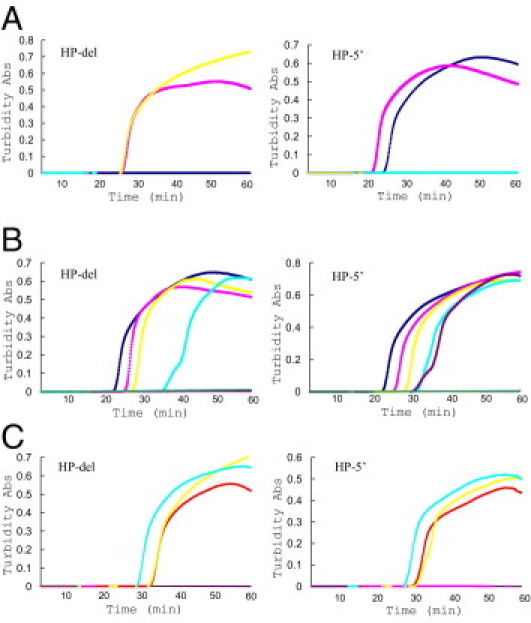
Results of the LAMP reaction for HPdel (left) and the 5′ region of HP exon 1 (right). A: Validation of specificity using DNA samples of known genotypes. Real-time amplification curves with turbidity detection of triplicate experiments with HP/HP, HP/HPdel, and HPdel/HPdel. The results are shown by navy (HP/HP), pink (HP/HPdel), and yellow (HPdel/HPdel). Negative control is shown by aqua. B: Validation of sensitivity using DNA samples of known genotypes. Amplification curves of triplicate experiments with various amounts of initial template DNA of HP/HPdel, 100, 10, 1, 0.1, 0.01, and 0.001 ng/reaction. The results are shown by navy (100 ng), pink (10 ng), yellow (1 ng), aqua (0.1 ng), purple (0.01 ng), and red (0.001 ng). A negative control is shown by green. C: Comparison of various preparations of blood samples. The results obtained from four different sample preparations are shown by red (1:2 dilution with water followed by heat), pink (1:10 dilution with water followed by heat), yellow (1:100 dilution with NaOH followed by heat), and aqua (1:100 dilution with NaOH). Negative control is shown by purple.
We then determined the sensitivity of the method by a dilution series of HP/HPdel DNA and found that the detection limit of HP-del was about 0.1 ng and that of HP-5′ was about 0.01 ng (Table 2 and Figure 2B). The threshold time (Tt) decreased roughly in proportion to the quantity of the initial template DNA for HP-del but not always for HP-5′. To determine analytical sensitivity, a total of 20 replicates were tested at each concentration (Table 3). Probit analysis showed that the analytical sensitivity for HP-del was estimated at 0.118 ng/reaction with a 95% confidence interval (CI) between 0.096 and 0.161 ng/reaction, and that for HP-5′, it was 0.021 ng/reaction with a 95% CI between 0.018 and 0.027 ng/reaction. No increase in turbidity was observed in the tubes of HP-del and Hp-5′ when using 100 ng/μL of DNA of HP/HP and HPdel/HPdel, respectively. Thus, the analytical specificity of the present method seems to be 100%.
Table 2.
Threshold Time Required for Turbidity of Solution to Exceed a Given Value with Various Templates
| Sample preparation | Mean (SD) Tt value (min) |
|
|---|---|---|
| HP-del | HP-5′ | |
| Detection limit of purified DNA | ||
| 100 ng | 24.9 (4.0) | 23.7 (2.4) |
| 10 ng | 26.8 (4.2) | 26.3 (2.8) |
| 1 ng | 30.9 (5.5) | 30.0 (4.0) |
| 0.1 ng | 39.5 (5.3) | 33.0 (5.4) |
| 0.01 ng | — | 32.7 (5.4) |
| 0.001 ng | — | — |
| Serial dilution by NaOH | ||
| 1:10 | * | * |
| 1:30 | 28.6 (1.5) | 29.9 (2.4) |
| 1:100 | 34.0 (3.7) | 32.6 (3.9) |
| 1:300 | 41.5 (10.1) | 37.4 (2.5) |
| 1:1000 | 45.8 (9.2) | 37.9 (7.9) |
| 1:3000 | * | * |
| Water- or NaOH-diluted with/without heat | ||
| ×2 with water and heat | 31.8 (3.3) | 33.3 (4.5) |
| ×10 with water and heat | — | * |
| ×100 with NaOH and heat | 35.7 (4.0) | 36.7 (5.4) |
| ×100 with NaOH | 31.4 (2.9) | 32.2 (5.1) |
| Serial proportion of NaOH | ||
| 1 | 33.5 (1.0) | 35.4 (6.4) |
| 2 | 35.4 (2.3) | 36.0 (3.9) |
| 4 | 42.4 (2.8) | 42.8 (8.4) |
| 8 | — | — |
Note: — represents the group in which turbidity was not observed, and asterisks represent the group in which turbidity was not consistently observed.
Tt, threshold time.
Table 3.
Limit of Detection Determination of LAMP Reactions for HP-del and HP-5′
| HP-del |
HP-5′ |
||||||
|---|---|---|---|---|---|---|---|
| Amount of DNA, ng | Number tested | Number positive | Positive rate, % | Amount of DNA, ng | Number tested | Number positive | Positive rate, % |
| 0.2 | 20 | 20 | 100.0 | 0.05 | 20 | 20 | 100.0 |
| 0.1 | 20 | 16 | 80.0 | 0.02 | 20 | 17 | 85.0 |
| 0.05 | 20 | 13 | 65.0 | 0.01 | 20 | 12 | 60.0 |
| 0.025 | 20 | 5 | 25.0 | 0.005 | 20 | 3 | 15.0 |
| 0.0125 | 20 | 3 | 15.0 | 0.0025 | 20 | 0 | 0.0 |
| 0 | 20 | 0 | 0.0 | 0 | 20 | 0 | 0.0 |
Recent studies have demonstrated that heat-treated blood samples are applicable for LAMP (LAMP-HB) as a template.16,17 This time- and cost-saving method is now in common use for LAMP analysis. On the other hand, a blood sample diluted with 50 mmol/L NaOH (and heated) is used as a template for our real-time PCR for diagnosis of HPdel and isothermal single nucleotide polymorphism genotyping.11,12,18 We first prepared serial dilutions to identify the suitable dilution range, and turbidity was consistently observed in the tubes with dilutions from 1:30 to 1:1000 for both amplifications. The Tt increased roughly in proportion to the dilution ratio for HP-del, but not for HP-5′ (Table 2). A failure of amplification or an irregular curve was sometimes observed in the tubes containing 1:10 diluted bloods. This may due to incomplete lysis of cells by NaOH and subsequent formation of heat-denatured blood component(s) such as hemoglobin during the reaction. Use of such low dilutions produces a risk of false-positive results. In addition, the color of the mixture itself is brownish, making it difficult to evaluate the result visually. We also did not always observe positive results using 1:3000 dilutions of bloods. The reaction mixture with a 1:1000 dilution is estimated to contain about 30 pg of DNA. This sensitivity is consistent with that obtained using purified DNA and is comparable to that of real-time PCR (data not shown). These results suggested that blood sample dilutions between 1:30 and 1:1000 provide a good template for this LAMP reaction. Thus, we used 1:100 dilution of blood samples for further studies. Further, no change was observed in the optimal temperature for both amplifications when using NaOH-diluted templates instead of purified DNA (data not shown). We also compared the results using frozen bloods with those of freshly drawn blood samples and observed similar amplifications in both types of samples (data not shown).
We next investigated the effect of diluents (water or NaOH) and heat treatment. As shown in Figure 2C and Table 2, the shortest Tt was obtained using 1:100 NaOH–diluted blood without heat treatment (LAMP-NaOH) and LAMP-HB (1:2). On the other hand, heat following NaOH dilution decreased the efficiency of the LAMP reaction.
We then tested the concentration of NaOH in the mixture, and the inhibitory effect of the NaOH concentration on enzyme activity was observed (Table 2). Less than 8 mmol/L NaOH seems desirable as a final concentration.
We then examined blood samples from 100 patients who were scheduled for blood transfusion at Kurume University Hospital. Before the LAMP amplification, detection assays for the HPdel were performed by the TaqMan-based and SYBR Green I–based real-time PCR methods, and the quality of the samples were checked. Seven of 100 samples were HP/HPdel, and 93 samples were HP/HP. In the TaqMan assay, the mean (SD) threshold cycle (Ct) values were 31.4 (0.8) for HP-5′ and 35.5 (0.8) for HP-del, and the 90th percentile for the Ct for HP-5′ was 32.4. The mean (SD) Tt value was 29.8 (2.9) minutes, and the 90th percentile for the Tt was 32.4 minutes for HP-5′ and 31.9 (3.0) minutes for HP-del in the LAMP amplification. The zygosity of the HPdel of every sample was fully consistent with real-time PCR results.11,12 Amplifications were inspected visually by not only the turbidity, but also the color with the addition of Fluorescent Detection Reagent (Eiken) to the reaction mix before the reaction without the risk of contaminating products by opening the tubes as required when using SYBR Green I.13 We also used a frozen sample of HPdel/HPdel blood and got a positive result only in the HP-del tube (Figure 3).
Figure 3.
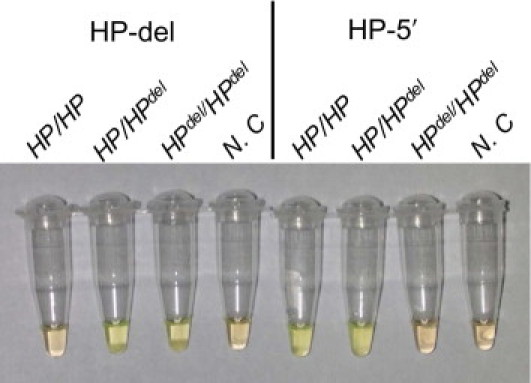
Evaluation of results by inspection of colors of reaction mixtures. One microliter of Fluorescent Detection Reagent (Eiken) was included in a 25-μL reaction mixture before the amplification reaction. Green and orange indicate positive and negative reactions, respectively. A 50 mmol/L NaOH-diluted blood sample of an HP/HP individual was green in the tube for HP-5′ and orange in the tube for the HP-del, that of HP/HPdel was green in both tubes, whereas that of HPdel/HPdel was green in the tube for HP-del and orange in the tube for the HP-5′. Negative controls (N.C) were orange in both tubes.
Discussion
We established rapid detection method of the HPdel by LAMP reaction using NaOH-diluted whole-blood samples. Although the results obtained by using both 1:100 NaOH-diluted blood without heat treatment (LAMP-NaOH) and LAMP-HB (1:2) were comparable, LAMP-NaOH was faster than LAMP-HB. Unlike PCR, which needs a heat-denaturing step, DNA amplification in the LAMP reaction is preceded by strand separation under isothermal conditions using betaine, which destabilizes the DNA helix. However, Njiru et al reported that preheating the template increased the efficiency of the assay because preheating produced a faster and/or a greater amount of strand separation, which translated into a far more rapid assay.14 Alkaline treatment also seems to promote single-stranded DNA formation and thus to facilitate the LAMP reaction. Accordingly, this treatment might be used in a wide variety of experiments as well as heat denaturation.
The two methods based on real-time PCR have two advantages over the LAMP method in the present study: we need only one tube per sample, and they are applicable to high-throughput analyses. Although two reaction tubes are needed for each sample, the LAMP method is cost effective and suitable for running only a few tests. Only about 1 hour after taking blood from the subject is required for allele determination, and this method seems to be better adapted to clinical diagnosis of patients before transfusion in clinical laboratories to prevent anaphylactic shock caused by anti-Hp antibody. In addition, the simple sample preparation using 50 mmol/L NaOH, LAMP-NaOH, may be suitable for various LAMP-based genotyping such as pharmacogenetics and even for diagnosis of infectious diseases.
Acknowledgments
We thank Ms. Katherine Ono for the English editing of this manuscript.
Footnotes
Supported by grants-in-aid for Scientific Research from the Ministry of Health, Labour and Welfare of Japan.
References
- 1.Eastlund T. Vasoactive mediators and hypotensive transfusion reactions. Transfusion. 2007;47:369–372. doi: 10.1111/j.1537-2995.2007.01154.x. [DOI] [PubMed] [Google Scholar]
- 2.Maeda N., Yang F., Barnett D.R., Bowman B.H., Smithies O. Duplication within the haptoglobin Hp2 gene. Nature. 1984;309:131–135. doi: 10.1038/309131a0. [DOI] [PubMed] [Google Scholar]
- 3.Carter K., Worwood M. Haptoglobin: a review of the major allele frequencies worldwide and their association with diseases. Int J Lab Hematol. 2007;29:92–110. doi: 10.1111/j.1751-553X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- 4.Koda Y., Soejima M., Yoshioka N., Kimura H. The haptoglobin-gene deletion responsible for anhaptoglobinemia. Am J Hum Genet. 1998;62:245–252. doi: 10.1086/301701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koda Y., Watanabe Y., Soejima M., Shimada E., Nishimura M., Morishita K., Moriya S., Mitsunaga S., Tadokoro K., Kimura H. Simple PCR detection of haptoglobin gene deletion in anhaptoglobinemic patients with antihaptoglobin antibody that causes anaphylactic transfusion reactions. Blood. 2000;95:1138–1143. [PubMed] [Google Scholar]
- 6.Teye K., Quaye I.K., Koda Y., Soejima M., Tsuneoka M., Pang H., Ekem I., Amoah A.G., Adjei A., Kimura H. A-61C and C-101G Hp gene promoter polymorphisms are, respectively, associated with ahaptoglobinaemia and hypohaptoglobinaemia in Ghana. Clin Genet. 2003;64:439–443. doi: 10.1034/j.1399-0004.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 7.Cox S.E., Doherty C., Atkinson S.H., Nweneka C.V., Fulford A.J., Ghattas H., Rockett K.A., Kwiatkowski D.P., Prentice A.M. Haplotype association between haptoglobin (Hp2) and Hp promoter SNP (A-61C) may explain previous controversy of haptoglobin and malaria protection. PLoS ONE. 2007;2:e362. doi: 10.1371/journal.pone.0000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soejima M., Koda Y., Fujihara J., Takeshita H. The distribution of haptoglobin-gene deletion (Hpdel) is restricted to East Asians. Transfusion. 2007;47:1948–1950. doi: 10.1111/j.1537-2995.2007.01467.x. [DOI] [PubMed] [Google Scholar]
- 9.Shimada E., Odagiri M., Chaiwong K., Watanabe Y., Anazawa M., Mazda T., Okazaki H., Juji T., O'Charoen R., Tadokoro K. Detection of Hpdel among Thais, a deleted allele of the haptoglobin gene that causes congenital haptoglobin deficiency. Transfusion. 2007;47:2315–2321. doi: 10.1111/j.1537-2995.2007.01473.x. [DOI] [PubMed] [Google Scholar]
- 10.Nishiki S., Hino M., Kumura T., Hashimoto S., Ohta K., Yamane T., Takubo T., Tatsumi N., Kitagawa S., Kamitani T., Watanabe Y., Shimada E., Juji T., Iida S. Effectiveness of washed platelet concentrate and red cell transfusions for a patient with anhaptoglobinemia with antihaptoglobin antibody. Transfus Med. 2002;12:71–73. doi: 10.1046/j.1365-3148.2002.00350.x. [DOI] [PubMed] [Google Scholar]
- 11.Soejima M., Koda Y. Rapid real-time PCR detection of HPdel directly from diluted blood samples. Clin Chem. 2008;54:1095–1096. doi: 10.1373/clinchem.2008.103747. [DOI] [PubMed] [Google Scholar]
- 12.Soejima M., Tsuchiya Y., Egashira K., Kawano H., Sagawa K., Koda Y. Development and validation of a SYBR Green I-based real-time polymerase chain reaction method for detection of haptoglobin gene deletion in clinical materials. Transfusion. 2010;50:1322–1327. doi: 10.1111/j.1537-2995.2009.02581.x. [DOI] [PubMed] [Google Scholar]
- 13.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Njiru Z.K., Mikosza A.S., Armstrong T., Enyaru J.C., Ndung'u J.M., Thompson A.R. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2:e147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori Y., Kitao M., Tomita N., Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Poon L.L., Wong B.W., Ma E.H., Chan K.H., Chow L.M., Abeyewickreme W., Tangpukdee N., Yuen K.Y., Guan Y., Looareesuwan S., Peiris J.S. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 17.Cheng S.H., Kwan P., Ng H.K., Ng M.H. New testing approach in HLA genotyping helps overcome barriers in effective clinical practice. Clin Chem. 2009;55:1568–1572. doi: 10.1373/clinchem.2009.127894. [DOI] [PubMed] [Google Scholar]
- 18.Mitani Y., Lezhava A., Kawai Y., Kikuchi T., Oguchi-Katayama A., Kogo Y., Itoh M., Miyagi T., Takakura H., Hoshi K., Kato C., Arakawa T., Shibata K., Fukui K., Masui R., Kuramitsu S., Kiyotani K., Chalk A., Tsunekawa K., Murakami M., Kamataki T., Oka T., Shimada H., Cizdziel P.E., Hayashizaki Y. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]


