Abstract
MicroRNAs (miRNAs) are small regulatory RNAs found in diverse eukaryotic lineages. In plants, a minority of annotated MIRNA gene families are conserved between plant families, while the majority are family- or species-specific, suggesting that most known MIRNA genes arose relatively recently in evolutionary time. Given the high proportion of young MIRNA genes in plant species, new MIRNA families are likely spawned and then lost frequently. Unlike highly conserved, ancient miRNAs, young miRNAs are often weakly expressed, processed imprecisely, lack targets, and display patterns of neutral variation, suggesting that young MIRNA loci tend to evolve neutrally. Genome-wide analyses from several plant species have revealed that variation in miRNA foldback expression, structure, processing efficiency, and miRNA size have resulted in the unique functionality of MIRNA loci and resulting miRNAs. Additionally, some miRNAs have evolved specific properties and functions that regulate other transcriptional or posttranscriptional silencing pathways. The evolution of miRNA processing and functional diversity underscores the dynamic nature of miRNA-based regulation in complex regulatory networks.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small RNAs found in plants, animals, and other diverse eukaryotes as well as a number of DNA viruses. Plant miRNAs range in size from 20 to 24 nucleotides and mediate gene silencing at the posttranscriptional level. Primary miRNA transcripts (pri-miRNAs) are generally RNA polymerase II transcripts that contain imperfect, self-complementary foldback regions. In animals, the pri-miRNA transcript is first processed by the RNase III domain–containing protein Drosha in association with the RNA binding protein encoded by DiGeorge syndrome Critical Region gene 8 (DGCR8), which binds to the lower stem region of MIRNA foldbacks (Carthew and Sontheimer, 2009). An average animal pri-miRNA contains a 33-bp stem with a terminal loop and two single-stranded flanking regions (Winter et al., 2009). The double-stranded stem and flanking regions are both important for DGCR8 binding and subsequent Drosha cleavage ~11 bp from the junction of single- and double-stranded RNAs (Zeng and Cullen, 2005; Zeng et al., 2005; Han et al., 2006; Kim et al., 2009). Processed miRNA precursors (pre-miRNA) are exported from the nucleus and are cleaved ~22 bp from the Drosha processing site by the RNase III domain–containing protein Dicer. By contrast, plants have no Drosha homolog; rather, the plant Dicer homolog, DICER-LIKE1 (DCL1), orchestrates both processing events within the nucleus, typically resulting in an ~21-nucleotide mature miRNA/miRNA passenger strand (miRNA*) duplex with two-nucleotide 3′ overhangs (Park et al., 2002; Reinhart et al., 2002; Xie et al., 2003; Kurihara and Watanabe, 2004; Kurihara et al., 2006). The specifics of miRNA processing in plants are somewhat less well understood. In the majority of cases, DCL1 first catalyzes cleavage of the foldback at the loop-distal side, and subsequently on the loop-proximal side, of the miRNA/miRNA* region, in a sequential manner like that in animals (Kurihara and Watanabe, 2004). These processing events are aided by other proteins, including the C2H2-zinc finger protein SERRATE (SE), the double-stranded RNA binding protein HYPONASTIC LEAVES1 (HYL1), and the RNA binding protein DAWDLE (Grigg et al., 2005; Kurihara et al., 2006; Yang et al., 2006; Dong et al., 2008; Laubinger et al., 2008; Yu et al., 2008; Szarzynska et al., 2009). After DCL1-mediated processing, the duplex is transported out of the nucleus, where the miRNA strand is bound by an ARGONAUTE (AGO) family protein. The miRNA “programs” the AGO protein to recognize target transcripts with complementarity to the miRNA.
The majority of plant miRNAs associate with AGO1 and bind target transcripts at highly complementary sites, usually one per target (Baumberger and Baulcombe, 2005; Mi et al., 2008; Cuperus et al., 2010a). In the majority of studied cases, targeting leads to slicing of at least a fraction of total target RNAs (Addo-Quaye et al., 2008; German et al., 2008; Mallory et al., 2008), although it is clear that repression of targets involves both degradative and nondegradative mechanisms (Voinnet, 2009). Genetic evidence suggests that AGO10 also controls several miRNA targets, although direct association with miRNAs has not been demonstrated (Mallory and Vaucheret, 2010). Animal miRNAs typically bind target transcripts at multiple sites with limited complementarity and repress target mRNAs through repression of translation and promotion of transcript destabilization through decapping and deadenylation pathways (Carthew and Sontheimer, 2009). miRNA target nodes are often part of larger regulatory networks that control plant development, nutritional responses, and other important processes. The importance of miRNA-mediated regulation is evident by the severe pleiotropic phenotypes in miRNA-deficient mutants (Park et al., 2002; Reinhart et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Flynt and Lai, 2008).
This review focuses on recent literature that sheds new light on the origins of plant MIRNA genes and on the diversification of miRNA biogenesis and functionality.
EVOLUTION OF MIRNA GENES
Old and Young Plant MIRNA Families
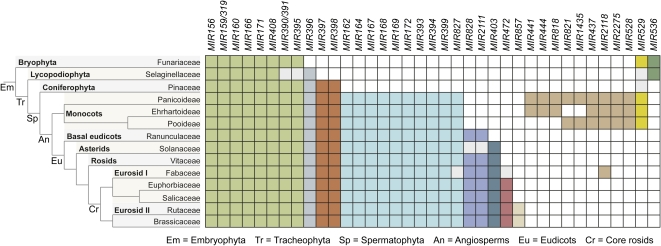
The majority of miRNAs that were discovered in early studies of Arabidopsis thaliana and rice (Oryza sativa) were from families that are conserved between the two species (Llave et al., 2002; Mette et al., 2002; Park et al., 2002; Reinhart et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). miRNAs from these loci were detected by low-depth cloning and sequencing or by computational predictions of foldbacks and complementary target sequences. These techniques resulted in a bias for detecting miRNA from abundantly expressed and ancient families (Axtell and Bartel, 2005). Combined with numerous data sets from high-throughput sequencing approaches, eight MIRNA families have been identified in the common ancestor of all embryophytes (Figure 1). The MIR396 family was present in the common ancestor of all tracheophytes (vascular plants), while the MIR397 and MIR398 families were acquired in the common ancestor of all spermatophytes (seed plants; Figure 1). Ten families are present in all angiosperm lineages, while all other families display more restricted taxonomic distributions. MIR828, MIR2111, and MIR403 are eudicot-specific families, although MIR403 may be restricted to core eudicot lineages (Figure 1). MIR472 is present in all core rosids, while MIR857 may be restricted to at least a subset of the eurosid II clade. At least nine families likely arose in the monocot lineage. Four of these are found in all three grass families, and five were lost in one lineage (Figure 1). Two additional family patterns suggest lineage-specific losses. MIR529 and MIR536 were present in the common ancestor of embryophytes but were lost in the common ancestors of eudicots and tracheophytes, respectively (Figure 1). No MIRNA families found in embryophyte plants have been found in the unicellular green alga Chlamydomonas reinhardtii (Molnár et al., 2007; Zhao et al., 2007). Despite this, two MIRNA annotated families (MIR854 and MIR855) were reported as conserved between the plant and animal lineages (Arteaga-Vázquez et al., 2006). However, loci from these families lie in retrotransposons. Analysis of deep sequencing data at these loci in Arabidopsis have failed to validate these as bona fide MIRNA; rather, they are loci that spawn heterogeneous, RNA-DEPENDENT RNA POLYMERASE2 (RDR2)/DCL3-dependent, 24-nucleotide short interfering RNAs (siRNAs; Lu et al., 2006; Rajagopalan et al., 2006; Fahlgren et al., 2007, 2010; Kasschau et al., 2007; Ma et al., 2010). Current evidence, therefore, suggests that known MIRNA families in plants and animals arose independently.
Figure 1.
Deeply Conserved MIRNA Families.
MIRNA families (columns) that are conserved between plant families (rows) for plant species represented in miRBase release 16 (Griffiths-Jones et al., 2008). Species- and family-specific MIRNA genes were omitted. Boxes are highlighted if a MIRNA family was identified in at least one species for each of the plant families listed or if the MIRNA family could be identified in the National Center for Biotechnology Information EST or whole-genome shotgun reads databases by BLAST (Altschul et al., 1997). Matches were identified as MIRNA if the sequences containing the putative mature miRNAs could be folded, using RNAfold (Hofacker, 2003), into a stem-loop structure containing the miRNA in the stem. Plant families that may have lost a MIRNA family, or where the family could not confidently be identified, are shaded gray. Groups of MIRNA families are highlighted different colors based on inferred taxonomic range.
Deep sequencing of A. thaliana small RNA populations revealed that, in addition to MIRNA genes that are conserved between at least two families (Figure 1), there are at least four times as many families that are not obviously conserved outside of the Brassicaceae (Lu et al., 2006; Rajagopalan et al., 2006; Fahlgren et al., 2007, 2010; Ma et al., 2010). A high proportion of species-specific or nonconserved MIRNA genes were also observed in Physcomitrella patens and Selaginella moellendorffii (Axtell et al., 2007), rice (Heisel et al., 2008; Lu et al., 2008; Sunkar et al., 2008; Zhu et al., 2008), Medicago truncatula (Szittya et al., 2008; Lelandais-Brière et al., 2009), and Glycine max (Subramanian et al., 2008). Data from additional plant genomes indicate that the majority of these are restricted to species or other subfamily lineages (www.mirbase.org; Griffiths-Jones et al., 2008). Given that a large number of MIRNA families are species-specific or restricted to closely related species, it is reasonable to suggest that plants harbor relatively large numbers of recently spawned MIRNA loci.
Origins of New MIRNA Genes
How Are New MIRNA Genes Formed?
Early sequence similarity searches revealed that the foldback arms of ath-MIR161, ath-MIR163, and ath-MIR822 (previously referred to as ASRP1729) shared extended similarity (outside of the miRNA/miRNA* regions) with their target genes (Allen et al., 2004). The foldback arms aligned in an inverted orientation, with the miRNA arm inverted relative to the target gene sequence. These data suggested that inverted duplication events could form self-complementary regions with the potential to spawn MIRNA genes. Initial duplication events would result in loci with perfect or near-perfect self-complementarity and produce siRNAs (Allen et al., 2004). Indeed, it was demonstrated experimentally that the ath-MIR822, ath-MIR839, and ath-MIR869 foldbacks are processed by DCL4, rather than DCL1, into miRNA-like siRNAs (Rajagopalan et al., 2006; Ben Amor et al., 2009). These MIRNA genes formed by an inverted duplication of a DC1 domain gene (MIR822), a P-glycoprotein gene (MIR839), or a SU(VAR)3-9 homolog gene (MIR869; Allen et al., 2004; Fahlgren et al., 2007, 2010) and thus represent good examples of young, transitional MIRNA genes. Over time, the accumulation of mutations in the foldback arms could result in an affinity for the miRNA biogenesis machinery and decreased similarity between the extended foldback arms and the locus of origin (Allen et al., 2004). Coupled with the coevolution of one or more target transcripts, a productive miRNA target node may arise and, in rare instances, be incorporated into new or existing regulatory networks.
Further evidence for the recent origin of some MIRNA genes was found by global analysis of genomic or transcript sequences with extensive similarity to MIRNA loci in A. thaliana, Arabidopsis lyrata, and P. patens (Rajagopalan et al., 2006; Axtell et al., 2007; Fahlgren et al., 2007, 2010; de Felippes et al., 2008). Twenty-seven and 18 MIRNA loci in A. thaliana and A. lyrata, respectively, contained sequence similarity with a putative locus of origin elsewhere in their respective genomes, with a large proportion of these in A. thaliana originating through duplications of transcribed, protein-coding gene sequences (Rajagopalan et al., 2006; Axtell et al., 2007; Fahlgren et al., 2007, 2010; de Felippes et al., 2008). However, other configurations were also detected. For some MIRNA, both arms of the foldback align to the putative foldback-originating locus in the same orientation or align to separate regions rather than one region, suggesting that a duplication event occurred at the originating locus before the duplication event that formed the MIRNA (Rajagopalan et al., 2006; Fahlgren et al., 2010).
Despite the large number of young A. thaliana and A. lyrata MIRNA genes with sequence identity to putative loci of origin, over half possess no identifiable MIRNA-related locus (de Felippes et al., 2008; Fahlgren et al., 2010). Where did these MIRNA loci come from? Conceivably, some of the originating loci were lost or rapidly diverged. For example, MIR161, MIR472, and MIR822 had significant matches with other loci in A. thaliana but not in A. lyrata, while MIR859 had a significant match in A. lyrata but not in A. thaliana (Fahlgren et al., 2010). On the other hand, any inverted duplication in a plant genome could be the starting point for a new MIRNA. Jones-Rhoades and Bartel (2004) identified 133,864 and 410,167 imperfect inverted repeats in the genomes of A. thaliana and rice, respectively. One source of inverted repeats are nonautonomous transposons that contain flanking terminal inverted repeats but that lack the internal sequences encoding genes required for transposition (miniature inverted-repeat transposable elements; Bureau and Wessler, 1992, 1994). A miniature inverted-repeat transposable element locus may have been the source of ath-MIR1888, which requires DCL1 for biogenesis but which also has similarity to a number of small inverted repeats that generate DCL3-dependent 24-nucleotide siRNAs (German et al., 2008). Several additional A. thaliana and rice miRNAs were proposed to be derived from transposable elements (Piriyapongsa and Jordan, 2008), although many of these are not bona fide MIRNA genes (see miR854 and miR855 above; Axtell and Bowman, 2008; Meyers et al., 2008). While the transposon origin model is plausible, transitionary (young) loci may be difficult to identify due to the intersection of miRNA and siRNA pathways at these loci (Chellappan et al., 2010).
A final possibility for the origin of young MIRNA genes that lack detectable MIRNA-related loci is that they were not formed by duplication events. Instead, new MIRNA genes could form through the accumulation of mutations within inverted repeats (de Felippes et al., 2008). If expressed, selection could conceivably act on mutations that modify miRNA processing efficiency and alter the affinity of the de novo–generated miRNA for a target if there were effects on existing regulatory networks.
What Is the Rate at Which MIRNA Genes Are Formed or Lost?
The abundance of species- or lineage-specific MIRNA families suggests that MIRNA genes are born and lost at a high frequency. The recent high-quality assembly of the A. lyrata genome (http://genome.jgi-psf.org/Araly1/Araly1.home.html) allowed for a thorough analysis of the shared and unique MIRNA contents of the A. thaliana and A. lyrata genomes (Fahlgren et al., 2010; Ma et al., 2010), providing the basis for the first MIRNA flux estimates. There are 102 and 116 identified MIRNA families in A. thaliana and A. lyrata, respectively, with 78 families shared between the two (www.mirbase.org; Griffiths-Jones et al., 2008; Fahlgren et al., 2010; Ma et al., 2010). Although the majority of MIRNA families are conserved between the two species, 24 to 33% of the families were gained or lost by A. thaliana or A. lyrata since they diverged ~10 million years ago (Koch et al., 2000; Wright et al., 2002; Ossowski et al., 2010). Additionally, preliminary mining of the Capsella rubella genome and small RNA sequencing data identified 43 and 42 MIRNA families conserved with A. thaliana or A. lyrata, respectively (Fahlgren et al., 2010). Given that C. rubella diverged from the Arabidopsis lineage ~20 million years ago (Koch et al., 2000; Wright et al., 2002; Ossowski et al., 2010), and allowing for errors in identifying MIRNA in the incomplete C. rubella genome, the net rate of flux (birth–death) for MIRNA genes in the Arabidopsis lineage was estimated to be from 1.2 to 3.3 genes per million years (Fahlgren et al., 2010). This estimate overlaps the 0.8 to 1.6 genes per million years estimated for the Drosophila lineage MIRNA flux rate (Berezikov et al., 2010).
Is the Formation of New MIRNA Genes a Selective or Neutral Process?
Young MIRNA genes formed by duplications of transcribed regions could yield miRNAs, or siRNAs, that have the potential to perturb existing regulatory networks. However, evidence from several plant species suggests that the vast majority of young miRNAs have few, if any, functions. First, whereas ~45% of A. thaliana targets for conserved miRNAs increase in abundance in multiple mutants with miRNA pathway defects (hyl1, hst, dcl1, hen1, and ago1; Allen et al., 2005; Ronemus et al., 2006), levels of target transcripts for nonconserved miRNAs are largely unaffected in miRNA biogenesis mutants, suggesting that most young miRNAs are not integrated into regulatory networks (Fahlgren et al., 2007). Alternatively, young miRNAs could function in restricted spatiotemporal patterns, in response to specific biotic or abiotic queues, or in targeting modes that do not result in target RNA degradation. Target conservation patterns between A. thaliana and A. lyrata also support a neutral regulatory role for most young miRNAs in these species. For MIRNA families conserved with non-Brassicaceae species, prediction and validation of miRNA targets were highly reliable and consistent between species (Ma et al., 2010). By contrast, target prediction and validation for miRNAs conserved between A. thaliana and A. lyrata, but not other plant families, were limited and highly species-specific (Ma et al., 2010). This supports the earlier finding that most young miRNAs in A. thaliana have few bona fide targets (Rajagopalan et al., 2006; Fahlgren et al., 2007).
Second, the abundance of conserved miRNAs, as a group, is substantially higher than the abundance of nonconserved miRNAs. Deeply conserved miRNAs consistently are sequenced at a higher frequency and cumulatively make up a larger portion of the total reads from all MIRNA families (Rajagopalan et al., 2006; Axtell, 2008; Ma et al., 2010). By contrast, Arabidopsis-specific miRNAs are generally less abundant and often expressed in only one of the two species (Ma et al., 2010). Data from a recent study that profiled MIRNA transcripts in miRNA biogenesis mutants and under stress conditions suggest that this bias is also present at the primary MIRNA transcript level (Laubinger et al., 2010). Over 90% of the families of MIRNA genes conserved between plant families had at least one member that was detectable at moderate to high levels, while less than half of the Brassicaceae-specific MIRNA families accumulated to the same levels (Laubinger et al., 2010). This could be the result of highly specific expression patterns in cells, tissues, or conditions that have not been studied. Alternatively, the majority of young MIRNA genes may lack regulatory elements that confer robust expression. The duplication that formed MIR163 was found to contain part of the promoter sequence of the originating target gene family (Wang et al., 2006), but similar patterns have not been reported for other young MIRNA genes. In addition to expression, young MIRNA genes are also processed less precisely. Those apparent transitional MIRNA families with transcripts that are processed by DCL4 exhibit the most striking examples of imprecise processing (see above; Rajagopalan et al., 2006), but processing precision is generally low for Brassicaceae-restricted MIRNA transcripts (Vazquez et al., 2008; Ma et al., 2010). Weak expression coupled with variable processing may partly explain the lack of experimental support for the majority of young MIRNA genes.
Finally, nucleotide divergence patterns between orthologous MIRNA genes in A. thaliana and A. lyrata are consistent with neutral evolution. For Arabidopsis MIRNA families conserved with non-Brassicaceae species, nucleotide divergence was highest in the loop and loop-distal stem regions and lowest in the miRNA and miRNA* regions (Ehrenreich and Purugganan, 2008; Warthmann et al., 2008; Fahlgren et al., 2010; Ma et al., 2010). Low divergence in the miRNA region likely reflects purifying selection to maintain complementarity between the mature miRNA and target RNAs, while the low divergence in the miRNA* region likely reflects purifying selection to maintain base pairing with the constrained miRNAs. By contrast, nucleotide divergence was much more uniform across the foldbacks of MIRNA genes not conserved with non-Brassicaceae species (Fahlgren et al., 2010; Ma et al., 2010). Although the pairwise differences were somewhat lower in the miRNA and miRNA* regions, divergence was significantly higher than that found for the deeply conserved MIRNA families (Fahlgren et al., 2010; Ma et al., 2010). While some young MIRNA genes may experience strong selection, these data suggest that selection is weak or neutral for the majority of these loci.
In contrast to deeply conserved MIRNA families, most young MIRNA genes are weakly expressed, processed imprecisely, more divergent, and tend to lack targets, suggesting that they may be evolving neutrally. The lack of verifiable targets may be due to the low accumulation and imprecise generation of mature miRNAs. For instance, imprecise excision of the mature miRNAs could result in functionally variable, or inert, miRNAs, as the miRNA sequence is important for selection of the miRNA over the miRNA* and sorting into functional AGO complexes (Mi et al., 2008; Montgomery et al., 2008a; Eamens et al., 2009; Ebhardt et al., 2010). Alternatively, young miRNAs could function primarily through nondegradative mechanisms that cannot be assayed by measuring target transcript abundance (Brodersen et al., 2008; Lanet et al., 2009). While this cannot be ruled out, it is difficult to explain why such a preference would exist. Additionally, the higher degree of nucleotide divergence observed for young MIRNA orthologs and the observed birth–death rate for Arabidopsis MIRNA support the idea that most are neutrally evolving, evolutionarily transient loci. New loci would have little detrimental impact on existing regulatory networks but could be sources of novel regulatory variations that are captured into networks on relatively rare occasions. The Brassicaceae-specific MIR824 may be such an example. miR824-guided cleavage of AGAMOUS-LIKE16 transcripts functions in a stomatal patterning developmental network (Kutter et al., 2007), and population variation at the ath-MIR824 locus shows significant departure from neutrality (de Meaux et al., 2008).
CONSERVATION AND DIVERSIFICATION OF miRNA PROCESSING
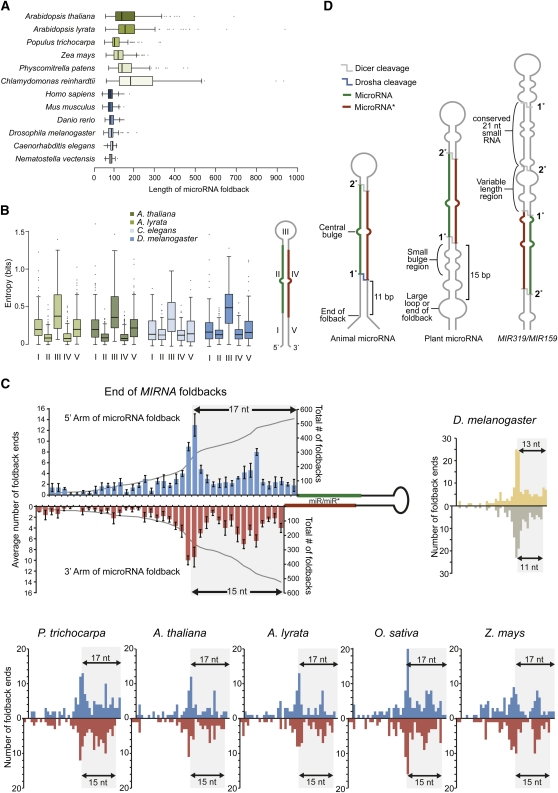
The transition of a young MIRNA locus from a nascent duplication product to one yielding a transcript that is recognized by the canonical MIRNA foldback processing machinery (involving DCL1, HYL1, or SE; Voinnet, 2009) presumably involves neutral mutations. It is clear that plant MIRNA foldbacks can qualify over a range of structures, as foldback sequence lengths in many species vary between less than 100 to over 900 nucleotides in length. This is in striking contrast to animal species foldback lengths, which are highly uniform and predominantly less than 100 nucleotides (Figure 2A). Several recent studies of P. patens and A. thaliana MIRNA have shed light on plant miRNA processing mechanisms. In this section, the “criteria” that qualify a transcript for miRNA processing in plants are discussed.
Figure 2.
miRNA Processing in Plants.
In the box plots shown in (A) and (B), boxes represent the 25th to 75th percentiles of the data range, and whiskers encompass the most extreme points that are no more than 1.5 times the interquartile range from the box.
(A) Predicted length of MIRNA foldbacks in plants and animals.
(B) Entropy of MIRNA foldbacks from five distinct regions. A foldback diagram (right) marks the locations of the five regions: I, 5′ arm, loop-distal; II, miRNA; III, loop, 5′ and 3′ arms, loop-proximal; IV, miRNA*; V, 3′ arm, loop-distal. Entropy was calculated per base using the program RNAfold (Hofacker, 2003). The lengths of regions I and V were either 17 and 19 nucleotides, respectively, for plants or 11 and 13 nucleotides, respectively, for Caenorhabditis elegans based on predicted differences in plant and animal miRNA processing from the foldback base.
(C) Distribution of large loops (three or more nucleotides [nt]) or ends of helices for foldbacks from 24 deeply conserved MIRNA families in five plant species. Both individual species and combined species data are shown (mean relative levels ± se). The 17/15-nucleotide region shows an enrichment of loops or helix ends in all plants shown, while Drosophila melanogaster large loops or helix ends are enriched at 13/11 nucleotides. The combined plant bar graph is overlaid with the cumulative number of predicted intact foldbacks remaining beyond each position.
(D) Illustration of cis-elements involved in animal and plant miRNA foldback processing. Boldface numbers represent initial and secondary processing sites leading to mature small RNAs.
MIRNA Foldback Recognition Features
Several recent studies revealed two structural features that confer optimized foldback processing characteristics. One feature was identified in mutants with defects in ath-MIR390a, ath-MIR172a, ath-MIR171a, ath-MIR167a, ath-MIR164c, and ath-MIR398a, with substitutions affecting the region 1 to 8 bp below the miRNA/miRNA* duplex and resulting in reduced or inaccurate miRNA accumulation (Cuperus et al., 2010b; Mateos et al., 2010; Song et al., 2010; Werner et al., 2010). For comparison, single nucleotide mutations in the loop-proximal region in many cases were considerably less deleterious, or had no effect, on miRNA accumulation (Mateos et al., 2010; Song et al., 2010; Werner et al., 2010). Mutations in the region below the miRNA/miRNA* duplex that maintained wild-type structure were generally neutral, while mutations that opened or closed predicted bulges reduced the accumulation or altered the processing of the mature miRNAs (Cuperus et al., 2010b; Mateos et al., 2010; Song et al., 2010; Werner et al., 2010). In plant MIRNA foldbacks, this region is characterized by relatively weak base pairing, with a high number of one- to three-nucleotide bulges (Figure 2B). In the case of MIR390a and MIR390b, which specify the identical miR390 sequence, natural variation in the strength of base-pairing in this region may account for differences in miR390 expression from each locus (Cuperus et al., 2010b). Additionally, the ath-mir390a-1 point mutation, which reduces miR390 accumulation and processing accuracy, is predicted to lead to enhanced base-pairing in the region just below the miR390/miR390* region (Cuperus et al., 2010b). Weak or flexible base-pairing within the loop-distal region of the foldback may be necessary for efficient miRNA processing in plants, although this may not be the case in animals (Figure 2B).
Genetic analyses suggested that positioning of the initial DCL1 processing event was dependent on a stem length equivalent to ~15 bp below the miRNA/miRNA* duplex (Mateos et al., 2010; Song et al., 2010; Werner et al., 2010). Foldbacks from A. thaliana (Mateos et al., 2010; Song et al., 2010; Werner et al., 2010) and several additional plant species (Figure 2C) tend to have large loops, or terminate at ~15 bp below the miRNA/miRNA* region, and this characteristic appears important for optimal processing. Drosophila melanogaster MIRNA foldbacks are strongly characterized by loss of base pairing at ~11 bp below the miRNA/miRNA* region (Figure 2C), and this confers recognition by the RNA binding protein DGCR8, which then positions the initial cleavage events by Drosha (Sashital and Doudna, 2010). Interaction between HYL1 and DCL1 is required for accurate processing of pri-miRNAs and pre-miRNAs (Dong et al., 2008). HYL1 may assist in positioning DCL1 for the initial processing step, although direct evidence for how this occurs is lacking.
Although loop-distal processing occurs first during miRNA maturation of most foldbacks in plants and animals, alternative primary processing events have been characterized (Okamura et al., 2007; Ruby et al., 2007; Addo-Quaye et al., 2009; Bologna et al., 2009; Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). The MIR159/MIR319 family of foldbacks is processed first by DCL1 on the loop-proximal side of the stem, and this is a deeply conserved property of the family (Addo-Quaye et al., 2009; Bologna et al., 2009). In fact, two small RNAs are processed from MIR159/MIR319 family members, a loop-proximal small RNA and mature miR159/miR319, which are separated by 20 to 30 bp (Bologna et al., 2009; Figure 2D). Both mature miR319 and the loop-proximal small RNA are sensitive to hyl1 mutations (Bologna et al., 2009), suggesting that they depend on similar processing recognition mechanisms.
Conserved Size Variation in Mature miRNAs
Cis-features of the foldback convey information to accurately position the first DCL1 processing events. The positioning of the second processing step to release the miRNA/miRNA* duplex from the foldback precursor is determined by the “molecular ruler” property of DCL1 (Sashital and Doudna, 2010). The PAZ domain binds to the two-nucleotide 3′ overhang that forms on the precursor after the initial processing events. The RNA duplex lays along the Dicer surface. The RNaseIII domains are positioned at a distance equivalent to ~21 bp away from the position of the bound end. Most plant foldbacks yield a dominant mature miRNA species, although most or all produce some positional and size variants of the miRNA and miRNA* sequences. These likely reflect misprocessing events at the first and/or second DCL1-catalyzed step. The majority of miRNAs are 21 nucleotides long, but several families in A. thaliana and rice accumulate abundant 22-nucleotide miRNAs (Johnson et al., 2009; Chen et al., 2010; Cuperus et al., 2010a). Accumulation of these miRNAs is dependent on DCL1, and not DCL2, which generates 22-nucleotide siRNAs from long double-stranded RNA (Park et al., 2002; Gasciolli et al., 2005; Xie et al., 2005; Bouché et al., 2006; Deleris et al., 2006; Lu et al., 2006; Montgomery et al., 2008b; Cuperus et al., 2010a). In the majority of cases, foldbacks that generate 22-nucleotide miRNAs contain a single asymmetric, nonpaired nucleotide on the miRNA side of the duplex. Indeed, removal of the asymmetric bulged nucleotide leads to the formation of 21-nucleotide miRNAs, and introduction of an asymmetric bulged nucleotide leads to the formation of 22-nucleotide forms (Chen et al., 2010; Cuperus et al., 2010a). Assuming that the asymmetric bulge does not extend the length of the helix, this explains most of the longer, 22-nucleotide miRNAs. The MIR163 family is an extreme example of miRNA size variation. In A. thaliana, three bulged nucleotides result in a 24-nucleotide mature miRNA, while the A. lyrata locus only has two bulges and yields a 23-nucleotide miRNA (Griffiths-Jones et al., 2008; Fahlgren et al., 2010). Functional consequences of miRNA size variation are discussed below.
FUNCTIONAL DIVERSIFICATION OF miRNA IN PLANTS
In addition to the repression of target mRNAs, some miRNAs have other specialized functions or confer unique properties to miRNA–AGO complexes.
miRNA-Triggered Production of 21-Nucleotide siRNAs
Trans-acting siRNAs (tasiRNAs) are RDR6- and DCL4-dependent products of a refined RNA interference pathway, and they function as repressors on specific, coevolved target mRNAs (Allen and Howell, 2010). TasiRNAs accumulate among arrays of siRNAs formed from noncoding RNA transcripts from TAS loci. The siRNA arrays form in phase, or in register, with the site of precise miRNA-guided cleavage of the primary TAS transcript (Allen et al., 2005; Axtell et al., 2006; Rajagopalan et al., 2006; Montgomery et al., 2008a). Phasing of TAS1/TAS2, TAS3, and TAS4 tasiRNAs is set by cleavage guided by miR173–AGO1, miR390–AGO7, and miR828–AGO1 complexes, respectively (Allen et al., 2005; Axtell et al., 2006; Rajagopalan et al., 2006; Montgomery et al., 2008a, 2008b). Thus, miR173, miR390, and miR828 function as activators, rather than repressors, of a siRNA pathway. The involvement of these miRNAs in tasiRNA biogenesis is a specialized function, as most plant miRNAs, at least those associated with AGO1, cannot effectively substitute in these pathways (Montgomery et al., 2008a, 2008b). A unique property of miR390 is its association with AGO7, which has extremely high specificity for miR390 (Montgomery et al., 2008a). miR390 is excluded from AGO1 due to the presence of a 5′ adenosine, as AGO1 has a strong preference for miRNAs with a 5′ uridine (Mi et al., 2008; Montgomery et al., 2008a). miR390–AGO7 complexes associate with TAS3 transcripts at two sites, only one of which is sliced (Axtell et al., 2006; Howell et al., 2007). Interaction of miR390–AGO7 at the noncleaved site is postulated to mark the transcript for recruitment of RDR6 (Montgomery et al., 2008a). Both TAS3 and miR390 are deeply conserved in plants, although in P. patens, both miR390 target sites are cleaved (Arazi et al., 2005; Axtell et al., 2006). In contrast to TAS3, TAS1/TAS2 and TAS4 transcripts are targeted and sliced at only one site (Allen et al., 2005; Rajagopalan et al., 2006). While substitution of the miR173 target site of TAS1 with other AGO1-associating miRNA target sites leads to cleavage, no TAS1 tasiRNAs are generated (Montgomery et al., 2008b). The only requirement for siRNA generation in the case of TAS1 is the presence of a miR173 target site. Addition of a miR173 target site to an unrelated mRNA-generating locus results in siRNA formation (Montgomery et al., 2008b). Interestingly, miR173 and miR828 are both 22 nucleotides long, rather than the more common 21-nucleotide length. Twenty-one-nucleotide versions of miR173 and miR828 guide the cleavage of target RNA but do not trigger the production of siRNA (Chen et al., 2010; Cuperus et al., 2010a).
In addition to tasiRNA from evolved noncoding loci, some miRNA-targeted mRNAs yield secondary siRNAs (Axtell et al., 2006; Ronemus et al., 2006; Howell et al., 2007; Chen et al., 2010; Cuperus et al., 2010a; Zheng et al., 2010). Some of these mRNAs produce phased RDR6/DCL4-dependent, 21-nucleotide siRNAs like TAS transcripts (Axtell et al., 2006; Chen et al., 2007, 2010; Howell et al., 2007; Cuperus et al., 2010b). In some cases, targeting of transcripts by miRNAs at multiple sites, or in combination with tasiRNAs, may trigger secondary siRNA formation (Axtell et al., 2006). In other cases, generation of secondary siRNAs is triggered by a single miRNA-targeting event, and removal or substitution of these target sites may result in a loss of siRNAs (Allen et al., 2005; Axtell et al., 2006; Ronemus et al., 2006; Howell et al., 2007; Vaucheret, 2009). Routing of miRNA-targeted transcripts into secondary siRNA-generating pathways appears to be regulated, as only ~20% of miRNAs or tasiRNA-targeted transcripts yield secondary siRNAs (Cuperus et al., 2010a). As with TAS1/TAS2 and TAS4 tasiRNAs, siRNA from these protein-coding transcripts is frequently associated with 22-nucleotide triggers in both Arabidopsis and rice (Johnson et al., 2009; Chen et al., 2010; Cuperus et al., 2010a). In rice, miR2118 mediates the recruitment of 21-nucleotide secondary siRNA-generating machinery, but miR2775-targeted transcripts generate 24-nucleotide siRNAs (Johnson et al., 2009). The function of 24-nucleotide secondary siRNAs is not clear, but similar 24-nucleotide phased siRNAs were also found in Brachypodium distachyon (International Brachypodium Initiative, 2010). Most 22-nucleotide miRNAs associate with AGO1, suggesting that 21- and 22-nucleotide miRNA–AGO1 complexes are functionally distinct (Chen et al., 2010; Cuperus et al., 2010a). The mechanisms underlying this distinction are not yet known, although it was postulated that miRNA size affects an AGO1 functional state that mediates the recruitment, either directly or indirectly, of RDR6 (Chen et al., 2010; Cuperus et al., 2010a).
miRNA-Directed DNA Methylation
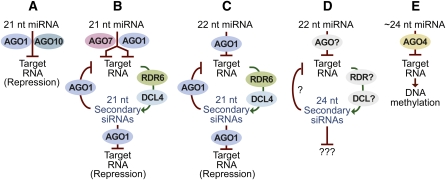
A subset of miRNA variants preferentially associate with AGO proteins involved in RNA-directed DNA methylation (Law and Jacobsen, 2010). In A. thaliana, AGO4, AGO6, and AGO9 typically associate with 24-nucleotide siRNAs containing a 5′ adenosine and use these siRNAs as guides to direct the methylation of target loci DNA (Qi et al., 2006; Mi et al., 2008; Havecker et al., 2010). Twenty-four-nucleotide siRNAs are generated by DCL3 from double-stranded RNAs produced through the activities of RDR2- and DNA-dependent RNA polymerase IV (Law and Jacobsen, 2010). AGO4–siRNA complexes may interact with targets through interaction with noncoding transcripts generated by a second plant-specific nuclear RNA polymerase, polymerase V (Wierzbicki et al., 2009). In rice, DCL3a also processes multiple MIRNA foldbacks, yielding 24-nucleotide, miRNA-like siRNAs (Wu et al., 2010). While biogenesis of these small RNAs is DCL3-dependent, some loci require initial processing by DCL1a and lead to the production of both 21- and 24-nucleotide small RNAs. In contrast to 21-nucleotide miRNA variants, the 24-nucleotide miRNA-like siRNAs preferentially associate with rice AGO4a and AGO4b, and like 24-nucleotide siRNAs, they can guide the methylation of target DNAs (Wu et al., 2010). Similarly, 23- to 27-nucleotide DCL3-dependent small RNAs that preferentially associate with AGO4 and direct methylation, and that originate from miRNA foldbacks, were identified in A. thaliana (Chellappan et al., 2010). Studies in P. patens also link miRNAs to transcriptional gene silencing through DNA methylation, as mutants with increased levels of miRNA target RNA duplexes exhibit elevated DNA methylation levels at the target locus (Khraiwesh et al., 2010). It is clear, therefore, that miRNA and miRNA–AGO complexes have evolved functions that are distinct from canonical mRNA repression and that miRNAs can function through crosstalk with both posttranscriptional and transcriptional silencing pathways (Figure 3).
Figure 3.
Functional Crosstalk of the miRNA Pathway in Plants.
Plant miRNA functions include repression of targets, triggering of siRNAs from targets, and triggering of RNA-directed DNA methylation. Pathways A, B, and E have been described in monocots, eudicots, and bryophytes, pathway C in monocots and eudicots, and pathway D only in monocots. nt, Nucleotides.
CONCLUSIONS AND FUTURE PROSPECTS
The rapidly growing collection of plant genomes and advances in high-throughput small RNA sequencing have enhanced our understanding of the evolution of MIRNA genes and their functions. High-throughput sequencing has been particularly informative in showing that most MIRNA families are lineage-restricted or species-specific. Deeply conserved MIRNA families are integral components of functional regulatory networks that orchestrate development, nutritional responses, stress responses, and other processes. By contrast, the young MIRNA genes, as a group, appear to be evolving neutrally and are prone to be lost over short evolutionary periods. Given that a large proportion of new MIRNA genes are formed by duplication events, are there conditions or genome evolution processes that favor the formation of new MIRNA genes? In A. thaliana, the youngest MIRNA genes were found in genomic regions that lacked orthologous positions in A. lyrata. Additionally, there was a weak association between the youngest A. thaliana MIRNA and the presence of flanking transposable elements, suggesting that some MIRNA-forming duplication events may be mediated by transposition or recombination events involving repetitive DNA (Fahlgren et al., 2010). Further research will be needed to understand transposition and recombination events that yield nascent MIRNA loci.
Does miRNA processing heterogeneity, or differential processing activity during development, represent a biologically relevant regulatory mechanism? Differential processing of miRNAs is well established in animals, but few examples have been demonstrated in plants (Newman and Hammond, 2010). Maize (Zea mays) zma-MIR166a transcripts accumulate in the tip of the shoot apical meristem, but no mature miR166 was detected by in situ hybridization (Nogueira et al., 2009). A. thaliana ath-MIR172b transcripts were detected in wild-type inflorescence tissues but not in seedlings, even though transcripts were detected in both tissues in dcl1 mutant plants (Laubinger et al., 2010). These examples suggest that processing efficiency may be modulated in a tissue- or precursor-specific manner. Further work is needed to understand the prevalence of differential or allele-specific processing and how this would be regulated.
Some miRNAs possess specialized features that trigger the production of secondary siRNAs from protein-coding transcripts. What is the biological role of these siRNAs? Secondary siRNAs might reinforce the silencing of transcripts from the miRNA target locus or conceivably expand the range of target repression to related family members that interact with the siRNAs. For example, targeting of members of the pentatricopeptide repeat (PPR) family by several miRNAs and tasiRNAs in A. thaliana triggers the production of secondary siRNAs with the theoretical potential to target other PPR transcripts in trans, although compelling evidence for the activity of the secondary siRNAs is lacking (Axtell et al., 2006; Chen et al., 2007; Howell et al., 2007; Addo-Quaye et al., 2008). Another fascinating possibility is that secondary siRNAs might function as non-cell-autonomous signals to form regulatory gradients or to silence transcripts or loci at distal sites (Chitwood et al., 2009; de Felippes et al., 2010; Dunoyer et al., 2010; Molnar et al., 2010).
Finally, additional diversified functions of miRNAs will likely emerge from studies of diverse plants. In-depth analyses of rice and Arabidopsis species have already revealed distinct flavors of miRNAs, effector pathways, and crosstalk between silencing pathways. As additional plant lineages are explored, novel innovations involving miRNAs will likely become apparent. Further analyses in diverse species should also illuminate how the evolution of MIRNA genes impacts the evolution of regulatory networks.
References
- Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Howell M.D. (2010). miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43: 837–848 [DOI] [PubMed] [Google Scholar]
- Arteaga-Vázquez M., Caballero-Pérez J., Vielle-Calzada J.P. (2006). A family of microRNAs present in plants and animals. Plant Cell 18: 3355–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. (2008). Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochim. Biophys. Acta 1779: 725–734 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Snyder J.A., Bartel D.P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Liu N., Flynt A.S., Hodges E., Rooks M., Hannon G.J., Lai E.C. (2010). Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat. Genet. 42: 6–9; author reply 9–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N.G., Mateos J.L., Bresso E.G., Palatnik J.F. (2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28: 3646–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Lauressergues D., Gasciolli V., Vaucheret H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Bureau T.E., Wessler S.R. (1992). Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4: 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau T.E., Wessler S.R. (1994). Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P., Xia J., Zhou X., Gao S., Zhang X., Coutino G., Vazquez F., Zhang W., Jin H. (2010). siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D., Wolfe S.A., Giraldez A.J. (2010). A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010a). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C. (2010b). Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes F.F., Ott F., Weigel D. (December 5, 2010). Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. http://dx.doi.org/10.1093/nar/gkq1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J., Hu J.Y., Tartler U., Goebel U. (2008). Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8994–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Dong Z., Han M.H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916 [DOI] [PubMed] [Google Scholar]
- Eamens A.L., Smith N.A., Curtin S.J., Wang M.B., Waterhouse P.M. (2009). The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt H.A., Fedynak A., Fahlman R.P. (2010). Naturally occurring variations in sequence length create microRNA isoforms that differ in argonaute effector complex specificity. Silence 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I.M., Purugganan M.D. (2008). Sequence variation of microRNAs and their binding sites in Arabidopsis. Plant Physiol. 146: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Carrington J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D., Carrington J.C. (2010). MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes F.F., Schneeberger K., Dezulian T., Huson D.H., Weigel D. (2008). Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A.S., Lai E.C. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008). miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg S.P., Canales C., Hay A., Tsiantis M. (2005). SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. (2006). Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel S.E., Zhang Y., Allen E., Guo L., Reynolds T.L., Yang X., Kovalic D., Roberts J.K. (2008). Characterization of unique small RNA populations from rice grain. PLoS ONE 3: e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. (2003). Vienna RNA secondary structure server. Nucleic Acids Res. 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C., Schöb H., Stadler M., Meins F., Jr, Si-Ammour A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Zeller G., Henz S.R., Buechel S., Sachsenberg T., Wang J.W., Rätsch G., Weigel D. (2010). Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc. Natl. Acad. Sci. USA 107: 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Brière C., Naya L., Sallet E., Calenge F., Frugier F., Hartmann C., Gouzy J., Crespi M. (2009). Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lu C., et al. (2008). Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Coruh C., Axtell M.J. (2010). Arabidopsis lyrata small RNAs: Transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A., Vaucheret H. (2010). Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Elmayan T., Vaucheret H. (2008). MicroRNA maturation and action—The expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol. 11: 560–566 [DOI] [PubMed] [Google Scholar]
- Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. (2010). Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 20: 49–54 [DOI] [PubMed] [Google Scholar]
- Mette M.F., van der Winden J., Matzke M., Matzke A.J. (2002). Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 130: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Molnár A., Schwach F., Studholme D.J., Thuenemann E.C., Baulcombe D.C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447: 1126–1129 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008a). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.A., Hammond S.M. (2010). Emerging paradigms of regulated microRNA processing. Genes Dev. 24: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T., Chitwood D.H., Madi S., Ohtsu K., Schnable P.S., Scanlon M.J., Timmermans M.C. (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J., Jordan I.K. (2008). Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M., Vaughn M.W., Martienssen R.A. (2006). MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., Bartel D.P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital D.G., Doudna J.A. (2010). Structural insights into RNA interference. Curr. Opin. Struct. Biol. 20: 90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Axtell M.J., Fedoroff N.V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Subramanian S., Fu Y., Sunkar R., Barbazuk W.B., Zhu J.K., Yu O. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhou X., Zheng Y., Zhang W., Zhu J.K. (2008). Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarzynska B., Sobkowiak L., Pant B.D., Balazadeh S., Scheible W.R., Mueller-Roeber B., Jarmolowski A., Szweykowska-Kulinska Z. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37: 3083–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Moxon S., Santos D.M., Jing R., Fevereiro M.P., Moulton V., Dalmay T. (2008). High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2009). AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 4: e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Blevins T., Ailhas J., Boller T., Meins F., Jr (2008). Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 36: 6429–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang Y., Hindemitt T., Mayer K.F. (2006). Significant sequence similarities in promoters and precursors of Arabidopsis thaliana non-conserved microRNAs. Bioinformatics 22: 2585–2589 [DOI] [PubMed] [Google Scholar]
- Warthmann N., Das S., Lanz C., Weigel D. (2008). Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol. Biol. Evol. 25: 892–902 [DOI] [PubMed] [Google Scholar]
- Werner S., Wollmann H., Schneeberger K., Weigel D. (2010). Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20: 42–48 [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. (2009). Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wright S.I., Lauga B., Charlesworth D. (2002). Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau K.D., Carrington J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C. (2010). Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 107: 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F., Dong A., Huang H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. (2005). Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 280: 27595–27603 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B.R. (2005). Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Li G., Mi S., Li S., Hannon G.J., Wang X.J., Qi Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Ryvkin P., Li F., Dragomir I., Valladares O., Yang J., Cao K., Wang L.S., Gregory B.D. (2010). Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Spriggs A., Matthew L., Fan L., Kennedy G., Gubler F., Helliwell C. (2008). A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 18: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]