This article reports the characterization of rice FH5/BENT UPPERMOST INTERNODE1 (BUI1), which encodes a class II formin that nucleates actin assembly from actin or actin/profilin and bundles actin filaments and consequently influences cell expansion and rice morphogenesis.
Abstract
The actin cytoskeleton is an important regulator of cell expansion and morphogenesis in plants. However, the molecular mechanisms linking the actin cytoskeleton to these processes remain largely unknown. Here, we report the functional analysis of rice (Oryza sativa) FH5/BENT UPPERMOST INTERNODE1 (BUI1), which encodes a formin-type actin nucleation factor and affects cell expansion and plant morphogenesis in rice. The bui1 mutant displayed pleiotropic phenotypes, including bent uppermost internode, dwarfism, wavy panicle rachis, and enhanced gravitropic response. Cytological observation indicated that the growth defects of bui1 were caused mainly by inhibition of cell expansion. Map-based cloning revealed that BUI1 encodes the class II formin FH5. FH5 contains a phosphatase tensin-like domain at its amino terminus and two highly conserved formin-homology domains, FH1 and FH2. In vitro biochemical analyses indicated that FH5 is capable of nucleating actin assembly from free or profilin-bound monomeric actin. FH5 also interacts with the barbed end of actin filaments and prevents the addition and loss of actin subunits from the same end. Interestingly, the FH2 domain of FH5 could bundle actin filaments directly and stabilize actin filaments in vitro. Consistent with these in vitro biochemical activities of FH5/BUI1, the amount of filamentous actin decreased, and the longitudinal actin cables almost disappeared in bui1 cells. The FH2 or FH1FH2 domains of FH5 could also bind to and bundle microtubules in vitro. Thus, our study identified a rice formin protein that regulates de novo actin nucleation and spatial organization of the actin filaments, which are important for proper cell expansion and rice morphogenesis.
INTRODUCTION
Rice (Oryza sativa) is a major food resource for nearly half of the world human population. Rice productivity is highly associated with its architectural pattern, including plant height, which is attributable mainly to stem internode elongation (Sasaki et al., 2002; Wang and Li, 2008). The uppermost internode is of particular importance for rice productivity, since the elongation of the uppermost internode promotes panicle emergence (Zhu et al., 2006). The phytohormones gibberellins (GAs) and brassinosteroids are the two major factors that affect rice internode length by modulating cell expansion (Wang and Li, 2008). The cytoskeleton, including microtubules and actin microfilaments, is also essential for plant development and morphogenesis by modulation of cell expansion. For example, loss of function of DWARF AND GLADIUS LEAF1, which encodes an ATPase katanin-like protein in rice, caused disorganization of microtubule arrays and inhibited cell elongation, resulting in a dwarf phenotype (Komorisono et al., 2005). However, the information about the functions of the actin cytoskeleton in cell elongation and rice morphogenesis is rather limited.
Pharmacological perturbation of actin organization indicates that the actin cytoskeleton is a major regulator of cell elongation in Arabidopsis thaliana and other plant species (Baluska et al., 2001; Collings et al., 2006). Simultaneous downregulation of ACTIN2 and ACTIN7 reduced cell elongation in Arabidopsis hypocotyls (Kandasamy et al., 2009). Misexpression of actin regulatory proteins, such as profilin and actin-depolymerizing factors, also perturbs cell elongation (Ramachandran et al., 2000; Dong et al., 2001; Kandasamy et al., 2009). In addition, the actin cytoskeleton plays pivotal roles in polar cell expansion and the establishment of cell division planes by governing cytoplasmic streaming, organelle movement, and vesicle transport (Martin et al., 2001; Staiger and Blanchoin, 2006). However, the molecular mechanisms by which actin regulates these physiological processes remain poorly understood.
The function of the actin cytoskeleton is tightly coupled with its dynamic properties (Traas et al., 1987). Actin dynamics include maintenance of the monomeric actin (G-actin) pool, nucleation, actin filament assembly and disassembly, actin bundle formation, and actin cable construction, which are modulated by a precise orchestration of the activities and functions of a plethora of actin binding proteins (Staiger and Blanchoin, 2006; Higaki et al., 2007). Nucleation is the rate-limiting step during spontaneous actin filament assembly (Pollard and Borisy, 2003). To date, several actin nucleation factors have been identified, including the Actin-Related Protein2/3 complex, formins, Spire, Cordon-bleu, Leiomodin, and Junction-Mediating and Regulatory protein, which allow the cell to determine when and where to polymerize actin filaments (Baum and Kunda, 2005; Quinlan et al., 2005; Ahuja et al., 2007; Chereau et al., 2008; Zuchero et al., 2009).
Formins, originally identified from a mouse limb deformity mutant, have been found to exist in many eukaryotic organisms, including animals, fungi, and plants, and are involved in many fundamental cellular processes, including cytokinesis, cell motility, and polarity (Woychik et al., 1990; Goode and Eck, 2007). Formins are multidomain-containing proteins, characterized by two highly conserved formin-homology domains, FH1 and FH2. Some formins in fungi and animals also share additional conserved domains such as the FH3 domain, the Rho binding domain, the Diaphanous-autoregulatory domain, and the Diaphanous-inhibitory domain, which confer functional regulation of these formins (Goode and Eck, 2007). The FH1 domain, consisting of several consecutive polyproline stretches, binds profilin or profilin/actin complexes to induce actin polymerization from the barbed end (Pruyne et al., 2002; Kovar et al., 2006). The number of polyproline stretches differs among formin proteins. The FH2 domain contains actin binding sites and acts as a dimer to nucleate new actin filaments (Pruyne et al., 2002; Xu et al., 2004; Otomo et al., 2005). General activities of formins include nucleating actin assembly and interacting with the barbed end of actin filaments (Kovar, 2006; Goode and Eck, 2007). Some formins also have severing and bundling activities (Harris et al., 2004; Michelot et al., 2005; Moseley and Goode, 2005; Harris et al., 2006). Along with their functions in regulating actin cytoskeleton organization, several animal formins, including mDia1, mDia2, Cappuccino (Capu), and Inverted Formin1 (INF1), have been shown to bind directly to microtubules, thus regulating their dynamic properties (Palazzo et al., 2001; Rosales-Nieves et al., 2006; Bartolini et al., 2008; Young et al., 2008; Miki et al., 2009). The plant formin At FH4 also associates with microtubules via the unique group Ie domain, which is located between the transmembrane domain and the FH1 domain (Deeks et al., 2010), suggesting that these formins may function in the crosstalk between microtubule and actin cytoskeleton systems.
Plant genomes encode a large family of formins that do not share recognizable autoregulatory domains found in animal and yeast formins (Grunt et al., 2008). Based on sequence similarity and conservation, plant formins are divided into two classes, referred to as class I and class II. Class I formins have an N-terminal membrane-anchoring domain followed by a transmembrane region and C-terminal FH1 and FH2 domains, whereas class II formins carry an N-terminal phosphatase and tensin-related (PTEN)-like domain besides the conserved FH1 and FH2 domains (Deeks et al., 2002; Cvrcková et al., 2004; Grunt et al., 2008). Functional analysis of class I formins has shown that they are involved in many fundamental physiological processes. For instance, a loss-of-function mutation of At FH5 leads to defective cytokinesis in seed endosperm (Ingouff et al., 2005). At FH5 was recently shown to stimulate actin assembly from the subapical domain of pollen tubes to facilitate pollen tube growth, and downregulation of FH5 inhibits tip growth of pollen tubes in Nicotiana tabacum (Cheung et al., 2010). Downregulation of AFH3 causes depolarized pollen tube growth, whereas overexpression of AFH1 arrests pollen tube tip growth, and overexpression of At FH8 disturbs root hair tip growth (Cheung and Wu, 2004; Yi et al., 2005; Ye et al., 2009). By contrast, functional studies of class II formins are relatively limited. Recent results from the moss Physcomitrella patens demonstrated that class II formins could elongate actin filaments at an amazingly rapid rate and are essential for polar cell expansion (Vidali et al., 2009). Most recently, a class II formin, AFH14, was reported to bind both microtubules and microfilaments and was shown to play important roles in cell division and microspore formation in Arabidopsis (Li et al., 2010). However, it remains a major challenge to explore the physiological functions of each formin protein in plants, particularly in the model cereal rice, for which no single formin has been functionally characterized.
We previously reported the identification of the rice ELONGATED UPPERMOST INTERNODE (EUI) gene encoding a P450 that deactivates bioactive GAs. Mutation of Eui led to increased bioactive GA levels and an extremely elongated uppermost internode (Zhu et al., 2006). To understand rice internode development better, we characterized a rice mutant with a bent uppermost internode, designated as bent uppermost internode1 (bui1). The bui1 mutant displayed multiple defects in plant architecture, including a bent uppermost internode, short culms, high tillering, wavy panicle rachises, and enhanced gravitropism. Map-based cloning revealed that BUI1 encodes the class II formin FH5. Cytological and biochemical analyses demonstrated that FH5/BUI1 plays essential roles in diffuse cell expansion and rice morphogenesis.
RESULTS
Phenotypic Characterization of the bui1 Mutant
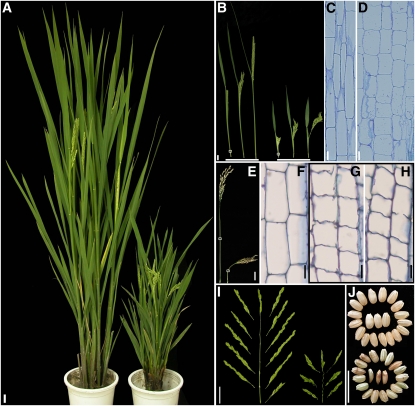
To understand better the cellular mechanism underlying rice internode development, we isolated a nonclassic dwarfed mutant from a γ-ray–induced mutation pool (Zhu et al., 2006). The mutant was named bui1 based on its bent uppermost internode phenotype (see below). Compared with wild-type plants, bui1 had more tillers but significantly reduced plant height (Figure 1A; see Supplemental Figures 1A and 1B online). During the early heading stage, the bui1 uppermost internodes were much shorter than those of the wild-type plants, and young panicles of bui1 could hardly break out of the flag leaf sheath (Figure 1B). Longitudinal section analysis revealed that the cells of the bui1 uppermost internode were markedly shorter and swelled compared with the wild-type cells (Figures 1C and 1D; see Supplemental Figure 1C online). These results indicate that cell elongation was inhibited by the bui1 mutation.
Figure 1.
Phenotypes of Wild-Type and bui1 Mature Plants.
(A) Wild-type (Zhejing 22; left) and bui1 (right) plants. Bar = 2 cm.
(B) Panicle exsertion of wild-type (left three) and bui1 (right three) plants. Bar = 2 cm.
(C) and (D) Longitudinal sections of the uppermost internodes of the wild type (C) and bui1 (D) at heading stage. The regions used for analysis are indicated with squares in (B). Bars = 100 μm.
(E) Panicles of the wild type (left) and bui1 (right) at the mature stage. Bar = 2 cm.
(F) to (H) Longitudinal sections of the uppermost internode regions (indicated with squares in [E]) of the wild type (F) and bui1 (G). Severely slanted cells of the same bui1 internode region are shown in (H). Bars = 50 μm.
(I) Panicle rachis of the wild type (left) and bui1 (right). Note the wavy rachis in bui1. Bar = 2 cm.
(J) Grains of the wild type (top) and bui1 (bottom). Bar = 0.5 cm.
Although the bui1 panicles could eventually protrude from the leaf sheath, they were small and loose, exhibiting a cockscomb shape (Figure 1B). The uppermost internode of bui1 did not grow upward; instead, it was bent almost perpendicularly to the vertical axis (Figure 1E). To understand fully the cellular basis underlying the bending phenotype, we analyzed cell morphology in the middle part of the uppermost internode, where shoot bending was most severe. Longitudinal section analysis revealed that, in addition to the shorter and slightly swollen cell shapes, bui1 also displayed other cellular phenotypes. In the wild-type plants, cell margins were even and connected tightly with each other (Figure 1F), whereas cell boundaries in bui1 were rough, with obvious protuberances along the margins and ruptures in cell-to-cell boundaries (Figure 1G). Compared with the rectangular cells in the wild type, most bui1 cells were slanted, and cell files were not aligned properly (Figure 1H). Taken together, these results indicate that polar cell expansion was also disrupted in bui1.
The bui1 mutant displayed other morphological defects besides the bent uppermost internode. All primary and secondary rachises of bui1 were wavy, and the number of rachis branches and spikelets were reduced compared with the wild-type plants (Figure 1I). The seeds of bui1 were smaller with irregular shapes and were not well filled (Figure 1J).
Mutation of BUI1 Affects Seedling Development and Gravitropism
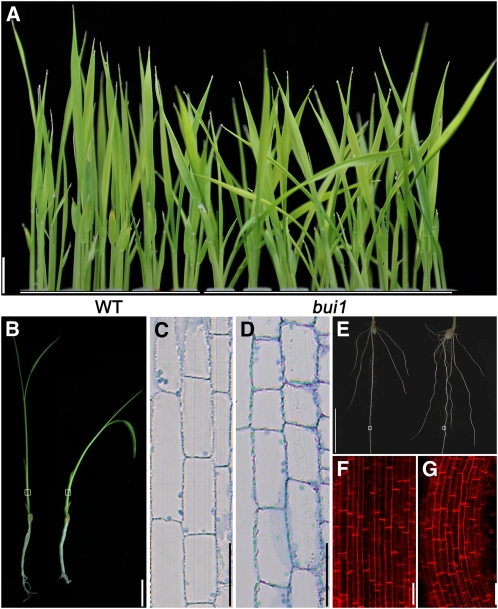
Wild-type young seedlings display erect upward growth due to negative gravitropism, whereas bui1 seedlings bend in all directions (Figure 2A), suggesting an altered gravitropic response in bui1. The bending of bui1 under normal growth conditions makes it difficult to analyze shoot gravitropism. We thus analyzed the gravitropic response of the roots instead. The kinetics of gravity-induced root bending of light-grown seedlings revealed that gravitropism in bui1 was strongly promoted, with an accelerated response to the gravitropic stimulus and an increased bending degree (see Supplemental Figure 2 online).
Figure 2.
Comparison of Wild-Type and bui1 Seedlings.
(A) Wild-type (WT) and bui1 seedlings (7 d old). Note that the bui1 shoots are bent. Bar = 2 cm.
(B) to (D) Longitudinal sections of seedling shoots of the wild type (C) and bui1 (D). (B) shows 9-d-old seedlings of the wild type (left) and bui1 (right). Bar in (B) = 2 cm; bars in (C) and (D) = 100 μm.
(E) Roots of the wild type (left) and bui1 (right) grown in half-strength medium. Bar = 1 cm.
(F) and (G) Cell morphology of wild-type (F) and bui1 (G) roots by propidium iodide staining. The regions used for analysis are indicated with squares in (E). Bars = 50 μm.
The shoots of bui1 seedlings were shorter than those of wild-type plants (Figure 2B; see Supplemental Figure 3A online). Longitudinal section analysis revealed that cell elongation was inhibited in the bui1 shoot (Figures 2C and 2D; see Supplemental Figure 3B online). Interestingly, when cultured in half-strength Murashige and Skoog medium, bui1 roots displayed a strong wavy phenotype, with root length only slightly reduced (Figure 2E; see Supplemental Figure 3C online). To gain insight into the cellular mechanism for the wavy-root phenotype, we analyzed the internal structure of bui1 and wild-type roots. These analyses revealed that cells in the bui1 roots were slanted with reduced cell length but little change in cell width (Figures 2F and 2G; see Supplemental Figure 3D online). Cell files corresponding to the wavy region also exhibited a wavy manner, but they still remained parallel to each other (Figure 2G), suggesting that this wavy growth behavior is different from the twisted growth in the Arabidopsis microtubule-related mutants (Ishida et al., 2007). We speculated that bui1 might be defective in normal root thigmotropism, as observed in the Arabidopsis mildew resistance locus o4 mutant (Chen et al., 2009). Based on these observations, we concluded that cell growth was inhibited in bui1 but to a lesser extent at the seedling stage.
BUI1 Encodes the Class II Formin FH5
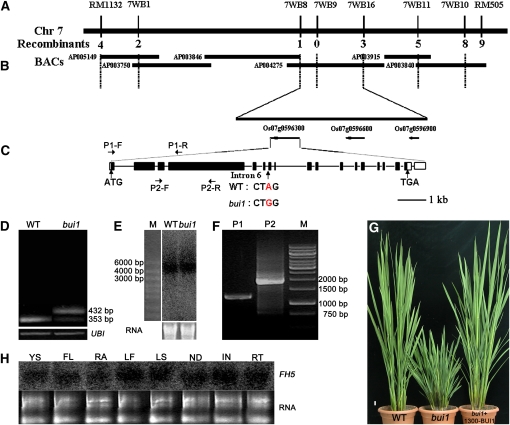
To understand fully the cellular mechanism by which the bui1 mutation inhibits cellular growth, we isolated the BUI1 gene using a map-based cloning strategy. An F2 population of ~10,000 plants was generated from a cross between bui1 and Zhenshan 97 (an indica cultivar) and was used for PCR-based mapping. The BUI1 locus was delimited to a genomic region on chromosome 7 between two simple sequence repeat (SSR) markers, RM1132 and RM505 (Figure 3A). Six insertion/deletion markers were developed between the two SSR markers, and BUI1 was further narrowed down to a 60-kb DNA region between 7WB8 and 7WB16 on a single BAC, AP004275. There are three open reading frames (ORFs) in this region (Figure 3B). Sequencing analysis revealed that bui1 had a substitution of A to G in the sixth intron of the gene FH5/Os07g0596300 (Figure 3C), changing the highly conserved 3′-intron end AG to GG. We suspected that this mutation interferes with the correct splicing of the sixth intron. Indeed, RT-PCR analysis revealed the presence of the 79-bp intron fragment in the predominantly amplified cDNA fragment of FH5/Os07g0596300 (Figure 3D).
Figure 3.
Map-Based Cloning of BUI1.
(A) The BUI1 locus was mapped on chromosome 7 between two SSR markers, RM1132 and RM505.
(B) Fine-mapping of BUI1. BUI1 was narrowed to a 60-kb region between two makers, 7WB8 and 7WB16, on a single BAC (AP004275), which contains three predicted genes.
(C) Sequence comparison revealed a substitution of A to G in one intron of the gene FH5/Os07g0596300 in bui1. WT, wild type.
(D) Detection of altered splicing in bui1 by RT-PCR analysis. Rice UBI1 was used as an internal control.
(E) RNA gel blot analysis to confirm the length of the FH5/BUI1 transcript. Total RNA (10 μg) extracted from the wild type and bui1 was used for the analysis (shown below the blot). M, RNA ladder.
(F) Confirmation of the full-length FH5/BUI1 transcript by RT-PCR. The primer pairs (P1 and P2) used for RT are indicated in (C).
(G) Complementation test of the FH5/BUI1 gene. One representative line (1300-BUI1) of complementation is shown. Bar = 2 cm.
(H) Expression pattern of FH5 revealed by RNA gel blot. YS, Young seedling; FL, flower; RA, rachis; LF, leaf; LS, leaf sheath; ND, node; IN, internode; RT, root.
RNA gel blot analysis was performed to determine the transcript length of FH5. Using a probe specific to its 3′ region, as supported by an isolated cDNA clone (GenBank accession number AK120222), we detected an ~5-kb-long transcript (Figure 3E), suggesting that the previously predicted full-length mRNA sequence (GenBank accession number NM_001066706) was a truncated one. The detected transcript size is consistent with the predicted protein of the FH5 locus (Q84ZL0, isoform 1, annotated by the UniProtKB/Swiss-Prot website), which is 1627 amino acids long. The 5′ region of the FH5 transcript was verified by RT-PCR using two sets of primers (Figures 3C and 3F) and sequencing. We thus generated a cDNA clone containing the entire ORF for the annotated FH5 polypeptide. This ORF and the native FH5 promoter were then cloned into a binary vector, and the resulting transgene was introduced into bui1 plants. Figure 3G shows that the FH5 minigene successfully complemented the mutant phenotypes. We thus concluded that the A-to-G substitution in the FH5 locus is responsible for the observed bui1 mutant phenotypes.
Plant formins are named on the basis of sequence similarity and conservation by phylogenetic analysis, and BUI1 corresponds to FH5, a class II formin (Cvrcková et al., 2004; Grunt et al., 2008). FH5 is predicted (http://hits.isb-sib.ch/cgi-bin/PFSCAN) to contain a PTEN-like domain (amino acids 198–336) at its N terminus, followed by a typical FH1 domain (amino acids 825–1170) and an FH2 domain (amino acids 1188–1588; see Supplemental Figure 4A online). The FH1 domain consists of 21 polyproline stretches (see Supplemental Figure 5 online), and the FH2 domain shares 31, 31, 29, 30, 61, and 24% amino acid similarity with those of the formins AFH1, AFH3, At FH5, At FH8, For2A, and Bni1p, respectively (see Supplemental Figure 4B online). Importantly, the key residues (Ile-1431 and Lys-1601) of Bni1p involved in actin nucleation and barbed end capping are also conserved in FH5 FH2 (see Supplemental Figure 4B online; Xu et al., 2004), implying that FH5 may perform conserved actin regulatory actions. Inclusion of the sixth intron possibly introduced a premature stop codon and could result in a truncated protein that contains the PTEN domain, the FH1 domain, and part of the FH2 domain in bui1, suggesting that a complete FH2 domain is essential for FH5 function. Because the mutation in the FH5/BUI1 gene is fully recessive, we proposed that the predicted truncated protein, which could accumulate predominantly in bui1, could interfere with the function of the less produced full-length protein.
RNA gel blot analysis revealed that FH5 is expressed ubiquitously in all tissues (Figure 3H). RT-PCR analysis demonstrated that most rice formins are expressed ubiquitously in all the examined tissues, with the exception of FH18 and FH12, whose transcripts could be detected only in the young panicles (see Supplemental Figure 6 online).
Actin Filament Organization in bui1
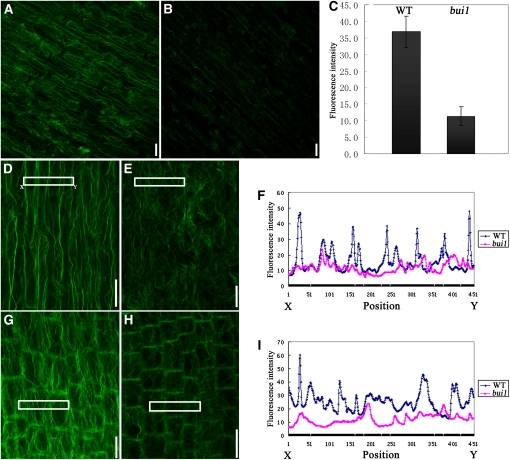
Because formins are known for their abilities to regulate actin cytoskeleton assembly and organization (Goode and Eck, 2007), we sought to determine the effect of bui1 mutation on actin organization. We examined the actin cytoskeleton organization in the wild type and bui1 directly by staining root cells with AlexaFluor488–phalloidin, which has been widely used for staining actin cytoskeleton in plants. We found that the overall fluorescence signal of the bui1 cells was much weaker than that of the wild-type cells under identical staining conditions and confocal settings (Figures 4A and 4B). Since phalloidin specifically binds to actin filaments (F-actin), the amount of F-actin is proportional to the fluorescence intensity. Therefore, we decided to compare the F-actin levels between the wild-type and bui1 cells by quantifying fluorescence intensity. As shown in Figure 4C, the average fluorescence intensity was reduced by nearly threefold in bui1. These results suggest that FH5/BUI1 plays an important role in maintaining the level of F-actin.
Figure 4.
F-Actin Organization in Wild-Type and bui1 Cells.
F-actin organization was visualized by AlexaFluor488–phalloidin staining. Each image is a maximum projection of the fluorescence signals.
(A) and (B) F-actin organization in the cortex cells of the root elongation regions of the wild type (A) and bui1 (B). Bars = 20 μm.
(C) Quantitative analysis of F-actin levels in wild-type (WT) and bui1 cells as detected in (A) and (B). Data shown are means ± se of fluorescence intensity of 144 cells in the wild type and bui1. P < 0.01, by t test.
(D) and (E) F-actin organization in the root elongation region cells of the wild type (D) and bui1 (E). Confocal settings for bui1 were increased to give clear signals. Bars = 20 μm.
(G) and (H) F-actin organization in the root transition region cells of the wild type (G) and bui1 (H). Confocal settings for bui1 were increased to give clear signals. Bars = 20 μm.
(F) and (I) Fluorescence intensities corresponding to the regions marked in (D)/(E) and (G)/(H), respectively.
When the detection settings for bui1 were increased so that both the wild-type and bui1 cells gave clear fluorescence signals, we found that actin filaments were severely disorganized in the bui1 cells. In wild-type plants, cells in the root elongation zone displayed a clearly organized actin cytoskeleton structure. Prominent actin cables oriented longitudinally through the whole cell cortex and were cross-linked by transversely or longitudinally oriented fine actin filaments (Figure 4D). However, longitudinal actin cables were barely detected in the bui1 cells. Although some thick actin cables were observed in bui1, they were quite short. In addition, the fine actin filaments were also shorter and arranged randomly in bui1 cells compared with those in the wild-type cells (Figure 4E). Measurement of fluorescence intensity revealed peaks in the wild-type cells, which correspond to the thick longitudinal actin cables, and such peaks were compromised in the bui1 cells (Figure 4F). We also examined the organization of actin cytoskeleton in the root transition region, where cells undergo rapid elongation. In the wild-type cells, actin cytoskeleton was composed mainly of longitudinal cables that connect with the actin-rich cell ends (Figure 4G), which were proposed to support vesicle trafficking and rapid cell expansion (Ketelaar et al., 2003). Such longitudinal actin cables were reduced in the bui1 cells, where thin actin filaments were arranged randomly in the cell cortex (Figure 4H). The decrease in the amount of actin cables in bui1 root transition region cells was also confirmed by analyzing the fluorescence intensity (Figure 4I). Similar results were obtained for the leaf sheath cells (see Supplemental Figure 7 online). To observe further the actin organization in living cells, we generated a construct with enhanced green fluorescent protein (EGFP) fused to both the C and N termini of the actin binding domain 2 (ABD2) of Arabidopsis fimbrin1 (Wang et al., 2008). The resulting EGFP-fABD2-EGFP construct was transformed into wild-type and bui1 plants. Both transformations gave more than 15 independent transgenic lines in which the actin filaments were labeled. Comparison of actin filament organization in the living cells of these transgenic plants gave similar results to those observed by AlexaFluor488–phalloidin staining (see Supplemental Figure 8 online). In summary, mutation of FH5/BUI1 resulted in severe disruption of actin filament organization in plant cells.
Both FH2 and FH1FH2 Domains of FH5 Nucleate Actin Polymerization
Our microscopic results showed that FH5 is required for the maintenance of F-actin levels and spatial organization of actin filaments in vivo. To investigate the biochemical activities of FH5 on actin cytoskeleton regulation, we attempted to generate recombinant proteins containing the FH2 domain and the FH1FH2 domain of FH5 (see Supplemental Figure 4C online). While the FH2 domain of FH5 was expressed and purified successfully as both a 6×His fusion and a glutathione S-transferase (GST) fusion (see Supplemental Figure 4D, lanes 2 and 3, online), the full-length FH1FH2 domain of FH5 failed to express in different Escherichia coli strains, yeast cells, or the baculovirus expression system after numerous attempts with varying induction schemes. This might be due to the high content of Pro. Indeed, FH5 has 21 polyproline stretches in the FH1 domain, which ranks the most among all rice formins (see Supplemental Figure 5 online). It was previously shown that the FH1 domain is required for formin to bind and elongate actin processively in the presence of profilin and that the number of polyproline stretches in the FH1 domain is proportional to the effect of profilin on increasing the elongation rate of barbed ends that are associated with the FH1FH2 domain (Kovar and Pollard, 2004; Paul and Pollard, 2008). Thus, the number of polyproline stretches in a given formin is related quantitatively but not qualitatively to its effect on actin assembly from profilin-bound actin. Therefore, we generated several FH5 expression constructs containing FH2 plus the FH1 domain with decreasing numbers of polyproline stretches at the N terminus, expressed them in E. coli, and succeeded in expressing and purifying a recombinant FH5 FH1FH2 protein carrying 16 polyproline stretches (see Supplemental Figure 4D, lane 4, online).
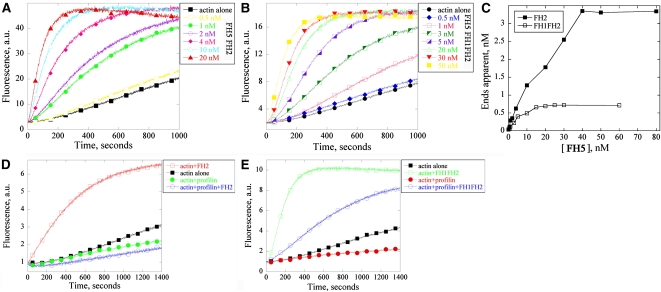
Because the nucleation activity is a general and basic feature of formins, we first examined the effects of FH5 FH1FH2 and FH5 FH2 on actin polymerization by the pyrene–actin assay and direct visualization of actin filaments using fluorescent light microscopy. In the kinetic pyrene–actin assays, both FH5 FH1FH2 and FH5 FH2 recombinant proteins decreased the initial lag phase of actin polymerization in a dose-dependent manner, indicating their active nucleation activity (Figures 5A and 5B). The FH2 domain alone was sufficient to nucleate actin assembly (Figure 5A). This quality is similar to that of AFH1 (Michelot et al., 2005) but is distinct from that of AFH5 and AFH3, which nucleate actin assembly only in the presence of the FH1 domain (Ingouff et al., 2005; Ye et al., 2009).
Figure 5.
FH5 Nucleates Actin Assembly from G-Actin and the Profilin/Actin Complex.
(A) and (B) Time course of actin polymerization in the presence of FH5 FH2 (A) or FH5 FH1FH2 (B) monitored by pyrene fluorescence. Various concentrations of FH5 FH2 or FH5 FH1FH2 were added to 2 μM 5% pyrene-labeled actin before the initiation of actin polymerization.
(C) Nucleation efficiency of FH5 FH2 and FH5 FH1FH2. The efficiency was calculated at half-maximal actin polymerization according to Blanchoin et al. (2000).
(D) FH5 FH2 was not able to nucleate actin from the profilin/actin complex. The reactions were conducted with different combinations as indicated at right.
(E) FH5 FH1FH2 was able to nucleate actin assembly from the profilin/actin complex. The reactions were conducted with different combinations as indicated at right.
Direct visualization of actin polymerization showed that addition of 100 nM FH2 or 100 nM FH1FH2 of FH5 reduced the length of actin filaments but increased the number of actin filaments in the observation field (see Supplemental Figures 9A–9C online), which is in perfect agreement with the pyrene polymerization curves (Figures 5A and 5B). Similar results were obtained with addition of 100 nM AFH1 FH1FH2 (see Supplemental Figure 9D online), indicating that these proteins indeed possess actin nucleation activity. Because the length of actin filaments is known to be inversely proportional to the number of nuclei, we also measured the length of actin filaments. As shown in Supplemental Figure 9E online, compared with that of actin alone, the actin filament lengths were reduced in the presence of 100 nM FH5 FH2 and 100 nM FH5 FH1FH2. Both recombinant proteins behaved quite similarly to the well-characterized AFH1 FH1FH2. These results demonstrated that both FH1FH2 and FH2 of FH5 could nucleate actin assembly in vitro.
FH5 Nucleates Actin Assembly from the Profilin/Actin Complex
Previous studies have shown that in some plant cells, the majority of actin monomers are sequestered by profilin, a highly abundant actin binding protein, to suppress spontaneous actin polymerization (Gibbon et al., 1997; Wang et al., 2005; Chaudhry et al., 2007). Several Arabidopsis formins have been shown to nucleate actin assembly from the profilin/actin complex (Deeks et al., 2005; Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009). We thus decided to test whether FH5 also possesses such a nucleation activity. As shown in Figure 5D, FH5 FH2 nucleated actin efficiently in the absence of profilin. However, when adding an equimolar amount of human profilin I, the initial lag time corresponding to the nucleation phase increased, suggesting that profilin strongly inhibited the FH5 FH2–facilitated actin nucleation. By contrast, FH5 FH1FH2 could reduce the initial lag phase, suggesting active nucleation (Figure 5E). We thus concluded that FH5 is capable of nucleating actin assembly from the profilin/actin complex, which absolutely requires the polyproline-rich FH1 domain.
We further analyzed the effect of FH5 on actin assembly from the profilin/actin complex by directly visualizing actin assembly via total internal reflection fluorescence microscopy (TIRFM). Actin polymerization was observed near the surface of the cover glass, which was coated with N-ethylmaleimide (NEM)–myosin alone or NEM–myosin plus FH5 FH1FH2 or FH5 FH2. The image was acquired as soon as the focal plane was found (typically less than 25 s after injection), and the interval time between subsequent images was 15 s. After the polymerization of 150 s, the number of actin filaments in the observation field increased in the presence of FH1FH2 and FH2 of FH5 (see Supplemental Figures 10A–10C online; see Supplemental Movies 1–3 online). Typically, there were three to four actin filaments per 3600 μm2 of cover slip for profilin/actin alone, and the number of actin filaments increased up to 500 in the same area in the presence of FH5 (see Supplemental Figure 10D online).
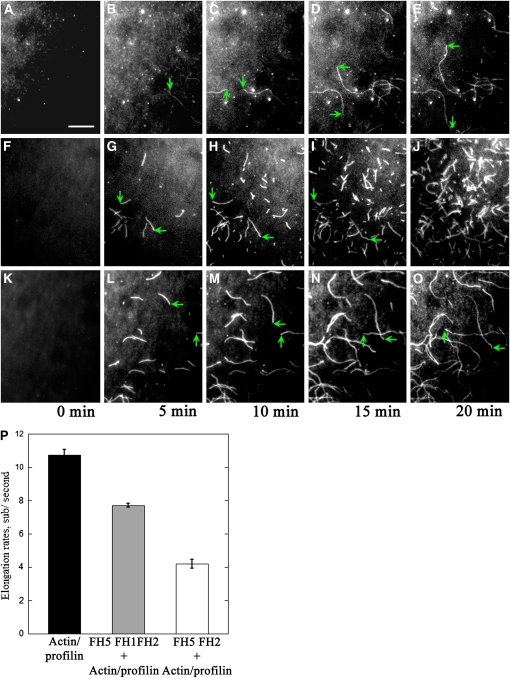
The TIRFM assay also allowed us to determine the effect of FH5 on the elongation of individual actin filaments. As shown in Figures 6A to 6E, individual actin filaments could grow to several micrometers long during the time course of observation (Figure 6P; see Supplemental Movie 4 online). Consistent with their nucleation activity, addition of 10 nM FH5 FH2 (Figures 6F–6J) or 5 nM FH5 FH1FH2 (Figures 6K–6O) resulted in more actin filaments appearing in the observation field (see Supplemental Movies 5 and 6 online) but substantially reduced the elongation rate of actin filaments (Figure 6P). This result suggested that the presence of FH5 FH2 or FH5 FH1FH2 retarded the addition of the profilin/actin complex into the barbed end of actin filaments. Based on the elongation rate of actin filaments in the presence of FH5 FH2 or FH5 FH1FH2, we calculated the nucleation efficiency according to the slope of the pyrene–actin polymerization curve at half-maximal fluorescence (Figures 5A and 5B). FH5 FH1FH2 generated a maximum of 0.118 barbed ends per molecule (Figure 5C). This is almost 5 times more efficient than Bni1p and AFH1 (Sagot et al., 2002; Michelot et al., 2005) and ~50 times more efficient than AFH5 (Ingouff et al., 2005) but about the same efficiency as Cdc12p and mDia1 (Kovar et al., 2003; Li and Higgs, 2003).
Figure 6.
Visualization of the Effect of FH5 on Profilin/Actin Polymerization by TIRFM.
Time-lapse evanescent wave microscopy was conducted with 1.5 μM ATP-Oregon-green-actin (100% labeled) and 5 μM human profilin I in the absence or presence of FH5. Images were acquired at various times as indicated below the panels. The green arrows indicate the ends of typical actin filaments during elongation. Conditions were as follows: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 50 mM DTT, 0.2 mM ATP, 50 mM CaCl2, 100 μg/mL Glc oxidase, 15 mM Glc, 20 μg/mL catalase, and 1.0% methylcellulose. Bar = 10 μm.
(A) to (E) Time-lapse micrographs of profilin/Oregon-green-actin polymerization. The profilin/Oregon-green-actin complex was perfused into the flow cell coated with NEM–myosin. See also Supplemental Movie 4 online.
(F) to (J) Time-lapse micrographs of profilin/Oregon-green-actin polymerization in the presence of FH5 FH2. The profilin/Oregon-green-actin complex with 10 nM FH5 FH2 was perfused into the flow cell coated with NEM–myosin. See also Supplemental Movie 5 online.
(K) to (O) Time-lapse micrographs of profilin/Oregon-green-actin polymerization in the presence of FH5 FH1FH2. The profilin/Oregon-green-actin complex with 5 nM FH5 FH1FH2 was perfused into the flow cell coated with NEM–myosin. See also Supplemental Movie 6 online.
(P) The mean elongation rate (±se) of actin filaments. The elongation rates were measured with profilin/actin alone (10.8 ± 0.3; n = 17), profilin/actin plus 5 nM FH5 FH1FH2 (4.2 ± 0.3; n = 11), and profilin/actin plus 10 nM FH5 FH2 (7.7 ± 0.1; n = 17). P < 0.01, by ANOVA test.
FH5 Binds to the Barbed End of Actin Filaments and Protects Actin Filaments from Dilution-Mediated Depolymerization
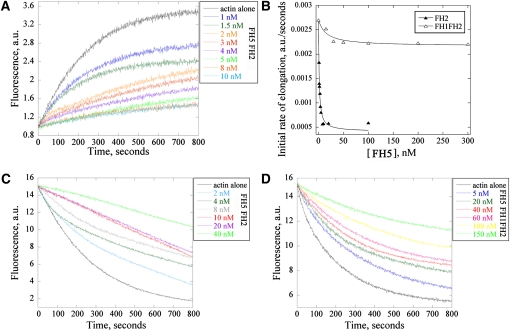
It is a general feature for formins to bind to the barbed end of actin filaments. In order to determine whether FH5 also binds to the barbed end of actin filaments and to measure the affinity for the interaction, we performed a seeded actin filament elongation assay (Blanchoin et al., 2000). As shown in Figures 7A and 7B, the initial rate of actin elongation was reduced by FH5 FH2 in a dose-dependent manner. Substoichiometric amounts of FH5 FH2 were sufficient to achieve this effect, strongly suggesting that FH5 FH2 capped actin filaments rather than sequestered actin monomers. The FH1FH2 of FH5 also reduced the initial rate of actin elongation in a similar fashion but required a much higher concentration to achieve the same effect. The effect was saturated at ~50 nM for FH1FH2. These results are in agreement with the direct TIRFM observation (Figure 6). From three independent experiments, mean Kd (n = 3) values of 2.1 ± 0.5 nM and 16.4 ± 2.1 nM were determined for FH2 and FH1FH2 of FH5, respectively. It seems that the presence of the FH1 domain substantially compromised the ability of FH2 to prevent the addition of actin to the barbed end of actin filaments, which is consistent with the leaky capping property reported previously for Bni1p and AFH1 (Evangelista et al., 1997; Michelot et al., 2005).
Figure 7.
FH5 Binds to the Barbed End of Actin Filaments and Inhibits Dilution-Mediated Actin Depolymerization.
(A) Kinetics of actin filament barbed end elongation. Preformed actin filaments (1.0 μM) were incubated with various concentrations of FH5 FH2 before the addition of 0.4 μM pyrene-labeled actin monomers. a.u., absorbance units.
(B) Plot of the initial rate of actin elongation versus the concentrations of FH5 FH2 and FH5 FH1FH2. The equilibrium dissociation constant was calculated by fitting the data with Equation 1 (see Methods). The representative Kd is 1.2 nM for FH5 FH2 and 14.6 nM for FH5 FH1FH2.
(C) and (D) Kinetics of actin depolymerization in the presence of various concentrations of FH5 FH2 (C) and FH5 FH1FH2 (D) monitored by the decrease in pyrene fluorescence. F-actin (5 μM) was incubated with various concentrations of FH5 FH2 or FH5 FH1FH2 for 5 min at room temperature, and actin depolymerization was initiated by diluting the mixtures 25-fold into buffer G.
To confirm further the capping effect of FH5 on actin filaments, an actin annealing assay was performed. Because this assay requires the presence of free ends of actin filaments, we sheared phalloidin-labeled actin filaments into short actin filaments (see Supplemental Figures 11A–11D online). In the absence of FH5, these short actin filaments annealed together to form long actin filaments (see Supplemental Figures 11E–11I online). Both FH5 FH2 and FH5 FH1FH2 inhibited annealing of actin filaments (see Supplemental Figures 11F, 11G, and 11I online). The effect of FH5 on actin filament annealing is similar to that of AFH1 FH1FH2 (Michelot et al., 2005; see Supplemental Figures 11H and 11I online). These results further supported that FH5 exhibits actin filament–capping activity.
As shown above, the capping activity of FH5 prevented the addition of actin subunits into the barbed end of actin filaments. This activity could also prevent the loss of actin subunits from the barbed end and consequently stabilize actin filaments under certain conditions. To test our prediction, a dilution-mediated actin depolymerization assay was performed. Pyrene-labeled actin filaments in the absence or presence of FH5 were diluted into buffer G to initiate depolymerization, and the depolymerization of actin filaments was tracked by monitoring the decrease in pyrene fluorescence. As expected, FH5 FH2 inhibited actin depolymerization in a dose-dependent manner (Figure 7C). Similar effects were observed for the FH5 FH1FH2 (Figure 7D). Our results indicated that FH5 prevented dilution-mediated actin depolymerization, implying that FH5 could function to stabilize actin filaments.
FH5 Binds and Bundles Actin Filaments in Vitro
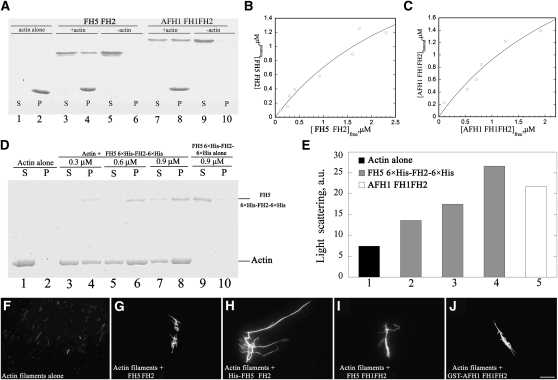
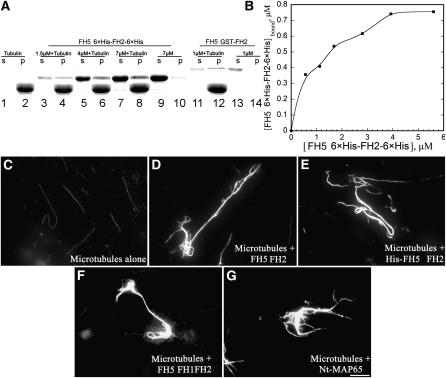
The inhibitory effect of FH5 on actin depolymerization could also be attributed to actin filament side binding activity and/or actin bundling activity. Indeed, several formins were reported to bundle actin filaments (Harris et al., 2004; Michelot et al., 2005; Moseley and Goode, 2005). We initially determined whether FH5 binds to the side of actin filaments by a high-speed cosedimentation assay. In the absence of actin, most of FH5 or AFH1, another well-characterized Arabidopsis formin (Michelot et al., 2005), stayed in the supernatant (Figure 8A, lanes 5 and 9). However, in the presence of actin, the amounts of FH5 or AFH1 cosedimented with actin increased substantially (Figure 8A, lanes 4 and 8), which suggests that FH5 can bind to actin filaments. To determine the binding affinity of FH5 to actin filaments, increasing amounts of FH5 were incubated with 3 μM F-actin and the amounts of bound FH5 were quantified by densitometry. The amounts of bound FH5 versus free FH5 were fitted with a hyperbolic function, as shown in Figure 8B for FH5 and Figure 8C for AFH1. From three independent experiments, the mean Kd values (n = 3) of 0.29 ± 0.07 μM and 0.27 ± 0.06 μM were determined for FH5 and AFH1, respectively. A stoichiometry at saturation of ~1:3 was calculated for FH5 binding to actin. To determine whether FH5 bundles actin filaments, a low-speed cosedimentation assay was employed. Our preliminary analysis showed that FH5 GST-FH2 could bundle actin filaments. To rule out the dimer formation due to the GST tag of the recombinant protein, we fused a 6×His tag to both N and C termini of FH5 FH2 and used the resulting fusion protein (6×His-FH2-6×His) to repeat the assay. In the absence of FH5 FH2, most of the actins stayed in the supernatant (Figure 8D, lane 1). However, addition of 6×His-FH2-6×His increased the amount of actin in the pellet in a dose-dependent manner (Figure 8D, lanes 4, 6, and 8), suggesting that 6×His-FH2-6×His organized actin filaments into high-order structures that could be pulled down by low-speed centrifugation. The ability of FH5 6×His-FH2-Cter-6×His to induce the formation of high-order actin structures was also confirmed by light-scattering assays, which have been used to characterize the bundling activity of VILLIN1 and AFH1 (Huang et al., 2005; Michelot et al., 2005). As shown in Figure 8E, FH5 6×His-FH2-Cter-6×His increased the absorbance of actin filaments, similar to AFH1 FH1FH2 (Michelot et al., 2005). The effect of FH5 FH2 on actin bundle formation was further confirmed by fluorescence light microscopy. In the absence of FH5 FH2, individual actin filaments could be easily observed (Figure 8F). However, in the presence of 500 nM GST-FH2, 6×His-FH2-6×His, or GST-FH1FH2 of FH5, actin filaments were organized into thick actin bundles (Figures 8G–8I), which is similar to GST-FH1FH2 of AFH1 (Figure 8J; Michelot et al., 2005). Taken together, these data unambiguously demonstrated that FH5 or its FH2 domain could efficiently bind to and bundle actin filaments in vitro.
Figure 8.
FH5 Binds and Bundles F-Actin in Vitro.
(A) A high-speed cosedimentation assay was performed to determine the binding between FH5 and F-actin. A mixture of 3 μM F-actin and 3 μM FH5 FH2 or 3 μM AFH1 FH1FH2 was centrifuged at 150,000g for 30 min at 4°C. Equal amounts of supernatant and pellet were separated by 10% SDS-PAGE and stained with Coomassie blue. S, Supernatant; P, pellet.
(B) Increasing concentrations of FH5 FH2 (0.2–3.5 μM) were cosedimented with 3 μM F-actin. The concentration of bound FH5 FH2 was plotted against the concentration of free FH5 FH2 and fitted with a hyperbolic function. The representative Kd value was calculated to be 0.30 μM for FH5 FH2.
(C) Identical experiments were performed with AFH1 FH1FH2 (0.33–3.3 μM). The representative Kd value was calculated to be 0.32 μM for AFH1 FH1FH2.
(D) Bundling activity of FH5 6×His-FH2-Cter-6×His was tested by low-speed cosedimentation assay. Lanes 1 and 2, Actin filaments (3 μM) alone; lanes 3 to 8, actin filaments with various concentrations of FH5 6×His-FH2-Cter-6×His; lanes 9 and 10, FH5 6×His-FH2-Cter-6×His alone.
(E) Light-scattering assays were performed to confirm the bundling activity of FH5 6×His-FH2-Cter-6×His. Lane 1, Actin filaments alone; lanes 2 to 4, actin filaments plus various concentrations of FH5 6×His-FH2-Cter-6×His (lane 2, 170 nM; lane 3, 340 nM; lane 4, 500 nM); lane 5, actin filaments plus 150 nM FH1FH2 of AFH1.
(F) to (J) Micrographs of actin bundles: actin filaments alone (F); actin filaments plus 500 nM FH5 FH2 (G); actin filaments plus 500 nM FH5 6×His-FH2-Cter-6×His (H); actin filaments plus 500 nM FH5 FH1FH2 (I); actin filaments plus 500 nM AFH1 FH1FH2 (J). Bar in (J) = 20 μm.
FH5 Binds to and Bundles Microtubules in Vitro, but Loss of Function of FH5/BUI1 Does Not Affect the Organization of Microtubules in Vivo
It was shown very recently that several Arabidopsis formins interact directly with microtubules (Deeks et al., 2010; Li et al., 2010), which prompted us to determine whether FH5 interacts with microtubules. To test this, we first performed microtubule cosedimentation assays. As shown in Figure 9A, in the absence of taxol-stabilized microtubules, most of the FH5 stayed in the supernatant (lanes 10 and 14). However, in the presence of taxol-stabilized microtubules, the amount of sedimented FH5 increased substantially (lanes 4, 6, 8, and 12). To determine the binding affinity, increasing amounts of FH5 were incubated with preassembled taxol-stabilized microtubules. After centrifugation, the amounts of FH5 in the supernatant and pellet were quantified by densitometry. As shown in Figure 9B, the amount of FH5 in the pellet increased in a dose-dependent manner, and the binding was saturated at about one FH5 molecule per six tubulin dimers. We tried to model these data by assuming it is one site binding, but it failed to fit well with a hyperbolic function. The estimation of 50% binding saturation from three independent experiments gave a Kd of ~0.8 μM for FH5 binding to microtubules. Given that some formins, such as AFH14, also bundle microtubules (Li et al., 2010), it was necessary to determine whether FH5 also bundles microtubules. Direct visualization by fluorescence light microscopy was performed. In the absence of FH5, only individual microtubules appeared in the field (Figure 9C). However, in the presence of 500 nM FH2, 500 nM 6×His-FH2-6×His, or 500 nM FH1FH2 of FH5, microtubule bundles appeared in the field, suggesting that FH5 is capable of bundling microtubules (Figures 9D–9F), which is similar to the presence of a well-characterized microtubule bundling factor, Nt MAP65 (Chang-Jie and Sonobe, 1993; Figure 9G). Taken together, these data suggest that FH5 binds to and bundles microtubules in vitro. We next wanted to know whether the organization of microtubules was altered in bui1 cells. The organization of cortical microtubules was revealed by immunostaining with anti-β-tubulin antibody in root cells. The results showed that the organization of cortical microtubules is rather normal in bui1 root cells (see Supplemental Figures 12I–12P online) compared with that in wild-type root cells (see Supplemental Figures 12A–12H online). This suggests that loss of function of FH5/BUI1 does not affect the organization of microtubules in root cells.
Figure 9.
FH5 Binds and Bundles Microtubules in Vitro.
(A) FH5 FH2 binds to taxol-stabilized microtubules. Taxol-stabilized microtubules (5 μM) were incubated with various concentrations of FH5 proteins for 30 min at room temperature, and the mixtures were then subjected to centrifugation at 25,000g for 30 min at 25°C. The supernatants (S) and pellet (P) were analyzed by SDS-PAGE. Lanes 1 and 2, Tubulin alone; lanes 3 to 8, tubulin with various concentrations of FH5 6×His-FH2-6×His; lanes 9 and 10, FH5 6×His-FH2-6×His alone; lanes 11 and 12, tubulin with 1 μM FH5 GST-FH2; lanes 13 and 14, 1 μM FH5 GST-FH2 alone.
(B) The amounts of FH5 6×His-FH2-6×His bound were plotted versus the total concentration of FH5. Bound represents the FH5 6×His-FH2-6×His protein in the pellet.
(C) to (G) Micrographs of microtubules in the absence or presence of FH5 proteins or Nt MAP65 as indicated: microtubules alone (C); microtubules plus 500 nM FH5 FH2 (D); microtubules plus 500 nM FH5 6×His-FH2-6×His (E); microtubules plus 500 nM FH5 FH1FH2 (F); microtubules plus 500 nM Nt MAP65 (G). Bar in (G) = 20 μm.
DISCUSSION
Formins constitute a large family of proteins that regulate actin dynamics to influence a wide range of cellular and developmental processes in animals and yeast (Goode and Eck, 2007). Although several formins were reported in Arabidopsis and moss, the function of formins in monocots remains virtually unknown. In this study, we identified FH5 as a functional formin in the model monocot rice by a forward genetics approach. Our cellular and biochemical assays demonstrate that FH5 is a prominent actin organization modulator that regulates rice morphogenesis by affecting diffuse cell expansion. Our study also reveals a previously unknown regulatory mechanism underlying the development of rice internodes, an important agronomic trait in crops.
BUI1 Encodes FH5, a Class II Formin Required for Rice Development
The actin cytoskeleton plays crucial roles in cell morphogenesis as well as in responses to external and internal stimuli (Hussey et al., 2006). The formation and reorganization of actin cytoskeletion are regulated by actin binding proteins, including formins. Despite the wealth of biochemical data obtained from studies on plant actin isovariants and actin binding proteins, genetic studies revealed only mild phenotypes restricted to certain tissues in some known actin-related mutants (Hussey et al., 2006; Staiger and Blanchoin, 2006). Overexpression or downregulation of formins in Arabidopsis led to only subtle growth defects in pollen tubes and root hairs (Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009). Recently, the class II formin AFH14 was reported to regulate cell division and microspore formation in Arabidopsis (Li et al., 2010). The failure to observe strong morphological/developmental defects in individual formin mutants is likely caused by functional redundancy and overlapping expression patterns of the plant formin genes (Deeks et al., 2002). Indeed, almost all Arabidopsis formins are expressed in multiple tissues (Cheung and Wu, 2004), and most rice formin genes are also expressed ubiquitously (see Supplemental Figure 6 online). Silencing of the moss class I formins resulted in a slight reduction in plant size with normal growth polarity, whereas strong developmental defects were observed when two class II formins were simultaneously silenced (Vidali et al., 2009). This study demonstrates that mutation of a single formin gene, FH5/BUI1, causes pleiotropic defects throughout the entire plant developmental cycle (Figures 1 and 2), indicating that FH5 plays an important role in rice growth and development.
FH5 was previously grouped into the class II formin family (Cvrcková et al., 2004; Grunt et al., 2008). The N-terminal PTEN-like domain is a characteristic feature of the class II family. Due to several mutations at the catalytic site, the PTEN-like domain of class II formins was predicted to be involved in protein localization rather than catalytic activity (Deeks et al., 2002; Cvrcková et al., 2004; Grunt et al., 2008). Indeed, the PTEN-like domain of the moss class II formin For2A is sufficient for targeting it to the apical region of growing cells (Vidali et al., 2009). Further studies are needed to determine the function(s) of the PTEN-like domain in FH5. Another distinguished feature of the class II formins is the long FH1 domain rich in polyproline stretches. It is worthy of note that FH5 contains more polyproline stretches than any other formin encoded in the rice genome (see Supplemental Figure 5 online), suggesting that FH5 might be an extremely efficient actin nucleator in the plant kingdom, which is supported by our nucleation assays.
FH5 Nucleates Actin Assembly and Bundles Actin Filaments and Microtubules
Our experiments indicated that FH5 is able to nucleate actin assembly from G-actin or profilin-bound G-actin (Figures 5 and 6; see Supplemental Figures 9 and 10 online). Unlike AFH3 and AFH5, whose actin nucleation activity requires the presence of the FH1 domain (Ingouff et al., 2005; Ye et al., 2009), FH5 FH2 is sufficient to nucleate actin assembly, suggesting that FH5 FH2 might be able to form stable dimers capable of binding and thus stabilizing actin monomers, leading to actin polymerization. Because the majority of actin is sequestered by equimolar profilin in some plant cells (Vidali and Hepler, 1997; Gibbon et al., 1999; Snowman et al., 2002; Wang et al., 2005), our finding that FH5 was able to nucleate actin assembly efficiently from the profilin-bound actin monomers suggested that FH5 could be a major promoting factor of de novo actin assembly in rice. Consistent with this hypothesis, the level of F-actin was decreased substantially in bui1 cells (Figure 4C; see Supplemental Figures 7 and 8 online).
FH5 could inhibit the elongation and annealing of actin filaments, demonstrating that FH5 has a capping activity (Figures 6 and 7; see Supplemental Figure 11 online). The presence of FH1 domain switches FH5 FH2 from a tight capper to a leaky capper, which is consistent with the leaky capping model proposed previously for Bni1p and AFH1 (Zigmond et al., 2003; Michelot et al., 2005). AFH3 also exhibits similar behavior (Ye et al., 2009). However, the moss class II formin For2A exhibits a different manner, elongating the barbed end almost 20 times faster than actin alone (Vidali et al., 2009). During the visualization of growing actin filaments on FH5 and NEM–myosin-coated cover glass, we never saw the “buckling” of actin filaments, which is the characteristic feature of a processive formin described previously (Kovar and Pollard, 2004). Therefore, we conclude that FH5 very likely is a nonprocessive formin.
FH5 could also bundle actin filaments in vitro (Figure 8). Similar to mammalian FRL1 and mDia2 (Harris et al., 2006), the FH2 domain of FH5 is sufficient to bundle actin filaments, distinguishing FH5 from AFH1, whose bundling activity requires the presence of the FH1 domain (Michelot et al., 2005). In support of the bundling activity of FH5, we discovered that the longitudinal actin cables in bui1 cells were dramatically reduced (Figures 4E and 4H; see Supplemental Figures 7 and 8 online), suggesting that FH5 also plays a prominent role in actin bundle assembly in vivo. Our detection of both capping and bundling activity of FH5 suggests that FH5 could play a role in stabilizing actin filaments, which was confirmed by dilution-mediated actin depolymerization assays (Figures 7C and 7D). Moreover, actin filament organization was severely disrupted in bui1 (Figure 4; see Supplemental Figure 8 online), further supporting the notion that FH5/BUI1 functions as an actin stabilizer in plant cells. Taken together, FH5 not only possesses the general activities of previously reported plant formins but also exhibits some previously unknown features, which might contribute to its biological significance in cell morphogenesis as well as growth and development in rice.
Despite the extensive studies uncovering the essential functions of formins in actin regulation, there is growing evidence that formins may also interact with microtubules. In animals, mDia2 and Capu bind to microtubules via the FH2 domains (Palazzo et al., 2001; Rosales-Nieves et al., 2006). INF1 binds to microtubules via its extended C-terminal microtubule binding domain but not its N-terminal FH1 and FH2 domains (Young et al., 2008). In plants, the Arabidopsis class I formin FH4 and the class II formin AFH14 were recently reported to associate with microtubules (Deeks et al., 2010; Li et al., 2010). We found that FH5 exhibits a similar feature that it can bind to microtubules directly via the FH2 domain. Both FH1FH2 and FH2 domains of FH5 can stimulate microtubule bundling in vitro, which prompted us to determine whether FH5 plays an important role in microtubule organization in vivo. However, microtubule organization in bui1 root cells was not affected (see Supplemental Figure 12 online), suggesting that the bui1 phenotypes were attributable mainly to the defects of the actin cytoskeleton. Certainly, we still cannot rule out the possibility that microtubule organization might be altered under certain conditions or in several specific cell types. Additionally, it could also be possible that microtubule dynamics are altered in bui1 cells. It deserves further characterization by measuring the parameters of microtubule dynamics, including microtubule growth and shrinking rate and the frequencies of catastrophe and rescue. Nonetheless, our study, together with other recent studies (Deeks et al., 2010; Li et al., 2010), supports the view that plant formins play important roles in the crosstalk between microtubules and actin filaments besides functioning as simple actin regulators.
FH5 Affects Diffuse Cell Expansion
Most plant cells grow by diffuse expansion, in which cell extension is dispersed over the entire cell surface and elongation occurs along one axis in a polar manner (Martin et al., 2001; Mathur, 2004). Cytological analyses reveal that FH5 affects rice morphogenesis by modulating cell expansion. FH5 affects mainly the expansion of diffusely growing cells. This is different from those previously reported Arabidopsis class I formins that are involved in tip growth of pollen tubes and root hairs (Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009; Cheung et al., 2010). During cell expansion, Golgi-derived vesicles containing membrane materials and cell wall matrix components move along the actin filaments to expansion sites and fuse with the plasma membrane to deposit the contents to the cell wall (Ketelaar et al., 2003; Kim et al., 2005; Smith and Oppenheimer, 2005; Hussey et al., 2006). The longitudinally oriented actin cables were previously proposed to be the primary tracks for vesicle movement, and disruption of the organization of such longitudinal actin cables always disturbs cell expansion (Dong et al., 2001; Hepler et al., 2001; Wasteneys and Yang, 2004; Deeks et al., 2007). It is conceivable that loss of longitudinal actin cables in bui1 probably led to insufficient vesicle transport, thus inhibiting longitudinal cell expansion. On the other hand, it appears that the level of longitudinal actin cables needs to be tightly regulated, since increasing actin cables by downregulation of Actin-Interacting Protein1 or overexpression of AFH1 also reduces cell length (Cheung and Wu, 2004; Ketelaar et al., 2004). Thus, FH5/BUI1 probably works together with other actin binding proteins to maintain the actin cables at an appropriate level to regulate cell expansion. Microtubules also play important roles in cell expansion by governing the movement of cellulose synthase complexes and microfibril deposition (Paradez et al., 2006; Paredez et al., 2006; Lloyd and Chan, 2008; Gutierrez et al., 2009). The high tensile microfibrils that are oriented transversely in the cell cortex have been supposed to resist lateral cell expansion (Lloyd and Chan, 2004). Mutation of Arabidopsis FRAGILE FIBER2 (FRA2), which encodes a plant katanin that severs microtubules, caused altered cortical microtubule organization and reduced cellulose and hemicellulose contents in cell walls, thus resulting in the short and wide cells in fra2 plants (Burk et al., 2001). As stated above, although we did not detect any disorganization of microtubules in bui1 root cells, we cannot rule out the possibilities that the organization of microtubules is altered in other cell types and/or under certain conditions or that microtubule dynamics are altered in bui1 cells, which escaped detection by immunostaining with anti-tubulin antibody. Therefore, the possibility still exists that the disturbance of microtubule cytoskeleton function may affect the deposition of cellulose microfibrils in bui1 cells, consequently inducing the release of lateral cell expansion restriction and causing swollen cells (Figures 1G and 1H; see Supplemental Figure 1C online).
The disorganized short actin filaments likely lead to misdelivery of new membrane and cell wall materials, thus resulting in rough margins in bui1 cells (Figures 1G and 1H). Additionally, cell alignment was also abnormal, with obviously slanted cells in bui1 shoots and roots (Figures 1G, 1H, and 2G). One possible explanation is that cell division plate determination is disrupted in bui1. During plant cell division, the actin-depleted zone region with weak cortical actin filaments has been shown to provide guidance for cell division plane determination (Hoshino et al., 2003; Sano et al., 2005; Van Damme et al., 2007; Panteris, 2008). Mutants such as fra7 and act7 with disrupted actin organization are known to cause defective cell morphogenesis and abnormal cell files, which was attributed to the misorientation of cell division planes (Gilliland et al., 2003; Zhong et al., 2005). It is thus tempting to speculate that the defective actin cytoskeleton organization in bui1 might interfere with normal cell division plane determination, causing the formation of abnormal cell files. Similar results come from the study by Zhang et al. (2011) in which RICE MORPHOLOGY DETERMINANT was also found to encode FH5. The detailed processes whereby FH5/BUI1 affects cell expansion remain a fascinating question for further study. It is interesting that gravitropism and thigmotropism were also changed in bui1, implying that FH5/BUI1 might participate in mechanical signaling transduction. However, the role of FH5/BUI1 in these cellular processes remains to be elucidated.
METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) bui1 mutant was isolated from γ-ray–induced mutations of a japonica cultivar (Zhejing 22). Plants were cultivated in an experimental field during natural growing seasons. For seedlings, sterile seeds were germinated in the dark for 2 d and then transferred to liquid medium in a growth chamber under growth conditions with 12-h days, 28°C, 80% RH followed by 12-h nights, 26°C, 60% RH. For statistical analysis to characterize the phenotypic differences between the wild-type and bui1 plants, we performed Student’s t test in the R programming language (version 2.11.0; http://www.R-project.org).
Gravitropism Analysis
Seeds were surface-sterilized and sown on half-strength Murashige and Skoog medium containing 0.45% phyto-agar. Four-day-old seedlings with radicles (~6 cm long) were used for gravitropism analysis. Seedlings were reorientated by 90°, and images of the roots were captured at each time point. The curvature degrees were measured from the digital images using ImageJ (version 1.38; http://rsbweb.nih.gov/ij/).
Histochemical Analysis
Tissues were fixed overnight at 4°C in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde and dehydrated in a graded ethanol series. Following substitution with xylene, the samples were embedded in Paraplast (Sigma-Aldrich) and sectioned at 8-μm thickness using a rotary microtome (Leica). Sections were stained with 0.05% toluidine blue and observed with a light microscope (Leica). For resin sections, samples were fixed with 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.2 M sodium phosphate buffer (pH 7.0) for 20 h at 48°C, rinsed with 0.1 M phosphate buffer (pH 7.0), and dehydrated in an ethanol series. The samples were embedded in Technovit 7100 resin (Hereaus Kulzer) and polymerized at 45°C. Transverse sections of 2 mm were cut using an Ultratome III ultramicrotome (LKB) and then stained with 0.25% toluidine blue O (Chroma Gesellshaft Shaud) and observed. To image root cells, roots were incubated in 10 g/mL propidium iodide for 10 min and subjected to confocal laser scanning microscopy using 530-nm excitation and 620-nm emission filters (Olympus).
Map-Based Cloning of BUI1 and Complementation
The bui1 mutant was crossed with Zhenshan 97 (indica) to generate an F2 mapping population. BUI1 was mapped to a 60-kb region on chromosome 7. Genomic DNA fragments of this region were amplified from bui1 and wild-type plants, sequenced, and compared using MegAlign (DNASTAR). For construction of the full-length ORF of FH5/BUI1, two fragments, 2826 and 2197 bp, were amplified using reverse-transcribed cDNA and the cDNA clone AK120222 as templates, respectively, and were fused using restriction sites as indicated. The primers used were CDNA-1-F (KpnI), CDNA-1-R (HindIII), CDNA-2-F (HindIII), and CDNA-2-R (SpeI; see Supplemental Table 1 online). The full-length ORF and the 3-kb promoter region of BUI1 were then cloned into the binary vector pCambia1300 (GenBank accession number AF234296) using the restriction sites SpeI, KpnI, and SacI (see Supplemental Table 1 online). The resulting construct, 1300-BUI1, was then introduced into the bui1 background via Agrobacterium tumefaciens–mediated transformation. More than 10 independent transgenic lines were produced that could successfully complement the mutant phenotypes.
Gene Expression Analysis
Total RNA was extracted from seedlings, young flowers, panicles, flag leaves, leaf sheaths, internodes, nodes, and roots using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For RNA gel blot analysis, 10 μg of RNA samples was separated on a 1% formaldehyde–agarose gel and then blotted onto Hybond-N+ membranes (Amersham). A 538-bp fragment of the FH5/BUI1 cDNA was amplified with primers northern-F and northern-R (see Supplemental Table 1 online). The product was labeled with [α-32P]dCTP using a random primer labeling kit (TaKaRa) for hybridization and autoradiography. For RT-PCR analysis, total RNA (2 μg) was reverse transcribed into cDNA and then used as template for PCR with gene-specific primers (northern-F and northern-R; see Supplemental Table 1 online). Thirty-six PCR cycles were performed for all class I formins and 30 PCR cycles for all class II formins. Rice UBI1 was used as an internal control with 30 PCR cycles using primers Rubi-F and Rubi-R (see Supplemental Table 1 online).
Observation of Microfilaments and Microtubules in Plant Cells
For observation of actin filaments, we used the method described by Vidali et al. (2007) with slight modifications. About 1-cm segments from root tips of 7-d-old seedlings or adaxial sheath epidermis of mature plants were carefully excised and incubated in PME buffer (100 mM Pipes, 10 mM EGTA, and 5 mM MgSO4, pH 6.8) containing 300 μM m-maleimidobenzoyl-N-hydroxysuccinimide ester, 1.5% glycerol, and 0.1% Triton X-100 with gentle agitation for 30 min. After cross-linking, samples were rinsed twice with PME buffer and then fixed in PME buffer containing 2% paraformaldehyde for 30 min. Samples were further rinsed with PME buffer and incubated in actin-staining buffer (PME, 1.5% glycerol, 0.1% Triton X-100, and 0.66 μM AlexaFluor488–phalloidin) at 4°C in the dark overnight. Samples were washed three times with PME and mounted in PME on glass slides for confocal fluorescence microscopy. We used more than 48 roots of wild-type and bui1 cells in one assay to quantify the F-actin level with the microscope (Zeiss LSM 510 META). The overall fluorescence signal of the wild-type and bui1 cells was obtained under identical staining conditions and confocal settings. Images were normalized to an equal grayscale, and three elongation region cells for each root were processed to determine the average pixel intensity with ImageJ (version 1.38; http://rsbweb.nih.gov/ij/) software. The region of interest size was 40 μm × 10 μm for each cell. The average pixel intensity was then plotted and analyzed in Microsoft Office Excel.
For construction of the EGFP-fABD2-EGFP fusion protein, the EGFP sequence was amplified using the plasmid DNA pEGFP (GenBank accession number U76561) and introduced into the vector pUN1301 (Wang et al., 2004) using primers N-EGFP-F and N-EGFP-R with the restriction sites BamHI and SmaI, resulting in the construct pUN1301-N-EGFP. ABD2 of Arabidopsis thaliana fimbrin1 was amplified from Arabidopsis cDNA using primers ABD2-F and ABD2-R. Another EGFP sequence was amplified using C-EGFP-F and C-EGFP-R and fused to the C termini of ABD2 using the restriction sites XbaI and SacI. The resulting ABD2-C-EGFP was introduced into pUN1301-N-EGFP using the restriction sites SmaI and SacI, and the resulting pUN1301-EGFP-ABD2-EGFP plasmid was transformed into wild-type and bui1 cells via A. tumefaciens–mediated transformation.
To reveal the organization of microtubules in vivo, root tips of 1 cm were cut off from 4-d-old rice seedlings grown in water under a light/dark cycle of 16/8 h at 32°C and subjected to fixation with 4% (w/v) paraformaldehyde in PME buffer1 (50 mM Pipes, 5 mM MgCl2, and 5 mM EGTA, pH 6.9) containing 0.05% (v/v) Triton X-100. After three washes in PME buffer1, the rice roots were digested with 1.5% (w/v) cellulase R-10 (Yakult Pharmaceutical Industry) and 1.5% (w/v) pectolase Y-23 (Yakult Pharmaceutical Industry) at 37°C for 1 h and then washed again with PME buffer1 three times. The roots were treated with ice-cold methanol at −20°C for 10 min and washed with PBS buffer (pH 6.9) at room temperature three times. After incubation in PBS buffer containing 50 mM Gly for 30 min, root tips were carefully placed onto cover slips and incubated in primary antibody (anti-β-tubulin; Sigma-Aldrich) at 4°C overnight, washed with PBS buffer three times, and then incubated with the secondary antibody (AlexaFluor488 goat anti-mouse IgG; Invitrogen) in the dark for 3 h at 37°C. Cover slips were washed twice and finally mounted with 50% (v/v) glycerol. o-Phenylenediamine (Sigma-Aldrich) was used to prevent fluorescence quenching when mounting. Microtubule observations were performed with a Leica TCS SP5 confocal laser scanning microscope, and the relevant fluorescent images were projections of Z-series sections at 0.45-μm steps.
Protein Production
The coding sequences of FH5 FH1FH2 and FH5 FH2 were amplified by RT-PCR using the primers GST-FH1FH2-F/GST-FH1FH2-R and GST-FH2-F/GST-FH1FH2-R (see Supplemental Table 1 online). The primers His-FH2-F and His-FH2-R were used to clone the 6×His fusion FH2 domain (see Supplemental Table 1 online). These fusion protein sequences were verified by sequencing and subsequently inserted into pGEX-4T-3 (GE Healthcare) and pET-32a (Novagen) vectors using the restriction sites BamHI and SalI and then transformed into Escherichia coli Tuner strain to produce the fusion proteins. The GST fusion proteins were purified with glutathione–Sepharose resin (Amersham Biosciences), and the 6×His fusion protein was purified with nickel resin (Novagen) according to the manufacturer’s instructions. The GST-purified proteins were dialyzed in 50 mM Tris-HCl, pH 8.0, and 6×His fusion proteins were dialyzed in 50 mM Tris-HCl, pH 8.0, 100 mM KCl. The purified proteins were flash-frozen in liquid nitrogen and stored at −80°C. All of these proteins were clarified further by centrifugation at 80,000g for 1 h before use. Protein samples were analyzed by 10% SDS-PAGE, the gel was stained with Coomassie Brilliant Blue R 250 (Sigma-Aldrich), and the concentrations of the proteins were determined by densitometry with ImageJ software using bovine serum as a standard. In order to remove batch-to-batch variability, more than four batches of FH5 FH1FH2, FH5 FH2, and FH5 6×His-FH2-6×His were purified according to the methods described above.
Actin was prepared from rabbit skeletal muscle as described previously (Spudich and Watt, 1971). G-actin was further purified by Sephacryl S-300 chromatography at 4°C in buffer G (0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, 0.1 mM imidazole, and 5 mM Tris-HCl, pH 8.0; Pollard, 1984). To trace the dynamics of actin polymerization and depolymerization, actin was labeled on Cys-374 with pyrene iodoacetamide (Pollard, 1984) and Oregon-green-488 iodoacetamide (Amann and Pollard, 2001), respectively. AFH1 FH1FH2 and human profilin I were purified according to the methods described previously (Michelot et al., 2005). His fusion Nt MAP65 was purified with nickel resin (Novagen) according to the manufacturer’s instructions. Porcine brain tubulin was prepared according to the method described previously by Castoldi and Popov (2003), and tubulin was labeled with N-hydroxysuccinimidyl–rhodamine according to the method described by Keating et al. (1997).
Actin Nucleation Assay
Actin nucleation assays were conducted according to the methods described previously (Higgs et al., 1999). Monomeric actin (2 μM; 5% pyrene-labeled) was incubated with various concentrations of FH5 FH2 or FH5 FH1FH2 for 5 min at room temperature. Polymerization of actin filaments was detected by pyrene fluorescence with a QuantaMaster Luminescence QM 3 PH fluorometer (Photo Technology International) immediately after the addition of one-tenth volume of 10× KMEI (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole-HCl, pH 7.0).
Affinity of FH5 FH2 and FH5 FH1FH2 for the Barbed End of Actin Filaments
To determine the affinity of FH5 for the barbed end of actin filaments, an actin elongation assay was performed according to the method of Michelot et al. (2005). Different amounts of FH1FH2 or FH2 of FH5 were incubated with 1 μM preformed actin filaments for 5 min at room temperature, and actin elongation was initiated by the addition of 0.4 μM G-actin (10% pyrene-labeled). The affinity of the fusion proteins for the barbed end of actin filaments was determined by plotting the initial actin elongation rate versus the concentration of FH5 proteins using the following equation:
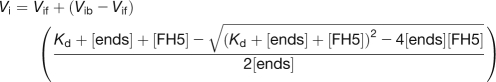 |
where Vi is the observed rate of elongation, Vif is the rate of elongation when all the barbed ends are free, Vib is the rate of elongation when all the barbed ends are capped, [ends] is the concentration of barbed end, and [FH5] is the concentration of FH5 protein. The data were modeled by Kaleidagraph software (version 3.6; Synergy Software).
Fluorescence Microscopy Observation of Actin Filaments and Microtubules in Vitro
Actin filaments labeled with rhodamine–phalloidin were observed in vitro as described previously (Blanchoin et al., 2000; Ye et al., 2009). Actin (4 μM) was polymerized with or without FH5 proteins or AFH1 FH1FH2 at room temperature in 1× KMEI at 25°C for 30 min and labeled with an equimolar amount of rhodamine–phalloidin (Sigma-Aldrich). The polymerized F-actin was then diluted to 10 nM in fluorescence buffer containing 10 mM imidazole-HCl, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 mg/mL Glc oxidase, 15 mg/mL Glc, 20 mg/mL catalase, and 0.5% methylcellulose. The effect of FH5 on microtubules was observed in vitro roughly according to the previously published methods (Huang et al., 2007; Li et al., 2010). N-hydroxysuccinimidyl–rhodamine-labeled tubulin was polymerized in PEM buffer (0.1 M Pipes, 1 mM EGTA, and 1 mM MgSO4, pH 6.9) containing 1 mM GTP (sodium salt hydrate; Sigma) at 35°C for 40 min. Taxol was added to a final concentration of 20 μM and incubated at 35°C for 20 min. The polymerized tubulin was centrifuged at 25,000g at 25°C for 20 min. Then, the pellets were resuspended gently with PEMT buffer (0.1 M Pipes, 1 mM EGTA, 1 mM MgSO4, and 20 μM taxol, pH 6.9). Taxol-stabilized microtubule was diluted with PEMT buffer to 100 nM and incubated with FH5 or Nt MAP65 for 30 min at room temperature before observation. A dilute sample (3 μL) was applied to a cover slip coated with poly-l-Lys (0.01%). Actin filaments and microtubules were observed by epifluorescence illumination with an IX71 microscope (Olympus) equipped with a 60×, 1.42-numerical aperture oil objective, and digital images were collected with a Retiga EXi Fast 1394 charge-coupled device camera (Qimaging) using Image-Pro Express software (version 6.3; Media Cybernetics). Filament length was measured using ImageJ. Actin filament annealing assays were performed according to Ye et al. (2009). Actin filaments (4 μM) labeled with equimolar rhodamine–phalloidin were sheared physically with a needle in the absence or presence of FH5 proteins or AFH1 FH1FH2. Actin filaments were then observed at various time points by epifluorescence illumination.
Depolymerization Assay
Dilution-mediated actin depolymerization was performed as described previously (Huang et al., 2003). F-actin (5 μM, 50% pyrene-labeled) was incubated with varying concentrations of FH5 FH1FH2 or FH5 FH2 at room temperature for 5 min. The depolymerization of actin filaments was initiated by diluting the mixtures 25-fold into buffer G at room temperature, and actin depolymerization was tracked by monitoring the changes in pyrene fluorescence.
TIRFM
TIRFM assays were performed essentially as described previously (Kovar and Pollard, 2004; Ye et al., 2009). The flow cell was made as described previously (Amann and Pollard, 2001). The slide was first coated with 100 nM NEM–myosin and then washed with BSA and 1× TIRF buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 50 mM DTT, 0.2 mM ATP, 50 mM CaCl2, 100 μg/mL Glc oxidase, 15 mM Glc, 20 μg/mL catalase, and 1.0% methylcellulose). Finally, 1.5 μM ATP-Oregon-green-actin (100% labeled) and 5 μM human profilin in the absence or presence of FH5 proteins were injected into the flow cell. The image was acquired as soon as the focal plane was found (typically less than 25 s after injection). Actin filaments were observed by epifluorescence illumination with a DMI6000CS microscope (Leica) equipped with a 100×, 1.46-numerical aperture HC PLAN objective. Digital images were collected with a Photometrics cascade II 512 charge-coupled device camera (Major Instruments) using LAS AF software. The interval time between subsequent images was 15 s. Filament length was measured using ImageJ. The elongation rates were converted from μm/s to subunits/s using 333 as the parameter of the number of actin monomers per micrometer.
Low-Speed Cosedimentation and Light-Scattering Assays
Low-speed cosedimentation assays were conducted to test the actin bundling properties of FH5 according to Huang et al. (2005). F-actin (3 μM) was incubated with varying concentrations of FH5 6×His-FH2-6×His protein at 25°C for 90 min. Samples were then centrifuged at 4°C, 13,600g for 30 min. Equal amounts of supernatant and pellet samples were separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue R 250.
Light-scattering assays were performed by monitoring 90° light scattering of unlabeled actin at 400 nm. Light-scattering change of actin filaments (2 μM) was recorded with a QuantaMaster Luminescence QM 3 PH fluorimeter (Photo Technology International). The data were then converted to bar graphs using Kaleidagraph software (version 3.6; Synergy Software).
Microfilament and Microtubule Cosedimentation Assays
High-speed cosedimentation assays were performed to determine the binding of FH5 to F-actin, roughly according to the recently published method (Zhang et al., 2010). Briefly, 3 μM F-actin was incubated with either FH5 FH2 or AFH1 FH1FH2 for 30 min at room temperature, and the mixtures were then subjected to centrifugation at 150,000g for 30 min at 4°C. To determine the binding affinity of FH5 to F-actin, 3 μM F-actin was mixed with various concentrations of FH5 FH2 (0.2–3.5 μM). Equal amounts of supernatant and pellet samples were separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue R 250. The Kd values for FH5 FH2 and AFH1 FH1FH2 were calculated by plotting the amount of bound protein in the pellet versus the amount of free protein in the supernatant to a hyperbolic function with Kaleidagraph software (Synergy Software).
Microtubule cosedimentation assays were adopted to determine the binding of FH5 to microtubules according to previously published methods (Huang et al., 2007; Li et al., 2010). Briefly, 5 μM preformed taxol-stabilized microtubules was incubated with FH5 FH2, FH5 6×His-FH2-6×His, or Nt MAP65 for 30 min at room temperature, and the mixtures were then subjected to centrifugation at 25,000g for 30 min at 25°C. To determine the binding affinity of FH5 to microtubules, 5 μM preformed taxol-stabilized microtubules was incubated with various concentrations of FH5 6×His-FH2-6×His. After centrifugation, equal amounts of supernatant and pellet samples were separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue R 250. The amount of FH5 in the pellet was determined by densitometry and plotted with the total concentrations of FH5.
Accession Numbers
Sequence data from this article can be found in GenBank databases under the following accession numbers: rice FH5/BUI1 (HQ123580), yeast Bni1p (NP_014128), Arabidopsis AFH1 (AAF14548), Arabidopsis AFH3 (ACQ91096), Arabidopsis At FH5 (AAK68741), Arabidopsis At FH8 (NP_177171), tobacco Nt MAP65 (AJ289864), and moss For2A (FJ997271).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Statistical Analysis of Wild-Type and bui1 Plants at the Heading Stage.
Supplemental Figure 2. Time Course of Root Gravitropic Curvature (after Reorientation).
Supplemental Figure 3. Statistical Analysis of Wild-Type and bui1 Seedlings.
Supplemental Figure 4. Alignment of the FH2 Domain of FH5 with FH2 Domains of Other Formins, and Purification of Fusion Proteins.
Supplemental Figure 5. Polyproline Stretch Number in the FH1 Domains of Rice Formins.
Supplemental Figure 6. RT-PCR Analysis of the Expression of Rice Formins in Different Tissues.
Supplemental Figure 7. F-Actin Organization in Leaf Sheath Adaxial Epidermis Cells.
Supplemental Figure 8. F-Actin Organization in Root Living Cells.
Supplemental Figure 9. Visualization of Actin Polymerization with a Fluorescence Light Microscope.
Supplemental Figure 10. Direct Visualization of Actin Nucleation from Profilin/Actin by TIRFM in the Presence of FH5 FH2 and FH5 FH1FH2.
Supplemental Figure 11. FH5 Prevents Annealing of Actin Filaments.
Supplemental Figure 12. The Organization of Microtubules Was Not Altered in bui1 Root Cells.
Supplemental Table 1. Primers Used in This Study.
Supplemental Movie 1. Time Lapse of Profilin/Oregon-Green-Actin Nucleation Corresponding to Supplemental Figure 10A.
Supplemental Movie 2. Time Lapse of the Effect of FH5 FH2 on Profilin/Oregon-Green-Actin Nucleation Corresponding to Supplemental Figure 10B.
Supplemental Movie 3. Time Lapse of the Effect of FH5 FH1FH2 on Profilin/Oregon-Green-Actin Nucleation Corresponding to Supplemental Figure 10C.
Supplemental Movie 4. Time Lapse of Profilin/Oregon-Green-Actin Polymerization Corresponding to Figures 6A to 6E.
Supplemental Movie 5. Time Lapse of the Effect of FH5 FH2 on Profilin/Oregon-Green-Actin Polymerization Corresponding to Figures 6F to 6J.
Supplemental Movie 6. Time Lapse of the Effect of FH5 FH1FH2 on Profilin/Oregon-Green-Actin Polymerization Corresponding to Figures 6K to 6O.
Acknowledgments
We are grateful to Zhenbiao Yang (University of California, Riverside) and Jianming Li (University of Michigan) for critical reading of the manuscript. We also thank Laurent Blanchoin (Centre National de la Recherche Scientifique) for providing AFH1 expression plasmid and Ming Yuan (China Agricultural University) for Nt MAP65-1 plasmid. This research was supported by grants from the National Natural Science Foundation of China (Grants 90817102 and 30721061 to Z.H; Grants 30821007 and 30771088 to S.H.) and by the Chinese Academy of Sciences through its Hundred Talent Program (to S.H.).
References
- Ahuja R., Pinyol R., Reichenbach N., Custer L., Klingensmith J., Kessels M.M., Qualmann B. (2007). Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 131: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann K.J., Pollard T.D. (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 98: 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F., Jasik J., Edelmann H.G., Salajová T., Volkmann D. (2001). Latrunculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev. Biol. 231: 113–124 [DOI] [PubMed] [Google Scholar]
- Bartolini F., Moseley J.B., Schmoranzer J., Cassimeris L., Goode B.L., Gundersen G.G. (2008). The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J. Cell Biol. 181: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Kunda P. (2005). Actin nucleation: Spire—actin nucleator in a class of its own. Curr. Biol. 15: R305–R308 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Castoldi M., Popov A.V. (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32: 83–88 [DOI] [PubMed] [Google Scholar]
- Chang-Jie J., Sonobe S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J. Cell Sci. 105: 891–901 [DOI] [PubMed] [Google Scholar]
- Chaudhry F., Guérin C., von Witsch M., Blanchoin L., Staiger C.J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18: 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Noir S., Kwaaitaal M., Hartmann H.A., Wu M.J., Mudgil Y., Sukumar P., Muday G., Panstruga R., Jones A.M. (2009). Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21: 1972–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. (2008). Leiomodin is an actin filament nucleator in muscle cells. Science 320: 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Niroomand S., Zou Y., Wu H.M. (2010). A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. USA 107: 16390–16395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings D.A., Lill A.W., Himmelspach R., Wasteneys G.O. (2006). Hypersensitivity to cytoskeletal antagonists demonstrates microtubule-microfilament cross-talk in the control of root elongation in Arabidopsis thaliana. New Phytol. 170: 275–290 [DOI] [PubMed] [Google Scholar]
- Cvrcková F., Novotný M., Pícková D., Zárský V. (2004). Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M.J., Cvrcková F., Machesky L.M., Mikitová V., Ketelaar T., Zársky V., Davies B., Hussey P.J. (2005). Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 168: 529–540 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Fendrych M., Smertenko A., Bell K.S., Oparka K., Cvrcková F., Zársky V., Hussey P.J. (2010). The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J. Cell Sci. 123: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Hussey P.J., Davies B. (2002). Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7: 492–498 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Rodrigues C., Dimmock S., Ketelaar T., Maciver S.K., Malhó R., Hussey P.J. (2007). Arabidopsis CAP1—A key regulator of actin organisation and development. J. Cell Sci. 120: 2609–2618 [DOI] [PubMed] [Google Scholar]
- Dong C.H., Xia G.X., Hong Y., Ramachandran S., Kost B., Chua N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M.S., Chow C.J., Adames N., Pringle J.R., Peter M., Boone C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276: 118–122 [DOI] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Ren H., Staiger C.J. (1997). Characterization of maize (Zea mays) pollen profilin function in vitro and in live cells. Biochem. J. 327: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland L.U., Pawloski L.C., Kandasamy M.K., Meagher R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33: 319–328 [DOI] [PubMed] [Google Scholar]
- Goode B.L., Eck M.J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76: 593–627 [DOI] [PubMed] [Google Scholar]
- Grunt M., Zárský V., Cvrcková F. (2008). Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harris E.S., Li F., Higgs H.N. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279: 20076–20087 [DOI] [PubMed] [Google Scholar]
- Harris E.S., Rouiller I., Hanein D., Higgs H.N. (2006). Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281: 14383–14392 [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Higaki T., Sano T., Hasezawa S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Higgs H.N., Blanchoin L., Pollard T.D. (1999). Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38: 15212–15222 [DOI] [PubMed] [Google Scholar]
- Hoshino H., Yoneda A., Kumagai F., Hasezawa S. (2003). Roles of actin-depleted zone and preprophase band in determining the division site of higher-plant cells, a tobacco BY-2 cell line expressing GFP-tubulin. Protoplasma 222: 157–165 [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Jin L., Du J., Li H., Zhao Q., Ou G., Ao G., Yuan M. (2007). SB401, a pollen-specific protein from Solanum berthaultii, binds to and bundles microtubules and F-actin. Plant J. 51: 406–418 [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey P.J., Ketelaar T., Deeks M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J.N., Guérin C., Robert H., Sørensen M.B., Van Damme D., Geelen D., Blanchoin L., Berger F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kaneko Y., Iwano M., Hashimoto T. (2007). Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2009). A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21: 701–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating T.J., Peloquin J.G., Rodionov V.I., Momcilovic D., Borisy G.G. (1997). Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA 94: 5078–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., de Ruijter N.C., Emons A.M. (2003). Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 15: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Park M., Kim S.J., Hwang I. (2005). Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17: 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komorisono M., Ueguchi-Tanaka M., Aichi I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M., Sazuka T. (2005). Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol. 138: 1982–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R. (2006). Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18: 11–17 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124: 423–435 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA 101: 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H.N. (2003). The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13: 1335–1340 [DOI] [PubMed] [Google Scholar]
- Li Y., Shen Y., Cai C., Zhong C., Zhu L., Yuan M., Ren H. (2010). The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22: 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2004). Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 5: 13–22 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Martin C., Bhatt K., Baumann K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4: 540–549 [DOI] [PubMed] [Google Scholar]
- Mathur J. (2004). Cell shape development in plants. Trends Plant Sci. 9: 583–590 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Okawa K., Sekimoto T., Yoneda Y., Watanabe S., Ishizaki T., Narumiya S. (2009). mDia2 shuttles between the nucleus and the cytoplasm through the importin-α/β- and CRM1-mediated nuclear transport mechanism. J. Biol. Chem. 284: 5753–5762 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. (2005). Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280: 28023–28033 [DOI] [PubMed] [Google Scholar]
- Otomo T., Tomchick D.R., Otomo C., Panchal S.C., Machius M., Rosen M.K. (2005). Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433: 488–494 [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Cook T.A., Alberts A.S., Gundersen G.G. (2001). mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3: 723–729 [DOI] [PubMed] [Google Scholar]
- Panteris E. (2008). Cortical actin filaments at the division site of mitotic plant cells: A reconsideration of the ‘actin-depleted zone’. New Phytol. 179: 334–341 [DOI] [PubMed] [Google Scholar]
- Paradez A., Wright A., Ehrhardt D.W. (2006). Microtubule cortical array organization and plant cell morphogenesis. Curr. Opin. Plant Biol. 9: 571–578 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Paul A., Pollard T. (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615 [DOI] [PubMed] [Google Scholar]
- Quinlan M.E., Heuser J.E., Kerkhoff E., Mullins R.D. (2005). Drosophila Spire is an actin nucleation factor. Nature 433: 382–388 [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Christensen H.E., Ishimaru Y., Dong C.H., Chao-Ming W., Cleary A.L., Chua N.H. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Nieves A.E., Johndrow J.E., Keller L.C., Magie C.R., Pinto-Santini D.M., Parkhurst S.M. (2006). Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. (2002). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4: 626–631 [DOI] [PubMed] [Google Scholar]
- Sano T., Higaki T., Oda Y., Hayashi T., Hasezawa S. (2005). Appearance of actin microfilament ‘twin peaks’ in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J. 44: 595–605 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., Kitano H., Matsuoka M. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger C.J., Blanchoin L. (2006). Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Traas J.A., Doonan J.H., Rawlins D.J., Shaw P.J., Watts J., Lloyd C.W. (1987). An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus. J. Cell Biol. 105: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D., Vanstraelen M., Geelen D. (2007). Cortical division zone establishment in plant cells. Trends Plant Sci. 12: 458–464 [DOI] [PubMed] [Google Scholar]
- Vidali L., Hepler P.K. (1997). Characterization and localization of profilin in pollen grains and tubes of Lilium longiflorum. Cell Motil. Cytoskeleton 36: 323–338 [DOI] [PubMed] [Google Scholar]
- Vidali L., Augustinea R.C., Kleinmanb K.P., Bezanillaa M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., Bezanilla M. (2009). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Yu Y., Chen Z.L., Xia G.X. (2005). Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Characterization of in vivo functions of a cotton profilin gene. Planta 222: 594–603 [DOI] [PubMed] [Google Scholar]
- Wang M., Chen C., Xu Y.Y., Jiang R.X., Han Y., Xu Z.H., Chong K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22: 409–417 [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Yoo C.M., Blancaflor E.B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Yang Z. (2004). New views on the plant cytoskeleton. Plant Physiol. 136: 3884–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik R.P., Maas R.L., Zeller R., Vogt T.F., Leder P. (1990). ‘Formins’: proteins deduced from the alternative transcripts of the limb deformity gene. Nature 346: 850–853 [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116: 711–723 [DOI] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Guo C., Chen D., Zhao B., Yang B., Ren H. (2005). Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 138: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.G., Thurston S.F., Copeland S., Smallwood C., Copeland J.W. (2008). INF1 is a novel microtubule-associated formin. Mol. Biol. Cell 19: 5168–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., Yuan Z., Hu J., Ren H., Zhang D. (2011). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Nairn C.J., Wood-Jones A., Morrison W.H., III, Ye Z.H. (2005). Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17: 1449–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H., Evangelista M., Boone C., Yang C., Dar A.C., Sicheri F., Forkey J., Pring M. (2003). Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Zuchero J.B., Coutts A.S., Quinlan M.E., Thangue N.B., Mullins R.D. (2009). p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]