Abstract
The sequential order of secondary structural elements in proteins affects the folding and activity to an unknown extent. To test the dependence on sequential connectivity, secondary structural elements were reconnected by their solvent-exposed ends, permuting their sequential order, called “re-wiring.” This new protein design strategy changes the topology of the backbone without changing the core sidechain packing arrangement. While circular and non-circular permutations have been observed in protein structures that are not related by sequence homology, to date no one has attempted to rationally design and construct a protein whose sequence is non-circularly permuted while conserving three-dimensional structure. Herein we show that Green Fluorescent Protein (GFP) can be rewired, still functionally fold, and exhibit wild-type fluorescence excitation and emission spectra.
Keywords: Protein engineering, GFP, re-wire, topology, protein folding, loop design
The topology of a protein, defined as the path of the protein backbone through space, is thought to be a primary determinant of the folding pathway (1). However, it has been shown that multiple backbone topologies can conserve the same core packing arrangement of secondary structure elements, where core packing arrangement refers to the spatial arrangement of contacting secondary structure elements (2). Yuan and Bystroff have catalogued many of these cases using non-sequential structure-based alignment, identifying for example that the non-homologous proteins alkaline phosphatase (1ALK) and glycogen phosphorylase (1C8K) conserve a 111-residue structural core after a triple permutation of the sequence order (3). Agrawal and Kishan identified a natural non-sequentially permuted structure between two functionally similar folds (Five stranded β-barrels), oligonucleotide/oligosaccharide-binding (OB) and Src homology 3 (SH3) folds, with distinct topologies (4). Abyzov and Ilyin noted that some recurrent packing arrangements in proteins conserve only the packing and not the sequence of secondary structural elements (5). The overall impression is that tertiary structure is dictated more by specific packing interactions than by topology, a theory also held by others (6). If this is true, then it should be possible to vary the topology of a protein in the loop regions, conserving the core side chains, and retaining the native three-dimensional structure.
Along these lines, Regan documented several studies involving loop redesign and circular permutations (7), observing that protein structures tolerate a remarkable number of insertions and other changes made at the topological level, yet are still able to fold to stable and active structures. For example, Nagi et al inserted loops of increasing size between two a helices of the four α-helix bundle protein, Rop, and observed a delay in the association of adjacent helices into the ordered packing of the folded state. These results highlight the relative importance of specific packing interactions in defining the fold of a protein. This is in opposition to the generally accepted model (8) where the fold is defined by non-specific forces of hydrophobic collapse combined with the topological constraints imposed by the ordering of secondary structural elements along the chain.
Many studies have shown that protein structural sub-domains can associate in vivo and in vitro without the aid of peptidic bonds (9). For example, β-galactosidase and LacZ, used in blue-white screening (10, 11), and the complementation of Green Fluorescent Protein (GFP) β-strand 11 to it’s fully folded counterpart, β-strands 1–10, used for protein tagging and folding applications (12–14). In addition, Huang et al has previously shown that circularly permuted GFP can undergo complementation with β-strand 7 (15), folding and binding simultaneously.
Based on this observed robustness of protein self-association, especially in GFP, we pose the following questions: 1. Can we change the topology of a protein without changing its core? 2. If so, how will such topological changes affect protein activity and stability? GFP has been chosen for the model system for this study having a high resolution crystal structure (16), a well studied folding pathway (17–19) and chromophore maturation reaction (20), and because its intrinsic fluorescence is a convenient reporter of its folded state (15, 18–23).
The model protein, GFP, is a highly stable β-barrel structure consisting of 11 β-strands connected by loops (24, 25) that harbors a distorted α-helix containing the fluorescent chromophore (Figure 1A and 1B). GFP variants with improved folding properties have been generated; for example, “cycle3” GFP with 3 mutations, “folding reporter” with 5 mutations, “superfolder” with 11 and “OPT” with 16 (22, 26). As a starting point for modeling we have used the latter. GFP-OPT (herein simply GFP) emits green light (λmax = 508 nm) under the excitation of cyan light (λmax = 485 nm). The chromophore in GFP is derived from posttranslational, intramolecular cyclization and oxidation of the tripeptide motif Thr65 or Ser65-Tyr66-Gly67 (27) by a proposed autocatalytic mechanism (28) consisting of four distinct steps: folding, cyclization, oxidation, and dehydration. Positionally conserved, well-ordered intramolecular water molecules are located in the interior cavity (28, 29), held in place by a proton relay system involving Asn146, His148, Arg168, Thr203, Ser205, and Glu222, all of which are part of the β-barrel. Mutations are tolerated at most positions in this network regarding protein folding, but any small change in this well-organized network usually results in a shift of the GFP excitation or the emission wavelengths (17, 23). The chromophore, once formed, does not revert to a tripeptide or undergo any other covalent changes upon denaturation, either using urea or by lowering the pH to 2. It is nonetheless completely quenched in the unfolded state, presumably through non-radiative decay of the excited state through the solvent. The return of fluorescence upon refolding is immediate and has been used as a probe of the refolding pathway (15, 17).
Figure 1.
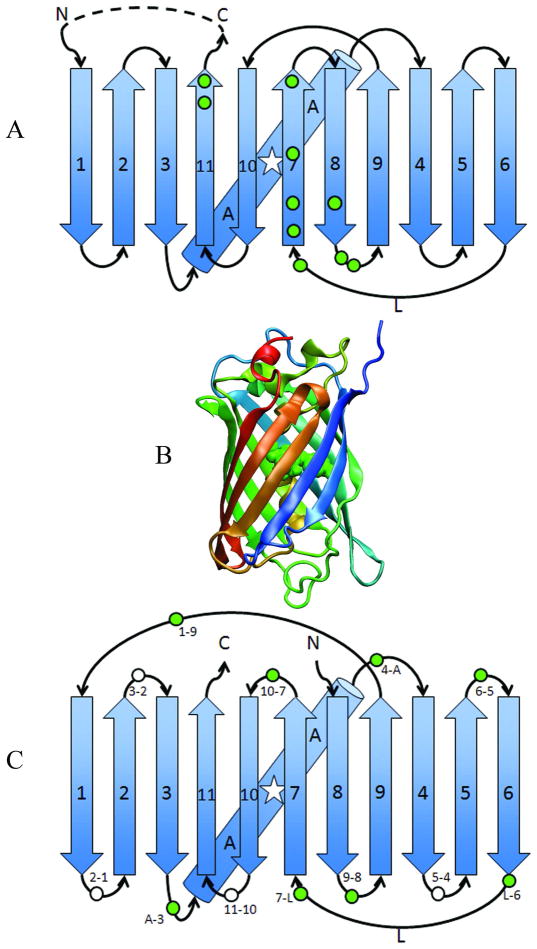
Structural makeup of GFP, rGFP3 and CP-rGFP3 (A) Strand order of GFP. β-strands are drawn as arrows and numbered in N-to-C order, and the a-helices are drawn as cylinders. The Baird et al study is summarized by the green circles that identify locations of allowed circular permutations in wt-GFP; the dotted line indicates the circular permutation introduced during this study (GGTGGS) (21). The peptidic chromophore is represented by a star. (B) Crystal structure of GFP (PDB 2b3q) in N (blue) to C (red) rainbow coloring with the chromophore shown in space fill green. Rendered in VMD (57). (C) Strand order of rGFP3 with strand numbering maintained from GFP. Circles indicate the location of circular permutants analyzed in this study, colored white for non- or weakly-fluorescent constructs and green for highly fluorescent constructs. Circles are labeled to indicate the name of that construct, in the format CP N-C from Table 2.
Non-sequential, non-circular sequence permutation by reconnecting loops, or “rewiring”, is a new protein engineering approach that potentially would allow one to explore the effects of topological changes on protein folding, stability, and activity without changing the core interactions. In this study, re-wiring GFP is shown to conserve the detailed core packing interactions as reported by the fluorescence spectrum. Other protein engineering and design techniques include rational single- and multiple-site mutations (30–34), combinatorial method (35, 36), in vitro evolution (37–40), and computational optimization of side-chain packing (41). But none of these methods has the capacity to permute the protein sequence. We expect our method to be widely applicable, since sequence rearrangements are observed in known protein structures (2, 3, 5, 42). Potentially, re-wiring may be used to engineer the stability and folding kinetics of proteins, since topological properties are known to influence the rates of folding (43) and possibly unfolding (44).
Results and Discussion
Characterization of rGFP1 and rGFP2
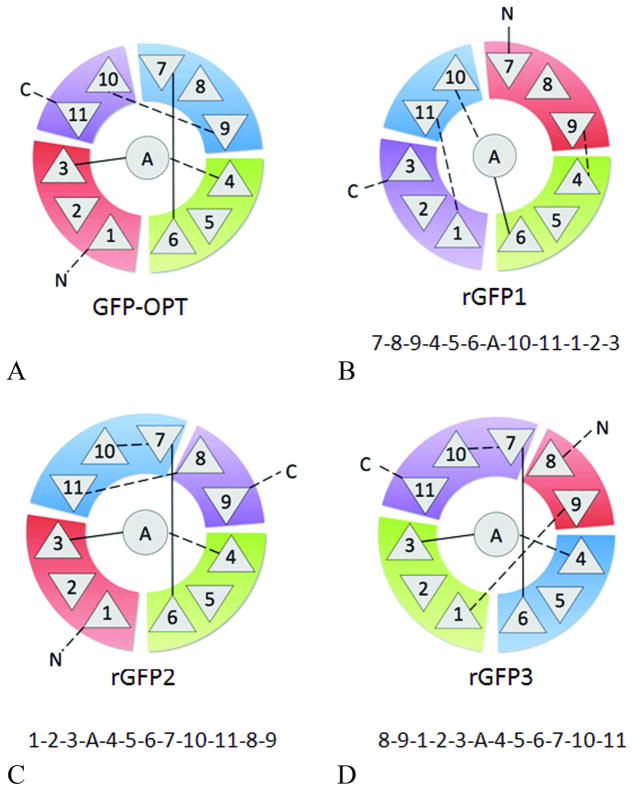
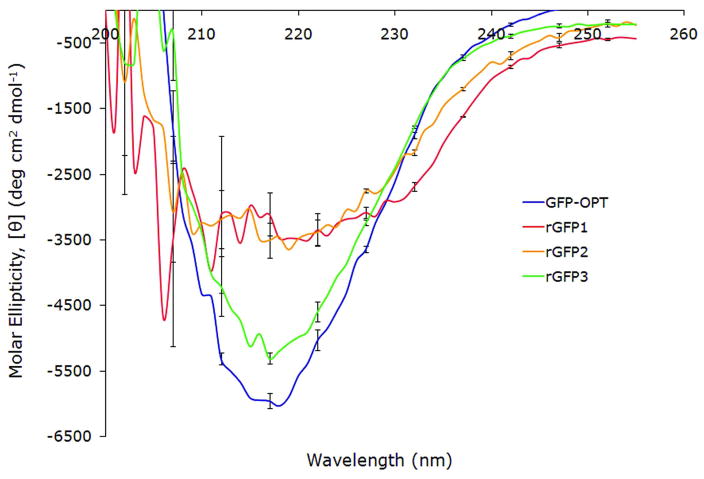
Rewiring GFP was initiated with a conservative design strategy wherein changes were restricted to short reconnections, without dividing the three β-meanders (β-meanders are three stranded antiparallel sheets) or the 10–11 hairpin turn, in order to preserve the wild-type supersecondary structure elements as much as possible (Figure 2A). The direction of the β-strands was also preserved. Given these constraints, only a single non-sequential permutation was geometrically possible (Figure 2B). rGFP1 was generated by removing four existing loops and adding four de novo linker sequences (Table 1). Modeling was carried out using the Molecular Operating Environment (MOE, CCG, Montreal) software (45). rGFP1 showed no fluorescence and no mature chromophore absorption peak at 380 nm when acid-denatured (data not shown). Circular dichroism (CD) of this construct shows ~50% less β-strand signal (218 nm) relative to GFP (Figure 3), suggesting a fast dynamic equilibrium between folded and unfolded states or misfolded state. A fast dynamic equilibrium would prevent efficient formation of chromophore, a slow auto-catalyzed reaction with a half-life on the order of minutes to hours. However, its expression as a soluble protein suggests that rGFP1 folds to a compact structure.
Figure 2.
TOPS (58) cartoons of wild-type GFP and three re-wired versions. β-strands are shown as triangles, with orientation indicating chain direction into (▽) or out of (△) the page. β-strands are grouped as three β-meanders and a hairpin turn in colored blocks, ordered from N-to-C as red-green-blue-purple. The α-helix is shown as a circle in the center.
Table 1.
Summary of loop design
| Construct | Loop Location (β-strands)a | Loop Sequence | Design comments |
|---|---|---|---|
| GFP-OPT | 1-2-3-A-4-5-6-L-7-8-9-10-11 | Native | |
| rGFP1 | 7-8-9-4-5-6-L-A-10-11-1-2-3 | FEGDTL | Same sequence as 5–6 loopb |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | MPGDG | I-sitesc motif, proline αp | |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | EKG | Design based on local interactionsd | |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | GGTGGS | Used in Baird et al 1999 | |
| rGFP2 | 1-2-3-A-4-5-6-L-7-10-11-8-9 | DGGV | Design based on local interactionse |
| 1-2-3-A-4-5-6-L-7-10-11-8-9 | G-GDGPKLVPD-S | 9–0 loop with changesf | |
| rGFP3 | 8-9-1-2-3-A-4-5-6-L-7-10-11 | SGTGSG | Loose, flexible loopg |
| 8-9-1-2-3-A-4-5-6-L-7-10-11 | GGSGGT | Loose, flexible loopg | |
| rGFP2b (CP1–9) | 1-2-3-A-4-5-6-L-7-10-11-8-9 | GGSGGT | Loose, flexible loopg |
| 1-2-3-A-4-5-6-L-7-10-11-8-9 | GGTGS | Loose, flexible loopg | |
| rGFP1b | 7-8-9-4-5-6-L-A-10-11-1-2-3 | GSGTGSG | Loose, flexible loopg |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | GGG | Loose, flexible loopg | |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | GGDGG | Loose, flexible loopg | |
| 7-8-9-4-5-6-L-A-10-11-1-2-3 | GGTGGS | Loose, flexible loopg | |
β-strands are labeled according to the native ordering of GFP and highlighted in the location of the loop sequence.
The loop FEGDTL was reused as the atom-to-atom distance between the final two amino acids on the associated β-strands was within 1Å. Codon changes were introduced to enable gene assembly by assembly PCR.
I-sites protein structure library, http://www.bioinfo.rpi.edu/bystrc/Isites2/ chosen based on atom-to-atom distances at the associated β-strands.
These short linkages were chosen by analysis in MOE based on the criteria that any loop should be as short as possible and closely associated to local structure, thus for EKG: Glu and Lys interact based on charge, negative and positive, respectively and the Gly faces outwards and is small and polar; For DGGV: two Gly, which are small and flexible, and Asp and Val, which are negatively charged and hydrophobic, respectively.
The loop GDGPVLLPDN was reused with changes to GGDGPKLVPDS based on local structural interactions in the new location as follows: (1) the Lys residue replaces the Val as a larger residue (by Van de Waals volume, Å3) as well as positively charged rather than hydrophobic (it faces outward in the new configuration), (2) the Val replaces the Leu as a smaller hydrophobic residue for fit, (3) the Ser replaces the Asn as a smaller residue, (4) and the Gly is inserted into the sequence to encourage the close fit of the new loop based on length.
For the final set of designs, we found that the sequence did not matter if the length of the loop was adequately long to inhibit interaction with the protein structure (>3Å from the surface after MOE energy minimization), and made of small, hydrophilic, and flexible residues (Ser/Thr/Gly) to encourage solubility.
Figure 3.
Circular dichroism of GFP and all re-wire constructs, obtained in 2 mM phosphate buffer, pH 7.4.
The loss of chromophore maturation in rGFP1 effectively refuted the assumption that retaining the supersecondary structures was a “conservative” design strategy. It suggested that larger structural units must be topologically conserved for folding and function. This was not unexpected, since previous studies have implicated connections and/or ordering of the first six strands (including the central helix) as required for foldability (46, 47). Demidov et al showed that a fragment of GFP containing only strands 1 through 6 is capable of forming a mature chromophore when expressed with the remaining strands as a separate chain (48). Furthermore, Baird et al have carried out exhaustive circular permutations of wild-type GFP, effectively locating all covalent linkages that can be cleaved without loss of foldability and fluorescence (21, 49), and they found no locations in strands 1 through 6 that could serve as the new termini for a functional circular permutant. A similar analysis by Pedelacq et al, using “folding reporter” GFP, found that placement of the termini only in positions located after strand 6 (specifically, between β-strands 6 and 7, and 8 and 9) yielded whole cell fluorescence greater than 10% that of the native protein (22). However, more recently Kent et al have shown that the helix, located between strands 3 and 4 in the sequence, can be left out entirely, and folding will occur when it is added exogenously (50). In rGFP1 we linked the interior helix to strand 10 instead of strand 4, breaking up the first six strands, resulting in a non-native state. This supports the requirement of a larger unpermuted unit, which may or may not include the helix.
In our next design, we focused on permuting only strands 7 through 11. Conserving strands 1–6 and the helix, and retaining all strand directions, there are a total of 12 possible topologies, including the wild-type. The most conservative of these, rGFP2 (Figure 2C) is an approximate mirror image of the wild-type topology (Figure 2A), with two new linkers (see Table 1 for loop details and Table 1S for all sequences) and a minimal change in the sequential strand order. However, like rGFP1, rGFP2 was soluble but not fluorescent (data not shown), consistent with a non-native or misfolded state, since a natively folded state would have fluorescence. Based on the CD spectra, rGFP1 and rGFP2 contain less beta secondary structure than the native state.
To ask whether the rGFP2 topology was intrinsically unstable or whether the designed loops led to instability, we took the approach of Nagi et al, increasing the linker loop lengths and making them more flexible (8). Rationally designed linker sequences were replaced with glycine-rich sequences, such as those used previously to link the termini in circular permutants of GFP (21, 22). In the new construct, rGFP3, the sequence order of rGFP2 was modified by circular permutation in which the terminal strands, 9 and 1, were connected by a flexible linker (SGTGSG), and the linker connecting strand 11 and 8 was cleaved to make the new termini (Figure 2D). Additionally, the designed three-residue tight turn between strands 7 and 10 in rGFP2 was relaxed into a more flexible six-residue loop (GGSGGT) in rGFP3 (Table 1). In this design, the linkers are assumed to be more unstructured over all than they were in rGFP1 and rGFP2. This construct was soluble and fluorescent. We conclude from this experiment that the rGFP2 topology is not intrinsically unstable but that the designed loops in rGFP2 favored uncharacterizable non-native states in the structure.
Characterization of rGFP3
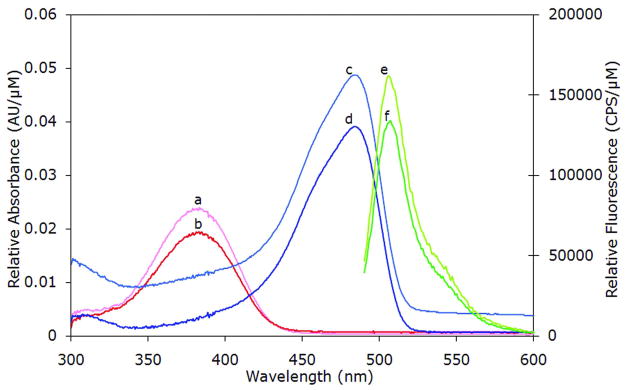
The re-wired permutant rGFP3 exhibits fluorescence excitation (485 nm) and emission (508 nm) maxima that are indistinguishable from GFP (Figure 4). The similar Stokes shift suggests that the barrel in rGFP3 is formed identically to that of the native GFP. Both unfold at low pH and renature upon return to neutral pH (Figure 1S). rGFP3 has approximately 88% of the CD signal at 218 nm (β-sheet, Figure 3), much higher than either rGFP1 or rGFP2. The relative quantum yield, calculated as the ratio of emission at 508 nm to absorbance at 485 nm at pH 7.5, is essentially the same for GFP and rGFP3, as is the ratio of chromophore absorbance at 485 nm (pH 7.5, folded) to absorbance at 380 nm (pH 2, unfolded). However, rGFP3 has between 80 and 90% of the GFP fluorescence when corrected for protein concentration (Figure 4). Combining these observations, we conclude that a small fraction (10 – 20%) of the rGFP3 molecules failed to form the chromophore. This uncharacterized misfolded state may be homo- or heterogeneous in nature. Andrews et al have described such a monomeric, misfolded state of GFP that is trapped during refolding and is kinetically stable (51). It is possible that we are seeing a similar trapped state in rGFP3, preventing chromophore maturation in a small fraction. Alternately, the missing chromophore signal could be due to chemical modifications, such as spontaneuos sidechain cleavage at Y66 (52).
Figure 4.
Absorbance and Fluorescence Characterization of rGFP3 and GFP. Absorbance (300–600 nm, 1 nm intervals, 2 nm slit width) and fluorescence emission (excitation at 485 nm, emission at 490–600 nm, 1 nm intervals, 3/1-nm excitation/emission slit widths) profiles of rGFP3 and GFP, normalized to concentration (10 μM or 0.1 μM for absorbance or fluorescence, respectively). (a and b) Absorbance of the denatured chromophore at pH 2, GFP and rGFP3, respectively. (c and d) Absorbance of the chromophore at pH 7.5, GFP and rGFP3, respectively. (e and f) Fluorescence emission by excitation at 485 nm at pH 7.5, GFP and rGFP3, respectively.
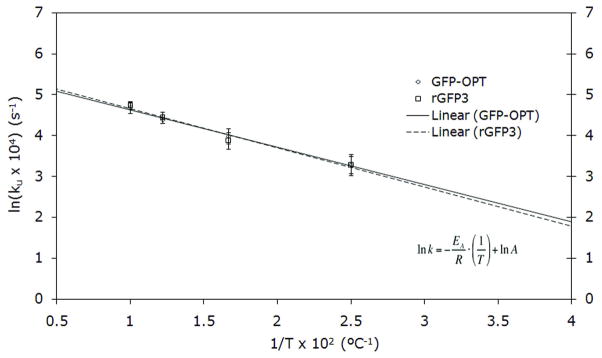
Thermal deactivation of GFP and rGFP3 was also indistinguishable (Figure 5), and given that it depends on the rate of unfolding, this suggests that there is no change in the hydrogen bonding within the β-barrel and no changes in core packing, either of which would affect the kinetic stability.
Figure 5.
Thermal deactivation of rGFP3 vs. GFP. Initial rates of fluorescence quenching were measured for four temperatures (40, 60, 80, 100ºC). The Arrhenius relationship, shown on the figure, was plotted to determine the difference in the activation energy term between the two species extrapolated to room temperature (1/T × 102 equaling 4.0 ºC−1).
Characterization of circular permutants of rGFP3
To study further the folding pathway of rGFP3 and GFP, comprehensive circular permutants (CPs) of rGFP3 were created and characterized, with new N and C termini placed at the intersection of each secondary structure in the chain, including breaks on either end of the long 6–7 loop (L in Table 2, Figure 1C). This resulted in twelve new constructs including one, CP1-9, that mirrors the topology of rGFP2 (Table 1S). Table 2 details the in vivo solubility and fluorescence for each construct. Two constructs with 0% solubility were solubilized with urea (Methods) and subsequently observed to be fluorescent. It is interesting to note that we found viable chain termini locations within the first seven secondary structure elements, strands 1–6, whereas Pedalacq et al, working in “folding reporter” GFP, did not (21, 22). Our template, GFP, includes a few stabilizing mutations beyond those in folding reporter (Table 1S), perhaps slowing the rate of aggregation for slow folding permutants. Alternatively, the non-sequential permutations and flexible linkers in rGFP3 may have altered off-pathway intermediates and misfolded states. Many of the permutants whose termini are within strands 1–6 are indeed marginally fluorescent or completely dark, including CPs 2–1, 3–2, and 5–4, however several, CPs A-3, 4-A, and 6–5 were strongly fluorescent (Figure 1C). Further study of the folding pathways of these constructs may reveal alternative folding pathways with altered kinetic phases.
Table 2.
Characterization of rGFP3 circular permutants
| Construct | Sequence Order | Fluorescent (+/−) | % Solubility |
|---|---|---|---|
| GFP-OPT | 1-2-3-A-4-5-6-L-7-8-9-10-11 | + | 80 |
| rGFP3 | 8-9-1-2-3-A-4-5-6-L-7-10-11 | + | 80 |
| CP 9–8 | 9-1-2-3-A-4-5-6-L-7-10-11-8 | + | 75 |
| CP 1–9 | 1-2-3-A-4-5-6-L-7-10-11-8-9 | + | 67 |
| CP 2–1 | 2-3-A-4-5-6-L-7-10-11-8-9-1 | + | 20 |
| CP 3–2 | 3-A-4-5-6-L-7-10-11-8-9-1-2 | + | 0 |
| CP A-3 | A-4-5-6-L-7-10-11-8-9-1-2-3 | + | 67 |
| CP 4-A | 4-5-6-L-7-10-11-8-9-1-2-3-A | + | 50 |
| CP 5–4 | 5-6-L-7-10-11-8-9-1-2-3-A-4 | − | 0 |
| CP 6–5 | 6-L-7-10-11-8-9-1-2-3-A-4-5 | + | 67 |
| CP 6-L | L-7-10-11-8-9-1-2-3-A-4-5-6 | + | 100 |
| CP L-7 | 7-10-11-8-9-1-2-3-A-4-5-6-L | + | 40 |
| CP 11–10 | 11-8-9-1-2-3-A-4-5-6-L-7-10 | + | 20 |
| CP 10–7 | 10-11-8-9-1-2-3-A-4-5-6-L-7 | − | 80 |
Summary of circular permutant expression data based on fluorescence data of purified constructs and visual inspection of SDS-PAGE of lysates (soluble) and pellets (insoluble) following French press (% soluble is defined by relative amount soluble to insoluble). In two cases, CP3-2 and CP5-4, the insoluble pellet protein was denatured and then allowed to refold on Ni-agarose column media (4 hrs at room temperature while his-tag immobilized on the column) before reassessing fluorescence (+/−). Sequence order refers to the secondary structure naming, based on the sequence of GFP, where numbers indicate β-strands going from N-to-C terminus, A is the α-helix, and L is the long unstructured loop connecting strands 6 and 7.
Surprisingly, the location that Cabantous et al utilized with great success in GFP-superfolder to create a complementation system (26), CP 11–10, yielded very little soluble protein in the context of rGFP3. Its misfolding signals a change in the folding pathway that is still not characterized. Possibly the non-specific association of the inserted 7–10 and 11–8 loops, both loose and long and on the same end of the barrel, confounds folding in this permutation.
Also surprising was that circular permutants involving the central α-helix formed soluble and fluorescent products (CP A-3 and 4-A), contrary to the circular permutations in “folding reporter” GFP reported by Pedelacq et al, who saw marginal fluorescence in whole cells with analagous CP-GFP constructs (22), but consistent with the results of Kent et al who showed that the helix can be left out and added exogenously reconstituting the fluorescent form (50).
Finally, CP 1–9, topologically identical to rGFP2, is fluorescent and soluble by the addition of loose linkers (Table 1), again pointing to overly constrained loops, not topology, as the main reason for its misfolding. A loose linker introduced between strands 7 and 10 replaced the short DGGV loop in rGFP2. The short linker may have introduced an early folding event, the formation of a 7–10 hairpin, that should have been late. By increasing the loop length and the corresponding entropy of loop closure, the association between strands 7 and 10 would be slowed, as shown in other systems (8), making it a later folding event and more consistent with the proposed native sequence of folding events.
After finding that unstructured loops overcame the folding difficulties of rGFP2 (CP 1–9), rGFP1 was similarly revisited by increasing the four loop lengths and by introducing flexible glycines into the loops (Table 1). This looser construct was again soluble but not fluorescent, indicating that loop inflexibility was not the cause of misfolding in this case. Instead, some aspect of the rGFP1 topology must have led to a compact and stable non-native state.
It should be noted that full loop modeling algorithms (such as MODELLER (53)) were not used in this study. As a result, some of the initial failures (rGFP2, not rGFP1) may have been due to the use of loops of the wrong structural propensities. Using glycine-rich flexible loops, having no specific structural tendencies, is naturally better than using loops of the wrong structure, but loops with the correct structural propensities would have been better still. Further, we base our major conclusions on the presence of green fluorescence, which requires the auto-catalyzed synthesis of the intrinsic chromophore to take place as in the native GFP. Our assumption is that the natively packed core residues are sufficient for this chromophore maturation reaction and that no aspect of the kinetics of folding, or the series of events which we call a folding pathway, are required for this maturation to occur.
Conclusions
In these case studies, we have explored rewired GFPs in which the connectivity of secondary structure elements was altered but the core packing arrangment was not disturbed. In one case (rGFP3 and some of its circular permutants) we saw that the native packing was attainable during folding, as signaled by green fluorescence, the CD spectrum, and other biophysical similarities. The same topology failed to fold if we used inflexible (or poorly chosen) linkers (rGFP2). Finally, a more severely altered topology did not fold correctly even if flexible linkers were used (rGFP1), as signaled by the lack of fluorescence and significant differences in the CD spectrum. The latter result suggests that for GFP’s particular spatial arrangement of secondary structure elements, there exists at least one rewired connectivity that has no pathway to the natively packed state.
This rewiring method is likely to be generalizable to other proteins. We propose that by studying which sequential rearrangements are possible and which are not, it is possible to deduce the folding pathways of proteins, and further, engineer them to have increased stability and improved folding kinetics.
Materials and Methods
Computational Modeling of rGFP
New structural models were constructed using Molecular Operating Environment (MOE), (CCG Inc., Montreal) including point mutations, sequence permutations, loop structure, and loop sequence design (45). Loop lengths were determined by estimating the distance to be spanned. Loop sequences were either selected based on a motif library (54), or set to a “loose linker” sequence such as GGSGGT, see Supplemental Materials for details. Database searches were used to build loose linker loop conformations. Loop structures, but not the conserved secondary structure elements, were energy minimized in vacuum using a molecular mechanics force field. Buried surface areas were calculated using MASKER (55).
Construction of genes and plasmids
Genes of re-wired constructs were constructed by assembly PCR (56), described here briefly. Overlapping oligomers for each gene were designed using the DNAworks 2.0 program (http://mcl1.ncifcrf.gov/dnaworks/dnaworks2.html) with the following parameters: 40nt oligo length, codon frequency threshold of 20, 50 mM Na+/K+, 200 nM oligo concentration, annealing temperature of 64ºC, number of solutions of 1, and 2 mM Mg2+. Assembly PCR was performed first with 0.2 ng/mL of each oligomer (www.idtdna.com), Pfu Turbo (Stratagene), with an annealing temperature of 59ºC (5º below set point temperature in DNAworks) and 35 cycles. Of this material, 1 μL was used as the template for amplification PCR with 0.2 μM of the forward and reverse primers (the outermost oligomers on each DNA strand) each, annealing temperature of 62ºC and 35 cycles.
Genes of circular permutants of rGFP3 were constructed by blunt end ligation (T4 kinase reaction to provide phosphate groups for ligation followed by T4 ligase reaction, NEB enzymes) of the rGFP3 gene to form a circular template for PCR. New 5′ and 3′ primers were designed for each circular permutant to include new EcoRI and NheI sites as well as stop codon followed by PCR performed as above (Pfu Turbo from Stratagene).
Completed genes with included NheI and EcoRI (enzymes from New England Biosciences, NEB) restriction sites were inserted into pET28a (Novagen) using T4 ligase per manufacturers instructions (NEB) and transformed into XL-1Blue or BL21 for DNA sequencing (www.mclab.com) and BL21 for expression (Invitrogen). DNA was collected using a Sigma GeneElute Plasmid Miniprep kit and buffer exchanged using a Promega Wizard SV Gel and PCR-cleanup System kit.
Protein expression and purification
Sequenced plasmids were transformed into BL21 cells and grown in LB media (Qbiogene) with 50 μg/mL kanamycin (Sigma) to 0.8–1.0 OD600 at 37ºC with 200rpm shaking before induction with 1 mM IPTG for further incubation at room temperature (20–22ºC) with 200 rpm shaking for 18–24 h. Cells were collected by centrifugation (10 min, 7,700 × g), washed and resuspended in 50 mM Na2HPO4, 50 mM Tris, 300 mM NaCl, 10 mM imidazole, pH 7.5 (NBB, 8 mL/g cells, chemicals from Sigma). French Press was used to lyse the cells (with addition of 1 μg/mL DNase I, Sigma) in a 30 mL cell, 1,800 PSI with two passes. Debris was removed by centrifugation at 4,000 rpm, 15 min, 4ºC. Supernatants were added and bound to a Ni-NTA column (2 mL resin/g cells, Invitrogen) equilibrated with NBB, washed with NWB (NBB with 20 mM imidazole) for 4 column volumes, and eluted into NEB (NBB with 200 mM imidazole). Insoluble pellets were resuspended in DBB (NBB with 8M Urea, no imidizole), bound to a pre-equilibrated Ni-NTA column, slowly washed with step dilutions of DWB (NWB with 8M urea, no imidizole) into NWB at room temperature, and finally eluted into NEB. Fluorescence was observed in some cases following these washes after at least 90min (chomophore maturation time). Protein was dialyzed into 50 mM Tris, 100 mM NaCl, pH 7.5 (TN, Sigma) using Pierce 1 mL, 10,000 MWCO Slide-dialyzer cassettes in 3.5 L buffer overnight at 4ºC with gentle stirring.
Concentration and fluorescence measurements
Protein concentration was determined using the BCA assay (Pierce). Fluorescence was measured using a Jorbin Yvon-Horiba Fluorlog-3 machine. Samples were diluted to 0.1 μM in TN and excitation wavelength was determined by scanning in 1 nm intervals between 200–500 nm with slit widths set at 3 and 1 nm for excitation and emission, respectively, monitoring emission at 508nm. After determining the dominant excitation wavelength, the profile of emission was measured by excitation at 485 nm and emission scanning at 490–600 nm, 1-nm intervals, with slit widths set at 3 and 1 nm for excitation and emission, respectively. Absorbance (both absorbance of denatured chromophore at 380 nm and absorbance of active chromophore at 485 nm) of rGFP3 and control GFP were characterized by diluting protein to 5.5 μM in TN buffer, and measurement of absorbance over 300 to 600 nm on a Shimadzu UV/Vis Spectrometer (2 nm slit width, 1-cm path length). HCl was used to lower the pH to 2 for mature chromophore determination over the same absorbance wavelengths with the same sample.
Circular dichroism
Samples for CD were prepared in 2 mM sodium phosphate buffer, pH 7.5 and analyzed on a (OLIS DSM-10 CD, Online Instruments) in a 10-mm path length quartz cuvette at concentrations between 30 and 300 μg/mL. Ellipticity was measured from 255 nm to 180 nm at room temperature. All measurements were performed in triplicate and normalized to concentration to determine molar ellipticity (degree cm2 dmol−1).
Thermal unfolding
Thermal unfolding was monitored by fluorescence. Initial rate of fluorescence quenching was measured for four temperatures (40, 60, 80, 100ºC) over 20 s in triplicate (485 nm/508 nm excitation/emission; 1.5 nm/1.5 nm excitation/emission slit widths; 0.1 μM protein in 25 mM Tris-HCl, 25 mM NaCl, pH 7, Sigma).
Supplementary Material
Acknowledgments
This work was supported by grants from NSF (DBI 0448072, CB), NIH (GM66712, JSD; GM88838, CB) and the New York State Foundation for Science, Technology and Innovation (NYSTAR, JSD).
Footnotes
Table 1S contains sequence information for all constructs used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Baker D. A surprising simplicity to protein folding. Nature. 2000;405:39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 2.Efimov AV. Structural trees for protein superfamilies. Proteins. 1997;28:241–260. doi: 10.1002/(sici)1097-0134(199706)28:2<241::aid-prot12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 3.Yuan X, Bystroff C. Non-sequential structure-based alignments reveal topology-independent core packing arrangements in proteins. Bioinformatics. 2005;21:1010–1019. doi: 10.1093/bioinformatics/bti128. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal V, Kishan RK. Functional evolution of two subtly different (similar) folds. BMC Struct Biol. 2001;1 doi: 10.1186/1472-6807-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abyzov A, Ilyin VA. A comprehensive analysis of non-sequential alignments between all protein structures. BMC Struct Biol. 2007;7:78. doi: 10.1186/1472-6807-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grantcharova V, Alm EJ, Baker D, Horwich AL. Mechanisms of protein folding. Curr Opin Struct Biol. 2001;11:70–82. doi: 10.1016/s0959-440x(00)00176-7. [DOI] [PubMed] [Google Scholar]
- 7.Regan L. Protein redesign. Curr Opin Struct Biol. 1999;9:494–499. doi: 10.1016/S0959-440X(99)80070-0. [DOI] [PubMed] [Google Scholar]
- 8.Nagi AD, Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Fold Des. 1997;2:67–75. doi: 10.1016/S1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 9.Morell M, Ventura S, Aviles FX. Protein complementation assays: approaches for the in vivo analysis of protein interactions. FEBS Lett. 2009;583:1684–1691. doi: 10.1016/j.febslet.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Ruther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178:475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- 11.Ullmann A, Jacob F, Monod J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J Mol Biol. 1967;24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 12.Cabantous S, Waldo GS. In vivo and in vitro protein solubility assays using split GFP. Nature Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh I, Hamilton AD, Regan L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. Journal of the American Chemical Society. 2000;122:5658–5659. [Google Scholar]
- 14.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 15.Huang YM, Bystroff C. Complementation and reconstitution of fluorescence from truncated circularly permuted green fluorescent protein. Biochemistry. 2009;48:929–940. doi: 10.1021/bi802027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood TI, Barondeau DP, Hitomi C, Kassmann CJ, Tainer JA, Getzoff ED. Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis. Biochemistry. 2005;44:16211–16220. doi: 10.1021/bi051388j. [DOI] [PubMed] [Google Scholar]
- 17.Enoki S, Saeki K, Maki K, Kuwajima K. Acid denaturation and refolding of green fluorescent protein. Biochemistry. 2004;43:14238–14248. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 18.Sanders JK, Jackson SE. The discovery and development of the green fluorescent protein, GFP. Chem Soc Rev. 2009;38:2821–2822. doi: 10.1039/b917331p. [DOI] [PubMed] [Google Scholar]
- 19.Zimmer M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 20.Craggs TD. Green fluorescent protein: structure, folding and chromophore maturation. Chem Soc Rev. 2009;38:2865–2875. doi: 10.1039/b903641p. [DOI] [PubMed] [Google Scholar]
- 21.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Nat Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 23.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 24.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 25.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 26.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nature Biotechnology. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 27.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical-Structure of the Hexapeptide Chromophore of the Aequorea Green-Fluorescent Protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 28.Rosenow MA, Huffman HA, Phail ME, Wachter RM. The crystal structure of the Y66L variant of green fluorescent protein supports a cyclization-oxidation-dehydration mechanism for chromophore maturation. Biochemistry. 2004;43:4464–4472. doi: 10.1021/bi0361315. [DOI] [PubMed] [Google Scholar]
- 29.Wachter RM. Chromogenic cross-link formation in green fluorescent protein. Acc Chem Res. 2007;40:120–127. doi: 10.1021/ar040086r. [DOI] [PubMed] [Google Scholar]
- 30.Bujnicki JM. Crystallographic and bioinformatic studies on restriction endonucleases: Inference of evolutionary relationships in the “midnight zone” of homology. Curr Protein Pept Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 31.Eijsink VGH, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, Vriend G. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Hansson MD, Karlberg T, Rahardja MA, Al-Karadaghi S, Hansson M. Amino acid residues His183 and Glu264 in Bacillus subtilis ferrochelatase direct and facilitate the insertion of metal ion into protoporphyrin IX. Biochemistry. 2007;46:87–94. doi: 10.1021/bi061760a. [DOI] [PubMed] [Google Scholar]
- 33.McKinney MK, Cravatt BF. Structure-based design of a FAAH variant that discriminates between the N-acyl ethanolamine and taurine families of signaling lipids. Biochemistry. 2006;45:9016–9022. doi: 10.1021/bi0608010. [DOI] [PubMed] [Google Scholar]
- 34.Sankpal UT, Rao DN. Mutational analysis of conserved residues in HhaI DNA methyltransferase. Nucleic Acids Res. 2002;30:2628–2638. doi: 10.1093/nar/gkf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz S, Patrick WM. Novel methods for directed evolution of enzymes: quality, not quantity. Curr Opin Biotechnol. 2004;15:291–297. doi: 10.1016/j.copbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Woycechowsky KJ, Vamvaca K, Hilvert D. Novel enzymes through design and evolution. Adv Enzymol Relat Areas Mol Biol. 2007;75:241–297. xiii. doi: 10.1002/9780471224464.ch4. [DOI] [PubMed] [Google Scholar]
- 37.Eijsink VGH, Gaseidnes S, Borchert TV, van den Burg B. Directed evolution of enzyme stability. Biomol Eng. 2005;22:21–30. doi: 10.1016/j.bioeng.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Farinas ET, Butler T, Arnold FH. Directed enzyme evolution. Curr Opin Biotechnol. 2001;12:545–551. doi: 10.1016/s0958-1669(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 39.Kaur J, Sharma R. Directed evolution: An approach to engineer enzymes. Crit Rev Biotechnol. 2006;26:165–199. doi: 10.1080/07388550600851423. [DOI] [PubMed] [Google Scholar]
- 40.Rubin-Pitel SB, Zhao HM. Recent advances in biocatalysis by directed enzyme evolution. Comb Chem High Throughput Screen. 2006;9:247–257. doi: 10.2174/138620706776843183. [DOI] [PubMed] [Google Scholar]
- 41.Dahiyat BI, Mayo SL. Protein design automation. Protein Sci. 1996;5:895–903. doi: 10.1002/pro.5560050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bujnicki JM. Sequence permutations in the molecular evolution of DNA methyltransferases. BMC Evol Biol. 2002;2:3. doi: 10.1186/1471-2148-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 44.Xia K, Manning M, Hesham H, Lin Q, Bystroff C, Colon W. Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE. Proc Nat Acad Sci U S A. 2007;104:17329–17334. doi: 10.1073/pnas.0705417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molecular Operating Environment software (MOE) Chemical Computing Group, Inc; 2008. [Google Scholar]
- 46.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 48.Demidov VV, Dokholyan NV, Witte-Hoffmann C, Chalasani P, Yiu HW, Ding F, Yu Y, Cantor CR, Broude NE. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2052–2056. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topell S, Hennecke J, Glockshuber R. Circularly permuted variants of the green fluorescent protein. FEBS Lett. 1999;457:283–289. doi: 10.1016/s0014-5793(99)01044-3. [DOI] [PubMed] [Google Scholar]
- 50.Kent KP, Oltrogge LM, Boxer SG. Synthetic control of green fluorescent protein. J Am Chem Soc. 2009;131:15988–15989. doi: 10.1021/ja906303f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews BT, Roy M, Jennings PA. Chromophore packing leads to hysteresis in GFP. J Mol Biol. 2009;392:218–227. doi: 10.1016/j.jmb.2009.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. The case of the missing ring: radical cleavage of a carbon-carbon bond and implications for GFP chromophore biosynthesis. J Am Chem Soc. 2007;129:3118–3126. doi: 10.1021/ja063983u. [DOI] [PubMed] [Google Scholar]
- 53.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bystroff C, Baker D. Prediction of local structure in proteins using a library of sequence-structure motifs. Journal of Molecular Biology. 1998;281:565–577. doi: 10.1006/jmbi.1998.1943. [DOI] [PubMed] [Google Scholar]
- 55.Bystroff C. MASKER: improved solvent-excluded molecular surface area estimations using Boolean masks. Protein Eng. 2002;15:959–965. doi: 10.1093/protein/15.12.959. [DOI] [PubMed] [Google Scholar]
- 56.Stemmer WPC, Crameri a, Ha KD, Brennan TM, Heyneker HL. Single-Step Assembly of a Gene and Entire Plasmid from Large Numbers of Oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 58.Westhead DR, Slidel TW, Flores TP, Thornton JM. Protein structural topology: Automated analysis and diagrammatic representation. Protein Sci. 1999;8:897–904. doi: 10.1110/ps.8.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.