Abstract
Purpose: By performing registration of preoperative multiprotocol in vivo magnetic resonance (MR) images of the prostate with corresponding whole-mount histology (WMH) sections from postoperative radical prostatectomy specimens, an accurate estimate of the spatial extent of prostate cancer (CaP) on in vivo MR imaging (MRI) can be retrospectively established. This could allow for definition of quantitative image-based disease signatures and lead to development of classifiers for disease detection on multiprotocol in vivo MRI. Automated registration of MR and WMH images of the prostate is complicated by dissimilar image intensities, acquisition artifacts, and nonlinear shape differences.
Methods: The authors present a method for automated elastic registration of multiprotocol in vivo MRI and WMH sections of the prostate. The method, multiattribute combined mutual information (MACMI), leverages all available multiprotocol image data to drive image registration using a multivariate formulation of mutual information.
Results: Elastic registration using the multivariate MI formulation is demonstrated for 150 corresponding sets of prostate images from 25 patient studies with T2-weighted and dynamic-contrast enhanced MRI and 85 image sets from 15 studies with an additional functional apparent diffusion coefficient MRI series. Qualitative results of MACMI evaluation via visual inspection suggest that an accurate delineation of CaP extent on MRI is obtained. Results of quantitative evaluation on 150 clinical and 20 synthetic image sets indicate improved registration accuracy using MACMI compared to conventional pairwise mutual information-based approaches.
Conclusions: The authors’ approach to the registration of in vivo multiprotocol MRI and ex vivo WMH of the prostate using MACMI is unique, in that (1) information from all available image protocols is utilized to drive the registration with histology, (2) no additional, intermediate ex vivo radiology or gross histology images need be obtained in addition to the routinely acquired in vivo MRI series, and (3) no corresponding anatomical landmarks are required to be identified manually or automatically on the images.
Keywords: mutual information, elastic registration, prostate cancer, in vivo, MACMI, magnetic resonance, histology
BACKGROUND AND MOTIVATION
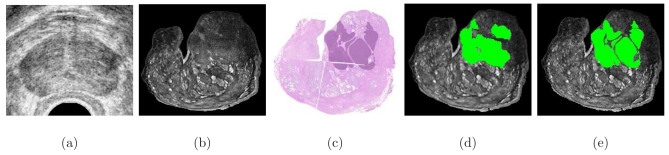
Recently, magnetic resonance (MR) imaging (MRI) has emerged as a promising modality for detection of prostate cancer (CaP), with several studies showing that 3 T endorectal in vivo T2-weighted (T2-w) imaging yields significantly higher contrast and resolution compared to ultrasound (U.S.).1 For example, Fig. 1a shows a typical in vivo U.S. image of a prostate, in which internal anatomical details, such as the urethra, ducts, and hyperplasia, are barely discernible, while in the segmented T2-w MR image shown in Fig. 1b, internal anatomical details within the prostate are clearly visible. An additional advantage offered by MRI is the ability to use different acquisition protocols to capture orthogonal sources of information, including functional [dynamic-contrast enhanced (DCE)], metabolic [magnetic resonance spectroscopy (MRS)], vascular [diffusion weighted imaging (DWI)], and structural (T2-w) attributes. Since multiple protocols can be acquired in the same scanning session, little additional setup time is required.
Figure 1.
(a) Ultrasound imagery of the prostate provides poor soft tissue resolution, while (b) high resolution MRI (ex vivo image shown) shows internal anatomical details of the prostate with greater clarity. Ground truth for CaP extent is obtained only through histopathologic analysis of (c) the corresponding hematoxylin and eosin stained tissue section. The histopathologic CaP extent on (c) can be mapped onto the MRI in (b) by either (d) manually labeling the MRI using histology as a visual reference or (e) automatically mapping CaP extent from (c) via image registration. Note that the morphology of the CaP extent is better preserved in the mapping from (c) onto (e) as compared to (d).
The use of multiprotocol MRI for CaP diagnosis has been shown to improve detection sensitivity and specificity compared to the use of a single MR imaging protocol.2, 3, 4 Previous studies have demonstrated improved CaP detection sensitivity and specificity by simultaneous use of multiple MRI protocols, including DCE and T2-w MRI,5 MRS and T2-w MRI,6 and DWI with both T2-w (Ref. 7) and DCE MRI.8 Since the current clinical diagnostic protocol involves no image-based detection of CaP, the ability to utilize in vivo multiprotocol diagnostic images for detection and localization of CaP in vivo would have clear implications for (1) noninvasive image-based screening, (2) targeted biopsies, and (3) conformal radiation therapy.
If the spatial extent for CaP on multiprotocol in vivo radiological imaging can be accurately delineated, it may then be possible to define specific imaging parameters with the greatest diagnostic accuracy in reliably characterizing CaP on in vivo clinical, radiologic images. The definition of such image signatures would be invaluable in building (a) a computer-assisted disease detection system6, 9, 10, 11 or (b) spatial disease atlases which could serve as training and educational tools for medical students, radiology residents, and fellows. However, direct annotation of disease extent on MRI is often challenging even for experienced radiologists. Thus, to reliably ascertain the extent of CaP on in vivo radiological images, it is necessary to utilize ex vivo tissue specimens, upon which “ground truth” estimates of CaP extent may be established by histopathologic inspection. [In the context of patients diagnosed with CaP and scheduled for radical prostatectomy (RP), in several centers in the United States, preoperative imaging is performed to identify presence of extracapsular spread.12] Figure 1c shows a whole-mount histology (WMH) section of a RP specimen on which cancerous tissue has been manually annotated, following microscopic examination of the excised gland.
Spatial correlation of diseased regions on histology and MRI may be performed by (a) visually identifying and labeling corresponding structures on each modality9, 13, 14, 15 or (b) using a semiautomated or fully automated image registration procedure.16, 17, 18, 19 For example, the spatial extent of CaP on MRI, obtained by manually labeling the in vivo image while visually referencing the histology, is shown in Fig. 1d. On the other hand, the disease extent established by the elastic registration of the MR and histology images in Figs. 1b, 1c is shown in Fig. 1e. Note that the shape of the disease mask mapped onto the MRI in Fig. 1e more closely resembles the histopathological ground truth for CaP extent in Fig. 1b compared to the manually annotated region shown in Fig. 1d. Thus, with an accurate registration technique, CaP extent on MRI can be established with greater accuracy, efficiency, and consistency compared to manual labeling.
The registration of images from different modalities such as histology and radiology is complicated on account of the vastly different image characteristics of the individual modalities. For example, the appearance of tissue and anatomical structures (e.g., hyperplasia, urethra, and ducts) on MRI and histology are considerably different, as may be appreciated from Figs. 1b, 1c. The shape of the WMH is also significantly altered due to uneven tissue fixation, gland slicing, and sectioning, resulting in duct dilation, gland deformation, and tissue loss. Traditional intensity-based similarity measures, such as mutual information (MI), are typically inadequate to robustly and automatically register images from two such significantly dissimilar modalities. There have been several efforts to complement intensity information with alternative image information, including image gradients,20 co-occurrence information,21 and color22 and image segmentations23 in conjunction with MI variants, specifically adapted to incorporate these additional channels of information. Similar to the use of calculated features to complement image intensity, it may also be advantageous to leverage additional imaging protocols that may be available. For instance, multiprotocol MR imaging is part of standard clinical practice at a number of medical centers for disease diagnosis and treatment.24, 25 These additional channels may provide complementary structural, metabolic, and functional data to complement image intensity for the registration process.
In this paper, we present an information theoretic approach, multiattribute combined mutual information (MACMI), to simultaneously utilize all available imaging channels, such as registered multiprotocol imagery (or image features calculated from the original images), in the registration of several images. In this work, we demonstrate the application of MACMI for establishing spatial extent of CaP on radiological imaging via registration of annotated ex vivo histology sections with corresponding multiprotocol in vivo MRI.
PREVIOUS WORK
The registration of prostate images is important for (a) the planning, guidance, and retrospective evaluation of radiotherapy procedures;26, 27, 28, 29 (b) the fusion of diagnostic images for improved CaP detection accuracy;30, 31, 32, 33 and (c) the automated annotation of radiological images via histopathologic correlation.18, 34, 35 Intramodality prostate image registration has been utilized by Bharatha et al.26 for image-guided prostate surgery via elastic registration of preoperative and intraoperative prostate MRI and by Foskey et al.27 for monitoring therapy related changes over time by registration of serial CT images. Lee et al.30 investigated the use of multimodal prostate images (MRI, CT, and SPECT) to characterize CaP, performing automated multimodal registration of MRI to SPECT via MI and manual registration of MRI to CT using a graphical user interface. Automated rigid registration of MRI with CT has been addressed by several groups,31, 32, 33 while automated elastic registration has only recently been addressed by Park et al.36 using a spatially constrained B-spline. Methods for the registration of MRI and 3D transrectal ultrasound for real-time MRI-guided prostate biopsy have been presented by both Xu et al.28 and Singh et al.29
The unique set of challenges associated with the registration of ex vivo histology and multiprotocol MRI of the prostate has begun to be addressed by several recent studies; these studies have, however, primarily been in the context of high resolution ex vivo MRI11, 19, 34, 37 utilized a thin plate spline (TPS) to model the elastic 2D deformations of histology to ex vivo MRI of prostate specimens using control points. However, “block face” photographs of thick tissue sections of the prostate, taken prior to microtome slicing and slide preparation, were used to facilitate correction of the nonlinear tissue deformations and in the creation of a histology volume. While these photographs allowed Park et al.34 to overcome the nonlinear deformations to histology and address the issue of slice correspondences (identifying a one-to-one relationship between histology sections and slices in the MRI volume), such photographs are not generally acquired as part of routine clinical practice. Recently, Ou et al.19 aligned ex vivo MRI and histology sections of a prostate specimen with the aid of precise cancer labels on both modalities to improve their objective function for registration. As part of an integrated registration and segmentation strategy, the required cancer extent on MRI was established via a pixelwise supervised classifier. The authors, however, did not address how classifier errors would affect registration accuracy. While high resolution (4 T) ex vivo MRI provided sufficient segmentation accuracy in Ref. 19, it is not clear that disease extent can reliably be established on in vivo clinical images. Zhan et al.37 also performed registration of ex vivo MRI and histology sections using pairs of automatically detected control points on each modality. However, automated identification of a large number of landmark pairs across ex vivo WMH and in vivo MRI (of lower image resolution and quality compared to the ex vivo MRI used in Ref. 37) may not be feasible.
The registration of clinical in vivo radiologic images with WMH of the prostate has also been recently addressed.18, 30, 35, 38 Lee et al.30 performed histopathologic validation of CaP estimates on CT and MRI by 2D elastic registration of histology sections with CT and MRI. Manually identified control points (anatomical landmarks), placed primarily along the gland boundary, were used to define a TPS interpolant. In a rat brain study, Meyer et al.35 also leveraged available block face photographs of the gland prior to sectioning (similar to Ref. 34) to generate a histology volume for 3D registration. Their approach also utilized an intermediate ex vivo MRI series, to which WMH was aligned via a TPS-based approach using manually selected initial control points, followed by MI-driven refinement of the coordinates of the control points. Subsequently, ex vivo MRI was registered to in vivo MRI, thus indirectly aligning the in vivo MRI and histology slices of the rat brains. Park et al.18 extended this approach to the human prostate and to include multiprotocol (T2-w and DWI) in vivo MRI and PET, again using block face photographs and ex vivo MRI as an intermediate. While the works of Park18 and Meyer35 successfully address the need to rely on approximate slice correspondences, neither block face photographs nor ex vivo MRI of prostate specimens are usually available in the course of the clinical workflow. This might also explain why only two patient studies were employed in Ref. 18 and one rat brain slide in Ref. 35.
Alignment of more than two images or volumes representing very different structural or functional attributes of the same object is not well studied. One approach to the registration of multiple images is to take a groupwise (GW) approach, whereby all images are simultaneously aligned, usually to some reference image or coordinate frame. The limitations of fully GW approaches are that they either (1) involve optimization problems with many degrees of freedom arising from multiple simultaneous transformations or (2) are limited to images with similar intensity and∕or deformation characteristics. In the GW registration method presented by Bhatia,39 all images contribute to the same histogram used for entropy calculation. This limits the technique to images of the same modality. On the other hand, the GW method of Studholme40 utilizes a high dimensional distribution suitable for multimodal data, but the use of a dense deformation field requires a constraint that penalizes deformations that deviate from an average deformation. However, in the context of large deformation fields, such as might be present between ex vivo and in vivo images, this technique is restrictive. Other methods require repeated refinement of individual transformations prior to convergence.41 More recently, Balci42 performed simultaneous interpatient registration of a large number (50) of brain MRI scans using a sum of univariate (1D) entropy values (“stack entropy”) calculated at every pixel location. However, since this cost function requires a large number of images to calculate entropy at each pixel location, it is suited only for the registration of very large populations of images from the same modality, as opposed to a smaller number of multimodal images from a single patient. Thus, while a GW approach is generally preferable to several pairwise (PW) registration steps, GW methods can be restrictive or computationally prohibitive.
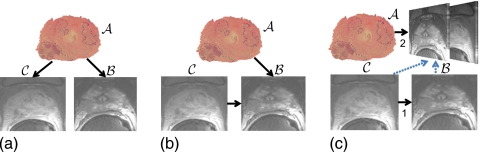
Given the challenges and constraints of GW registration approaches, simple sequential PW registration steps are most commonly performed to bring multiple images from different modalities into alignment. This was the method of choice for the prostate work in Refs. 30, 38, where several modalities are registered in steps using two images at a time. Figures 2a, 2b illustrate two possible approaches to PW registration of an ex vivo prostate histology (A) image to corresponding in vivo T2-w MR (B) and in vivo T1-w (single time point of a DCE series) (C) MR images. Figure 2a illustrates the case where image A is independently registered to both B and C. Figure 2b illustrates a scenario where the multiprotocol MR images are coregistered by the alignment of C to B and A is registered to just B, thus bringing A into alignment with both B and C. For this particular set of multimodal images, the approach illustrated in Fig. 2b is preferable to that shown in Fig. 2a since the alignment between multiprotocol MRI is less complicated compared to the multimodal alignment of ex vivo histology and in vivo MRI. The latter approach involves dealing with highly elastic deformations and dissimilar intensities. In both instances [illustrated in Figs. 2a, 2b], the two registration steps are independent and utilize only two images at a time.
Figure 2.
Registration of an ex vivo prostate histology (A) image to corresponding in vivo T2-w (B) and T1-w (C) MR images can be achieved in different ways. Two possible approaches using PW image registration involve (a) PW alignment of histology to each individual MRI protocol (A→B and A→C) or (b) alignment of multiprotocol MRI (C→B) and alignment of histology to just T2-w MRI (A→B). In the latter case, T1-w MRI would be in implicit alignment with histology at the end of the two registration steps. Alternatively, (c) a multiattribute image registration scheme involves initial PW alignment of images from the same modality (T1-w and T2-w MRI) as in (b), followed by alignment of histology to a multiattribute image comprising the registered multiprotocol MRI via a similarity measure specifically defined for high dimensional data.
NOVEL CONTRIBUTIONS AND SIGNIFICANCE
Both PW approaches illustrated in Figs. 2a, 2b consider only two images at a time. Hence, they exploit only a fraction of the available data in driving each registration step. Further, in subsequent alignment steps, it is necessary to select only a single image from the set of coregistered images for use as a reference. A more effective approach is to exploit all the information acquired from prior alignment steps to drive the subsequent registration operations. As illustrated in Fig. 2c, following registration of C to B, both the newly aligned images could be considered in unison (as a “multiattribute” image) to drive the registration with A. Such an approach would exploit the fact that C and B are (a) in implicit alignment and (b) represent different and informative image attributes (in this case, structural and functional). This approach is akin to previous studies that have used additional textural and gradient feature images20 to complement image intensity in order to improve image registration.
Multivariate formulations of MI have been shown to be useful in incorporating multiple image attributes (e.g., texture).21, 38 Thus, multivariate MI may also be applied in the context of applications where multiprotocol imaging (e.g., T2-w and T1-w MRI) needs to be registered to another modality (e.g., histology).
The novel contribution of this work is a formal quantitative image registration framework, which we refer to as MACMI. MACMI allows for incorporation of multiple modalities, protocols, or even feature images in an automated registration scheme, facilitated by the use of multivariate MI. MACMI is distinct from previous GW approaches in that it handles images that can significantly vary in terms of image intensities (e.g., multimodal data) and deformation characteristics (e.g., in vivo to ex vivo). Additionally, it involves a simple (low degree of freedom) optimization procedure whereby individual image transformations are determined in sequence. The use of an information theoretic similarity measure is central to the ability of MACMI to handle (1) multimodal data, which may contain nonlinear relationships between the image intensities of different modalities, and (2) high dimensional (multiattribute) observations, which may contain redundancies between the attributes that can be discounted via joint entropy. Finally, by employing a sequential approach to the alignment of multiple images within the multiattribute representation, each successive optimization procedure remains as simple (in terms of degrees of freedom) as with the conventional PW registration.
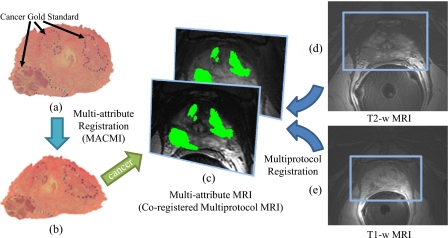
In this work, we evaluate MACMI in the context of a clinical problem involving multiprotocol (T2-w and DCE) MRI of the prostate and WMH sections, the sections being digitized following gland resection. Registration of WMH to corresponding multiprotocol MRI is then performed to map CaP extent from ex vivo WMH (previously delineated by pathologists) onto in vivo MRI. This procedure involves (1) initial affine alignment of the T2-w and DCE (T1-w) images [Figs. 3d, 3e] using MI to generate a multiattribute MR image [Fig. 3c], followed by (2) multimodal elastic registration of WMH with the multiattribute MRI.
Figure 3.
Establishing disease signatures on in vivo multiprotocol MRI using MACMI for registration of (a) WMH sections to corresponding (d) T2-w and (e) T1-w MRI. Alignment of the T2-w and T1-w MRI allows generation of (c) the multiattribute MRI comprised of coregistered multiprotocol MRI. MACMI is used to align (a) WMH to (c) the multiattribute MRI. CaP extent on the (b) elastically registered WMH is mapped directly onto both MR images in (c).
Our scheme for registration of in vivo MRI and ex vivo WMH of the prostate is distinct from previous related efforts18, 30, 35 in that (1) information from all in vivo imaging protocols is being utilized simultaneously to drive the process of automated elastic registration with histology; (2) no additional, intermediate ex vivo radiology or gross histology images need be obtained in addition to the clinically acquired in vivo MRI series; and (3) no point correspondences are required to be identified manually or automatically.
For the registration of 150 corresponding sets of prostate images from 25 patient studies with T2-w and DCE MRI, we quantitatively compare MACMI to a PW registration approach using conventional MI [Fig. 2b]. For 15 patients for which apparent diffusion coefficient (ADC) MRI was also acquired, we further demonstrate MACMI for including the third MR protocol in the elastic registration of histology with all three MRI series for 85 sets of images (in vivo T2-w, DCE and ADC MRI slices, and ex vivo WMH sections). We also quantitatively evaluate MACMI on a synthetic brain MRI study from BrainWeb,43 whereby T1-w and T2-w MR images are registered to PD MRI, where both the T1-w and T2-w protocols are simultaneously considered. MACMI is compared to PW MI-based approaches where T1-w and T2-w MR images are individually registered with PD MRI.
METHODS
Theory on mutual information and MACMI
Mutual information between scalar-valued images
Equation 1 below is a common formulation of MI for a pair of images (or random variables) A1,A2 in terms of Shannon entropy.
| (1) |
where I2(A1,A2) describes the interdependence of two variables or intensity values of a pair of images.20 As I2(A1,A2) increases, the uncertainty about A1 given A2 decreases. Thus, it is assumed that the global MI maximum will occur at the point of precise alignment, when maximal uncertainty about intensities of A1 can be explained by A2.
Mutual information between high dimensional (multiattribute) images
The conventional MI formulation can be extended to high dimensional observations by combining the multiple dimensions or attributes via high order joint entropy calculations. We refer to this application of MI as MACMI to distinguish it from conventional applications of MI and higher order MI and denote it as . Unlike the more familiar higher order MI (In,n≥2), the goal of MACMI is not to measure only the intersecting information between multiple sources (A1,…,An), but to quantify the combined predictive value of one multivariate source (e.g., [A1,…,An]) with respect to another (e.g., [B1,…,Bn]). Here we introduce the notion of an image ensemble as the concatenation of n intensity-valued images (I1,…,In) into an n-dimensional (multiattribute) image, denoted as [I1,…,In]. In the simplest case, the MI that a single image A1 shares with an ensemble of two other images, B1 and B2, is
| (2) |
By considering B1 and B2 as simultaneously measured semi-independent variables in the multidimensional ensemble [B1,B2], any dependence that exists between B1 and B2 may be discounted and remains bounded by the smaller of S(A1) and S(B1,B2). The generalized form of MI between the n-dimensional ensemble with the m-dimensional ensemble is
| (3) |
Thus, MACMI accomplishes fusion of the multiple dimensions of a multiattribute image, allowing only intersecting information between two such images (e.g., and ) to be quantified. Calculation of is discussed in Sec. 4C.
Framework for registration of multiple images using MACMI
In Sec. 4B1, we present a generalized algorithm (MACMIreg) for performing registration of m images Z1,…,Zm in a specific order. The order is specified using a hierarchical organization of the images within a family of sets Z and by progressively aligning and accumulating the registered images into an single ensemble ε. In Sec. 4B2 we illustrate the operation of the algorithm for four images (Z1,Z2,Z3,Z4), where Z is structured to register Z1 with Z2 and Z3 with Z4, prior to the alignment of the two resulting ensembles.
Algorithm
Consider a family of sets Z that contains m≥2 images Z1,…,Zm distributed throughout n≤m ordered subsets Zj(j∊I), where I={1,…,n}, (i.e., ∪j∊IZj={Z1,…,Zm} and ∩j∊IZj=∅). Each subset Zj (j∊{1,…,n}) may also be a family (i.e., have subsets of its own) or simply an ordered set of registered images. For example, if , we define Zj as a family of |Zj|=3 subsets, containing a total of k=∥Zj∥=4 images. We further denote the ensemble of all k images in Zj as . By organizing the m images into a hierarchy of subsets within the family Z, the order in which the images are registered and combined into multiattribute images is determined. The procedure for alignment of all images (within and between each Zj) into a single ensemble ε of registered images is described in the following recursive algorithm MACMIreg. Here we define the notation ε←εd as the expansion of an n-dimensional multiattribute image (ensemble) ε into an (d+n)-dimensional ensemble by concatenation with a d-dimensional ensemble εd. We also denote Zj⇐ε as the assignment of each of the m dimensions (intensity images) in ε to the existing m total members of Zj (independent of the organization of images within the family structure), thus replacing or updating Zj with the contents of ε.
Algorithm.
MACMIreg
| Input: Z={Z1,…,Zn},n≥1. |
| Output:ε. |
| Auxiliary Data Structures: Index k,j,α; Image ensemble ε0. |
| begin |
| 0. forj=1 to ndo |
| 1. k=|Zj|; |
| 2. ifk>1 then |
| 3. Obtain ensemble ε0=MACMIreg(Zj); |
| 4. Update Zj⇐ε0; |
| 5. endif; |
| 6. endfor; |
| 7. Initialize ε as an empty ensemble; |
| 8. ε←[Z1,…,Zk],k=∥Z1∥; |
| 9. α=k+1; |
| 10. forj=2 to ndo |
| 11. k=∥Zj∥; |
| 12. ε0=[Zα,…,Zα+k]; |
| 13. Obtain ; |
| 14. Obtain ; |
| 15. ; |
| 16. α=α+k+1; |
| 17. endfor; |
| end |
Lines 1–6 of MACMIreg use recursive calls to MACMIreg to register the images within each Zj containing more than one image. When MACMIreg(Zj) is executed on line 3, the algorithm is recursively instantiated in order to coregister the images within the subset Zj and any of its subsets, returning the registered images within ensemble ε. Line 4 then updates each Zj by replacing its constituent elements with the coregistered member images contained within ε. Lines 7–17 of MACMIreg perform the registration between the multiattribute images generated from each Zj, each of which now comprise only coregistered images (or a single image) following lines 1–6 of the algorithm. A spatial transformation T of the current moving image ensemble ε0 into alignment with the stationary growing ensemble ε is determined on line 13. The registered ensemble , obtained via T on line 14, is then combined with ε on line 15. The algorithm continues to align each subsequent Zj with the expanding reference ensemble ε.
Instance of MACMI for registration of four images
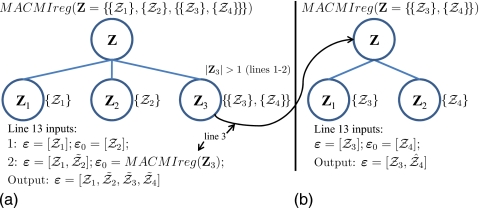
The operation of the MACMI algorithm is illustrated in Fig. 4 for a scenario involving the registration of four images (Z1,Z2,Z3,Z4), of which Z3 and Z4 are designated to be coregistered prior to alignment with Z1 and Z2. In this generic example, Z1 and Z2 could represent images from two different modalities, such as CT and PET, and Z3 and Z4 could represent multiprotocol images from the same modality such as T1-w and proton density (PD) MRI that may be in proximal alignment through hardware configuration or through prior application of a registration routine.
Figure 4.
(a) Graphical representation of the organization of four images (Z1,…,Z4) within a family of image sets (Z={{Z1},{Z2},{{Z3},{Z4}}}) and the application of the MACMIreg algorithm for alignment of all four images. Since only Z3 contains subsets (i.e., |Z3|=2), line 3 of MACMIreg in (a) begins a new instance of the algorithm in (b) with Z={{Z3},{Z4}} as the input. The instance in (b) brings Z3 and Z4 into alignment and returns the ensemble to the instance in (a). The instance in (a) first brings Z1 and Z2 into alignment and then align the ensemble of registered images from Z3 with the registered images of Z1 and Z2. At each registration step (line 13), a transformation T is determined by and ε is then expanded by (lines 14 and 15 of MACMIreg). The output of (a), containing all of the coregistered images in Z, is .
The operation of MACMIreg(Z) for Z={{Z1},{Z2},{{Z3},{Z4}}} begins by registration of images within each Zj(j∊{1,2,3}), where only Z3={{Z3},{Z4}} contains more than one image. Thus, as in Fig. 4b, MACMIreg(Z3) is called to register Z3 to Z4 and update Z3 with ensemble (lines 3–4 of MACMIreg). Having registered the images within each Zj (lines 1–6 of MACMIreg), all images in Z are registered as in Fig. 4a in two steps (lines 7–17 of MACMIreg). At each registration step, an optimal spatial transformation T of ε0 to ε is determined by (line 13 of MACMIreg) and ε is then expanded by (lines 14–15 of MACMIreg). Thus, Z2 is first registered to Z1, where ε=[Z1] and ε0=[Z2], and . Next, Z3 is registered to Z1 (and implicitly Z2), where and [the output of MACMIreg(Z3)], and . The final output is , comprising all of the coregistered images in Z.
The use of both Z3 and Z4 (and both Z1 and Z2) in the final registration step has the following benefits: (1) Avoids potential ambiguity in choosing between Z3 and Z4 (between Z1 and Z2) and (2) potentially provides improved alignment versus use of just Z3 or Z4 (Z1 or Z2) individually. The advantage of MACMI is that it yields cumulative incorporation of all images, while allowing flexibility to choose the order of multiattribute image construction. Implementation of MACMI within a complete registration framework is described in Sec. 4C below.
Requirements for implementation of MACMI
MACMI can be utilized to leverage multiple image sources in nearly any registration application by selecting the following components based on domain requirements:
-
(1)
MI estimation for high dimensional data: The most straightforward approach to estimating is to formulate the density estimates from high dimensional histograms. While histogram-based techniques are feasible and effective for up to four-dimensional observations with appropriate bin size, as demonstrated in Refs. 23, 21 higher dimensionality necessitates an alternate estimate of entropy or MI, such as those based on entropic spanning graphs or related quantities such as α-MI.44
-
(2)
Image transformation model(s): Since MACMI only dictates the construction of the objective function, MACMI is agnostic to the deformation model. Further, different deformation models may be used for each image since the individual image transformations are performed in independent steps.
-
(3)
Optimization scheme to find a maximum of : If the analytical gradient can be derived, as demonstrated for α-MI in Ref. 45, an efficient stochastic gradient descent method can be used. In the absence of analytical gradients of , methods including direct search (e.g., downhill simplex), quasi-Newton (e.g., Broyden–Fletcher–Goldfarb–Shanno), and other finite difference-based schemes can be employed.
Specific implementation details employed in this work are described in the following section.
EXPERIMENTAL DESIGN
A summary of the clinical prostate and synthetic brain image data sets investigated in this study is presented in Table 1.
Table 1.
Summary of the synthetic and clinical data sets registered by MACMI. Pixels are square.
| Data set | Description | Modalities | Dimensions | Studies (images) |
|---|---|---|---|---|
| Ss | Synthetic multiprotocol brain MRI from BrainWeb | T2-w, T1-w, PD MRI | 181×217 (1 mm pixel) | 1 (20) |
| Clinical multiprotocol MRI and histology of prostate | In vivo T2-w, DCE MRI, ex vivo WMH | T2-w MRI: 512×512 (0.230–.280 mm pixel) | 25 (150) | |
| Clinical multiprotocol MRI and histology of prostate | ADC, T2-w, DCE MRI, ex vivo WMH | T2-w MRI: 512×512 (0.230–.280 mm pixel) | 15 (85) |
Synthetic multiprotocol brain MRI
Data description
To quantitatively evaluate the performance of MACMI, we consider a synthetic registration task using a data set Ss comprising 20 2D multiprotocol (T1-w, T2-w, and PD) MRI slices from the BrainWeb simulated brain database.46 We denote the T1-w, T2-w, and PD MRI slices as T1, T2, and P, respectively.
Registration Experiment
Since synthetic brain MRI volumes T1, T2, and P are initially in alignment, we apply a known nonlinear deformation (Tap) to P to generate an image Pd, for which misalignment from T1 and T2 is known. The objective of the registration task is to recover the initial correct alignment via a corrective deformation (Tco). We denote the recovered P slice as Pr. Image transformation is implemented using an elastic free form deformation (FFD) model with a hierarchical mesh grid spacing scheme, as described in Ref. 21. Three mesh grid levels were defined for vertex spacings of approximately {36, 27, 18} mm at each level (nx,y∊{(6,5), (8,7), (12,10)}moving control points on zero-padded images). MACMI is performed in a manner similar to the scenario illustrated in Fig. 2c, whereby Pd (instead of WMH) is registered to the multiattribute image comprising the coregistered sections T1 and T2 via the recovered transformation
| (4) |
where ε(T1T2) represents the ensemble ε of T1 and T2. In order to compare to MACMI, registration is also performed using objective functions defined by the MI of (1) Pd with T1 and (2) Pd with T2. Thus, two additional Pr images are obtained by and by . Estimation of I2 and was achieved using 2D and 3D probability density estimates obtained using histograms with 128 and 62 gray level bins, respectively. The number of bins in each case were chosen empirically for reliable optimization of Eq. 4 using a Nelder–Mead simplex algorithm.47
Registration evaluation
For the synthetic data, quantitative evaluation of registration accuracy can be performed easily since the correct coordinate transformation Tap is known. The magnitude of error in the transformation Tco determined by registration can be quantified in terms of mean absolute difference (MAD) [FMAD(Tco)] and root mean squared (RMS) error [FRMS(Tco)] from Tap. Both MAD and RMS error are computed over the N total image pixels c in the common coordinate frame C of T1, T2, and P and can be expressed as
| (5) |
| (6) |
where N=|C| and ∥.∥L2 is the L2-norm of a coordinate vector. Further, the original P is compared directly to the resulting Pr using L2 distance (DL2) as the similarity measure.
Clinical multimodal prostate MRI and histology
Data description
We address the registration of two prostate data sets, and , comprising multimodal (3 T in vivo MRI and histology) and multiprotocol (T2-w, DCE, and ADC MRI) images (see Ref. 24 for details on acquisition). Set comprises 150 corresponding ex vivo WMH sections with CaP delineated and their closest corresponding 3 T in vivo T2-w and DCE MRI slices over all 25 patient studies. Set is a subset of 15 patients (85 image sets) in , for which DWI was also acquired and ADC maps calculated. We denote the T2-w MRI, DCE MRI, ADC MRI, and WMH images as S, F, D, and H, respectively. Each DCE MRI series comprises a sequence of T1-w gradient echo MRI volumes, acquired following bolus injection of the gadopentetate dimeglumine contrast agent at 0.1 mmol∕kg of bodyweight. Two precontrast and five postcontrast T1-w MRI volumes were obtained at a temporal resolution of 95 s. Maximal enhancement was generally obtained at the third postcontrast time point (denoted as F3), which was designated for use in the registration routines. No ex vivo MRI or gross pathology photographs were acquired.
Following RP and prior to sectioning, the excised prostate was embedded in a paraffin block while maintaining the orientation to keep the urethra perpendicular to the plane of slicing. This procedure facilitates the identification of a corresponding in vivo 2D axial MRI slice for each 2D histology slice. Preparation of the digitized WMH sections proceeds as follows: (1) The excised prostate is cut into sections that are 3–4 mm thick by slicing axial sections from the paraffin block using a circular blade, (2) a microtome is used to further cut the sections into thin slices that are about 5 μm thick, and (3) a single thin slice from each 3–4 mm thick section is chosen and digitally scanned. Each slide is then examined under a light microscope using up to 40× apparent magnification to identify and delineate the regions of CaP. As a result of this slide preparation process, spacing of the digital WMH slides is both coarse and irregular, ranging from 3–8 mm. Further, the nonlinear tissue deformations and artifacts introduced during microtome slicing are independent between slices and no block face photographs or fiducial markers are available to correct the distortions. Therefore, it is not feasible to accurately construct a 3D histology volume suitable for a 3D registration procedure without significant modifications to the established clinical routine.48, 49 Thus, for each WMH slice with disease, the closest corresponding T2-w MRI slice was visually identified by an expert radiologist.
Registration experiments
As previously described, the goal of this task is to register WMH to each MRI protocol in order to map CaP extent onto MRI. Since each MRI series was acquired in sequence and with minimal movement, 3D affine registration of the DCE to T2-w MRI volume was performed for each of the 25 patients in (and ) via MI between the 3D T2-w MRI volume and the 3D T1-w MRI volume corresponding to the third postcontrast time point of the 4D DCE MRI volume, using 256 gray level bins for the joint histogram. For each axial slice of the registered 4D DCE MRI volume, a multiattribute image representation was generated, as illustrated in Figs. 3c, 3d, 3e. Prior to 2D elastic registration of each H to each , the prostate capsule on the corresponding S images (six slices on average) for each patient was roughly delineated and the extraneous tissue masked out during registration. This was done to facilitate a rough global localization of the prostate on S relative to H.
For data set , automatic FFD registration of WMH to the MRI by MACMI is then performed for each of the 150 H slices using , resulting in a warped WMH , as shown in Fig. 3b. The CaP extent is then mapped onto S and by Te, as shown on Fig. 3c. In addition to MACMI, the elastic registration of H to the coordinate frame of S is also performed using (1) the conventional MI of H with S [I2(S,Te(H))] and (2) the conventional MI of H with . We refer to these PW registration approaches as PW-T2 and PW-DCE, respectively, and denote corresponding transformations as , , and .
For data set , the 3D ADC volume for each patient is also registered using a 3D affine transformation to the coregistered T2-w and DCE MRI volumes via a multiattribute volume [i.e., the volume composed of axial images ], hence generating a registered ADC volume comprising slices . Automated FFD registration of WMH to MRI is then performed for each set of corresponding H and S slices by MACMI, which also considers and via . CaP extent is then mapped to each of S, , and . PW registration using just H and D is not performed as the DWI protocol provides insufficient spatial resolution and anatomical detail for multimodal correlation based on image intensities alone.
Registration evaluation
Since ground truth for alignment of the clinical prostate data is not known or easily determinable, evaluation is performed by calculating similarities of both S and with , , and . To exclude the extraneous tissue outside the prostate in registration evaluation, similarity is calculated in terms of the MI [Eq. 1] for only the area in the image containing the prostate. Qualitative evaluation is also performed by visually comparing the CaP extent mapped from histology onto T2-w MRI via , , and . An estimate of CaP extent manually established by a radiologist on select slices of the T2-w MRI is used as the ground truth. This is done for only those specific MR images where CaP extent could be reliably delineated. It is important to note that these slices are not representative of all T2-w MR images with CaP, since easily delineated lesions are generally associated with dense tumors (i.e., those with compact cellular arrangements)50 and those occurring only in the peripheral zone.51 Further, delineation of the CaP boundary is more challenging than discerning the presence of a lesion.
RESULTS AND DISCUSSION
Synthetic brain registration
Table 2 presents a comparison of the evaluation measures FMAD, FRMS, and DL2 for transformations obtained in elastic registration of the n=20 multiprotocol MRI slices using MACMI and both PW registration approaches ( and ). The values of FMAD were compared between and using a paired t-test under the null hypothesis that there was no difference in FMAD between and . The values of FRMS and DL2 were also compared between and . Similarly, the values of FMAD, FRMS, and DL2 were also compared between and (second row of table). MACMI achieves better performance in terms of each measure, with significantly lower error (p<0.05 for n=20) compared to one or both PW methods.
Table 2.
Comparison of elastic registration accuracy for MACMI and pairwise MI alignment of n=20 pairs of synthetic PD MRI with coregistered T1-w and T2-w MRI brain images. The measures illustrated below correspond to (i) error of recovered deformation field (in mm) in terms of FMAD and FRMS and (ii) distance (DL2) between the undeformed and recovered PD MRI. MACMI results are significantly more accurate compared to either PW approach (p-values for both tests shown). The best values are indicated in bold.
| FMAD | FRMS | DL2 | |
|---|---|---|---|
| (T1-PD) | 0.9117 | 2.1407 | 1.83×10+03 |
| (T2-PD) | 0.9506 | 2.0248 | 2.35×10+03 |
| [ε (T1T2)-PD] | 0.8348 | 1.9307 | 1.71×10+03 |
| p ( vs ) | 0.0817 | 0.0578 | 0.0174 |
| p ( vs ) | 0.0013 | 0.2020 | 1.8×10−10 |
Clinical prostate registration
Mapping CaP extent from WMH onto in vivo T2-w and T1-w MRI
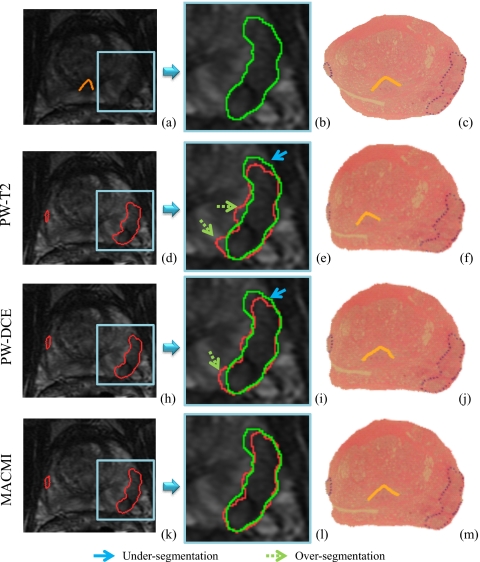
Figure 5 illustrates the registered WMH images and the corresponding T2-w MRI along with the contours of the mapped CaP extent and the urethra (verumontanum), for a single set of corresponding images in (and ). The S slice is shown in Fig. 5a with the region containing the dominant intraprostatic lesion (DIL) shown in the box and the urethra outlined. An expert delineation of the DIL is shown in Fig. 5b, while the original WMH image with CaP ground truth (dotted line) is shown in Fig. 5c. As described in Sec. 4B, affine registration of T2-w and DCE MRI protocol volumes (containing images S with F) is performed prior to elastic registration of H. Elastic registration of the H slice [shown in Fig. 5c] to S in Fig. 5a is then performed individually using each method (PW-T2, PW-DCE, and MACMI). The elastically warped H, obtained using only S (PW-T2), is shown in Fig. 5f and the CaP extent is shown mapped onto S in Figs. 5d, 5e. obtained using only F (PW-DCE) is shown in Fig. 5j and the CaP extent is again shown mapped onto S [Figs. 5h, 5i]. Finally, obtained using both S and F (MACMI) is shown in Fig. 5m and the CaP extent is again shown on S [Figs. 5k, 5l]. The contours of the mapped CaP extent on MRI suggest that (1) accurate elastic registration of WMH directly to corresponding in vivo MRI is feasible using the described FFD framework and (2) MACMI outperforms PW application of MI using data from only one of the several available MRI protocols. Note again that while the DIL shown in Fig. 5 is useful for qualitative evaluation, identifying such a clearly bounded lesion on T2-w MRI is rare. The positions of the urethra on in Figs. 5f, 5j, 5m also illustrate improved alignment of the image interior via MACMI compared to PW-T2 and PW-DCE. For example, note the improved urethral positioning via MACMI in Fig. 5m compared to the left misalignment by PW-T2 in Fig. 5f.
Figure 5.
(a) 3T in vivo T2-w MRI of a prostate with a clearly visible DIL and magnified in (b) with a manual estimate of CaP extent. (c) Closest corresponding WMH slice with CaP ground truth (dotted line) and urethra. [(d) and (e)] T2-w MRI with estimate of CaP extent as mapped from (f) WMH via elastic registration using only T2-w MRI. [(h) and (i)] T2-w MRI with CaP estimate from (j) WMH registered to DCE (T1-w) MRI (coregistered to T2-w MRI). [(k) and (l)] Registration using both T2-w and DCE MRI via MACMI results in closer agreement of the registration-derived CaP extent and the manual estimate. The verumontanum of the urethra is also shown on the registered WMH images in (f), (j), and (m).
The results in Fig. 5 qualitatively demonstrate that the general FFD-based registration paradigm described in this paper is capable of generating good alignment between in vivo MRI and ex vivo WMH prostate sections without the use of any additional ex vivo MRI series or gross histology (block face) photographs. Table 3 quantitatively illustrates the advantage of using MACMI over a single MRI protocol. For each of the n=25 patient studies in , registration accuracy for each of MACMI, PW-T2, and PW-DCE was approximated by the total MI of the elastically registered histology slices with all of the corresponding (1) S and (2) slices, both of which are in the same coordinate frame as S. Table 3 lists the average MI value over n=25 patients of all registered WMH slices obtained by MACMI, PW-T2, and PW-DCE (columns) with either S or (rows). The values of [and ] were compared between MACMI and PW-T2 using a paired t-test under the null hypothesis that there was no difference in [and ] between MACMI and PW-T2. The comparisons of and were also made between MACMI and PW-DCE. In all comparisons, MACMI demonstrated significant (p<0.05) improvement over both PW-T2 and PW-DCE, despite these PW methods using MI as their objective function.
Table 3.
Comparison of elastic registration accuracy for MACMI, PW-T2, and PW-DCE for n=25 patient studies. The measures illustrated below correspond to mean similarity in terms of the total MI of all registered WMH slices , obtained by MACMI, PW-T2, or PW-DCE (columns of table), with either T2-w MRI or DCE MRI (rows of table).
| Mean MI with WMH | Registration objective function | p (vs MACMI) | |||
|---|---|---|---|---|---|
| MR protocol | MACMI | PW-T2 | PW-DCE | PW-T2 | PW-DCE |
| T2-w: | 0.3378 | 0.3339 | 0.3297 | 0.0099 | 0.0006 |
| DCE: | 0.3155 | 0.3085 | 0.3102 | 4.0×10−4 | 0.0014 |
Mapping CaP extent from WMH onto ADC, T2-w and T1-w MRI
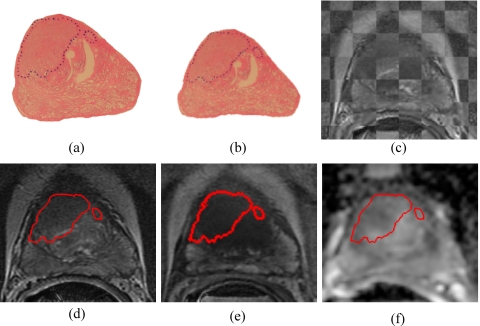
For the m=15 patient studies in for which ADC maps were also obtained, MACMI was applied in both (1) the 3D affine alignment of D to and (2) the 2D elastic alignment of H to . Figures 6a, 6b show the original H and the warped H[Te(H)], following elastic alignment with . Figure 6c shows a checker board visualization of the two coregistered S and slices [shown in Figs. 6d, 6e] after the 3D registration of the T2-w and DCE MRI volumes performed for the studies in . For each of the studies in , the second 3D affine multiprotocol registration step was performed to align the ADC volume to the T2-w and DCE MRI volumes. This enables the generation of aligned images [Fig. 6f] for each S (and ). S and the registered and images are shown in Figs. 6d, 6e, 6f with the contour of the mapped CaP extent from Te(H) [Fig. 6b]. It was observed that the inclusion of in had little effect on the resulting alignment with H when compared with the results obtained using only S and as in Sec. 6B1 above (comparison not shown).
Figure 6.
Using MACMI to include ADC MRI in the elastic registration of (a) histology to each of (d) T2-w, (e) DCE (T1-w), and (f) ADC MRI. Prior to elastic registration of histology, [(d) and (e)] T2-w and T1-w MRI were first successfully aligned via MI, as seen by (c) the checkerboard overlay of T2-w MRI and registered T1-MRI. (e) ADC was then registered to both T2-w and T1-w MRI via MACMI. (b) Elastically registered histology was obtained using the coregistered multiprotocol MRI via MACMI and CaP extent was mapped onto [(d)–(f)] MRI.
CONCLUDING REMARKS
Signatures for disease on multimodal in vivo imaging may be used to develop systems for computer-assisted detection of cancer or to assist in the training of medical students, radiology residents, and fellows. To establish in vivo radiological imaging signatures for prostate cancer, an accurate estimate of ground truth for cancer extent on each of the imaging modalities is necessary. In the context of certain anatomic regions and diseases, the spatial extent of disease may be obtained by spatial correlation or registration of histology sections with corresponding in vivo images. In this paper, we presented a new method termed MACMI within an automated elastic FFD registration framework for alignment of images from multiple in vivo acquisition protocols with corresponding ex vivo WMH sections. Our approach to registration of in vivo multiprotocol radiology images and ex vivo WMH of the prostate using MACMI is distinct from previous related efforts18, 30, 35 in that (1) information from all in vivo image sources is being utilized simultaneously to drive the automated elastic registration with WMH; (2) no additional, intermediate ex vivo radiology or gross histology images need to be obtained (this approach does not disrupt routine clinical workflow); and (3) no point correspondences are required to be identified manually or automatically. This last advantage is particularly relevant in the context of in vivo MR images where visual identification of anatomical landmarks is a challenge even for experts.
MACMI performs registration of several images by incorporating multiple image sources using an information theoretic approach. These may include different modalities, acquisition protocols, or image features. For the clinical application discussed in this paper, MACMI facilitates the use of all available in vivo prostate imaging protocols acquired during the standard clinical routine in order to perform automated elastic registration with the ex vivo WMH. Unlike fully groupwise registration techniques, the optimization problem remains simple while accommodating both highly dissimilar modalities and large deformations of variable magnitude.
We demonstrated the use of MACMI for registration of 150 multimodal (WMH, T2-w, and DCE MRI) prostate image sets from 25 patients; 85 sets of WMH, T2-w, ADC, and DCE MRI from 15 patients; and 20 sets of synthetic T1-w, T2-w, and PD MR brain images. Statistically significant improvement in registration accuracy was observed in using MACMI to simultaneously register PD MRI to both T1-w and T2-w MRI, compared to pairwise registration of PD to T1-w or T2-w MRI, for the synthetic data set. Qualitative examination of alignment between multiprotocol clinical prostate MRI and histology suggested improved performance via MACMI over pairwise MI. The inclusion of ADC MRI in the multiattribute registration had little effect on the resulting alignment with WMH when compared to the results obtained using only T2-w and T1-w MRI (Sec. 6B1). Nevertheless, it is possible that the use of MACMI in the registration of ADC to the ensemble of registered T2-w and T1-w MRI helped achieve more consistent multiprotocol MRI alignment than if either protocol were used alone. We intend to investigate this application of MACMI further in the future.
While we utilized histograms for density estimation, other techniques, such as entropic graphs,44 can be applied for larger numbers of images. However, independent of the implemented estimation method, MACMI affects information theoretic fusion of multiple image sources by computing multivariate MI between multiattribute images constructed as ensembles of coregistered images. It is important to note that in the absence of a predetermined order for combining images, MACMI may still be applied by combining images in a completely arbitrary order. Even in this scenario, MACMI still represents an improvement over fully pairwise registration by utilizing all registered images. Future work will investigate the influence of the order of multiattribute image construction on alignment accuracy.
ACKNOWLEDGMENTS
This work was made possible via grants from the Department of Defense Prostate Cancer Research Program (Grant No. W81XWH-08-1-0072), Wallace H. Coulter Foundation, National Cancer Institute (Grant Nos. R01CA136535-01, R01CA140772-01, and R03CA143991-01), and The Cancer Institute of New Jersey.
References
- Bloch B. N., Furman-Haran E., Helbich T. H., Lenkinski R. E., Degani H., Kratzik C., Susani M., Haitel A., Jaromi S., Ngo L., and Rofsky N. M., “Prostate cancer: Accurate determination of extracapsular extension with high-spatial-resolution dynamic contrast-enhanced and T2-weighted MR imaging–initial results,” Radiology 245, 176–185 (2007). 10.1148/radiol.2451061502 [DOI] [PubMed] [Google Scholar]
- Kurhanewicz J., Vigneron D., Carroll P., and Coakley F., “Multiparametric magnetic resonance imaging in prostate cancer: Present and future,” Current Opinion Urology 18, 71–77 (2008). 10.1097/MOU.0b013e3282f19d01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeks C. M. A., Fütterer J. J., Somford D. M., van Oort I. M., Huisman H., and Barentsz J. O., “Multiparametric MRI for prostate cancer screening,” Ned Tijdschr Geneeskd 153, B487-1–B487-7 (2009). [PubMed] [Google Scholar]
- Langer D. L., van der Kwast T. H., Evans A. J., Trachtenberg J., Wilson B. C., and Haider M. A., “Prostate cancer detection with multi-parametric MRI: Logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI,” J. Magn. Reson Imaging 30, 327–334 (2009). 10.1002/jmri.21824 [DOI] [PubMed] [Google Scholar]
- Viswanath S., Bloch B. N., Rosen M., Chappelow J., and Toth R., “Integrating structural and functional imaging for computer assisted detection of prostate cancer on multi-protocol in vivo 3 Tesla MRI,” Proc. SPIE 7260, 72603I-1–72603I-12 (2009). 10.1117/12.811899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P., Rosen M., and Madabhushi A., “A hierarchical spectral clustering and nonlinear dimensionality reduction scheme for detection of prostate cancer from magnetic resonance spectroscopy (MRS),” Med. Phys. 36, 3927–3939 (2009). 10.1118/1.3180955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider M. A., van der Kwast T. H., Tanguay J., Evans A. J., Hashmi A.-T., Lockwood G., and Trachtenberg J., “Combined T2-weighted and diffusion-weighted MRI for localization of prostate cancer,” AJR, Am. J. Roentgenol. 189, 323–328 (2007). 10.2214/AJR.07.2211 [DOI] [PubMed] [Google Scholar]
- Kozlowski P., Chang S. D., Jones E. C., Berean K. W., Chen H., and Goldenberg S. L., “Combined diffusion-weighted and dynamic contrast-enhanced MRI for prostate cancer diagnosis—Correlation with biopsy and histopathology,” J. Magn. Reson Imaging 24, 108–113 (2006). 10.1002/jmri.20626 [DOI] [PubMed] [Google Scholar]
- Madabhushi A., Feldman M. D., Metaxas D. N., Tomaszeweski J., and Chute D., “Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI,” IEEE Trans. Med. Imaging 24, 1611–1625 (2005). 10.1109/TMI.2005.859208 [DOI] [PubMed] [Google Scholar]
- Tiwari P., Kurhanewicz J., Rosen M., and Madabhushi A., “Semi supervised multi kernel (SeSMiK) graph embedding: Identifying aggressive prostate cancer via magnetic resonance imaging and spectroscopy,” in Proceedings of MICCAI, Vol. 6363, 666–673 (2010). [DOI] [PMC free article] [PubMed]
- Chappelow J., Viswanath S., Monaco J., Rosen M., Tomaszewski J., Feldman M., and Madabhushi A., “Improving supervised classification accuracy using non-rigid multimodal image registration: Detecting prostate cancer,” Proc. SPIE 6915, 69150V-1–69150V-12 (2008). 10.1117/12.770703 [DOI] [Google Scholar]
- Bartolozzi C., Menchi I., Lencioni R., Serni S., Lapini A., Barbanti G., Bozza A., Amorosi A., Manganelli A., and Carini M., “Local staging of prostate carcinoma with endorectal coil MRI: Correlation with whole-mount radical prostatectomy specimens,” Eur. Radiol. 6, 339–345 (1996). 10.1007/BF00180606 [DOI] [PubMed] [Google Scholar]
- Vos P. C., Hambrock T., van de Kaa C. A. H., Fütterer J. J., Barentsz J. O., and Huisman H. J., “Computerized analysis of prostate lesions in the peripheral zone using dynamic contrast enhanced MRI,” Med. Phys. 35, 888–899 (2008). 10.1118/1.2836419 [DOI] [PubMed] [Google Scholar]
- Ozer S., Langer D. L., Liu X., Haider M. A., van der Kwast T. H., Evans A. J., Yang Y., Wernick M. N., and Yetik I. S., “Supervised and unsupervised methods for prostate cancer segmentation with multispectral MRI,” Med. Phys. 37, 1873–1883 (2010). 10.1118/1.3359459 [DOI] [PubMed] [Google Scholar]
- Liu X., Langer D. L., Haider M. A., Yang Y., Wernick M. N., and Yetik I. S., “Prostate cancer segmentation with simultaneous estimation of Markov random field parameters and class,” IEEE Trans. Med. Imaging 28, 906–915 (2009). 10.1109/TMI.2009.2012888 [DOI] [PubMed] [Google Scholar]
- Chappelow J. and Madabhushi A., “Multi-attribute combined mutual information (MACMI): An image registration framework for leveraging multiple data channels,” in Proceedings of the IEEE International Symposium on Biomedical Imaging, 2010.
- Vos P. C., Hambrock T., Barenstz J. O., and Huisman H. J., “Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI,” Phys. Med. Biol. 55, 1719–1734 (2010). 10.1088/0031-9155/55/6/012 [DOI] [PubMed] [Google Scholar]
- Park H., Piert M. R., Khan A., Shah R., Hussain H., Siddiqui J., Chenevert T. L., and Meyer C. R., “Registration methodology for histological sections and in vivo imaging of human prostate,” Acad. Radiol. 15, 1027–1039 (2008). 10.1016/j.acra.2008.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y., Shen D., Feldman M., Tomaszewski J., and Davatzikos C., “Non-rigid registration between histological and MR images of the prostate: A joint segmentation and registration framework,” in Computer Vision and Pattern Recognition Workshops (IEEE Computer Society, Los Alamitos, 2009) pp. 125–132.
- Pluim J., Maintz J., and Viergever M. A., “Image registration by maximization of combined mutual information and gradient information,” IEEE Trans. Med. Imaging 19, 809–814 (2000). 10.1109/42.876307 [DOI] [PubMed] [Google Scholar]
- Rueckert D., Clarckson M., and Hawkes D. J., “Non-rigid registration using higher-order mutual information,” Proc. SPIE 3979, 438–447 (2000). [Google Scholar]
- Boes J. and Meyer C., “Multi-variate mutual information for registration,” in Proceedings of MICCAI, Vol. 1679, pp. 606–612, 1999.
- Studholme C., Hill D., and Hawkes D. J., “Incorporating connected region labeling into automatic image registration using mutual information,” in Mathematical Methods in Biomedical Image Analysis, Vol. 3979, pp. 23–31, 1996.
- Bloch B. N., Lenkinski R. E., and Rofsky N. M., “The role of magnetic resonance imaging (MRI) in prostate cancer imaging and staging at 1.5 and 3 Tesla: The Beth Israel Deaconess Medical Center (BIDMC) approach,” Cancer Epidemiology Biomarkers and Prevention 4, 251–262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurhanewicz J., Swanson M. G., Nelson S. J., and Vigneron D. B., “Combined magnetic resonance imaging and spectroscopic imaging approach to molecular imaging of prostate cancer,” J. Magn. Reson Imaging 16, 451–463 (2002). 10.1002/jmri.10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharatha A. et al. , “Evaluation of three-dimensional finite element-based deformable registration of pre- and intraoperative prostate imaging,” Med. Phys. 28, 2551–2560 (2001). 10.1118/1.1414009 [DOI] [PubMed] [Google Scholar]
- Foskey M., Davis B., Goyal L., Chang S., Chaney E., Strehl N., Tomei S., Rosenman J., and Joshi S., “Large deformation three-dimensional image registration in image-guided radiation therapy,” Phys. Med. Biol. 50, 5869–5892 (2005). 10.1088/0031-9155/50/24/008 [DOI] [PubMed] [Google Scholar]
- Xu S., Kruecker J., Turkbey B., Glossop N., Singh A. K., Choyke P., Pinto P., and Wood B. J., “Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies,” Comput. Aided Surg. 13, 255–264 (2008). 10.3109/10929080802364645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Kruecker J., Xu S., Glossop N., Guion P., Ullman K., Choyke P. L., and Wood B. J., “Initial clinical experience with real-time transrectal ultrasonography-magnetic resonance imaging fusion-guided prostate biopsy,” BJU Int. 101(5), 841–845 (2008). 10.1111/j.1464-410X.2007.07348.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Z., Sodee D. B., Resnick M., and Maclennan G. T., “Multimodal and 3D imaging of prostate cancer,” Comput. Med. Imaging Graph. 29, 477–486 (2005). 10.1016/j.compmedimag.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Parker C. C., Damyanovich A., Haycocks T., Haider M., Bayley A., and Catton C. N., “Magnetic resonance imaging in the radiation treatment planning of localized prostate cancer using intra-prostatic fiducial markers for computed tomography co-registration,” Radiother. Oncol. 66, 217–224 (2003). 10.1016/S0167-8140(02)00407-3 [DOI] [PubMed] [Google Scholar]
- Huisman H. J., Fütterer J. J., van Lin E. N. J. T., Welmers A., Scheenen T. W. J., van Dalen J. A., Visser A. G., Witjes J. A., and Barentsz J. O., “Prostate cancer: Precision of integrating functional MR imaging with radiation therapy treatment by using fiducial gold markers,” Radiology 236, 311–317 (2005). 10.1148/radiol.2361040560 [DOI] [PubMed] [Google Scholar]
- Vidakovic S., Jans H. S., Alexander A., and Sloboda R. S., “Post-implant computed tomography—Magnetic resonance prostate image registration using feature line parallelization and normalized mutual information,” J. Appl. Clin. Med. Phys. 8(1), 21–32 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Kwee S., Thibault G., Stack R., Sesterhenn I., Potter K., and Meyer C., “Registration methods for histological slides and ex vivo MRI of prostate,” in IEEE Nuclear Science Symposium Conference Record, 2007, Vol. 4, pp. 3102–3104, 2007.
- Meyer C. R., Moffat B. A., Kuszpit K. K., Bland P. L., Chenevert T. L., Rehemtulla A., and Ross B. D., “A methodology for registration of a histological slide and in vivo MRI volume based on optimizing mutual information,” Mol. Imaging 5, 16–23 (2006). [PMC free article] [PubMed] [Google Scholar]
- Park S. B., Rhee F. C., Monroe J. I., and Sohn J. W., “Spatially weighted mutual information image registration for image guided radiation therapy,” Med. Phys. 37, 4590–4601 (2010). 10.1118/1.3463609 [DOI] [PubMed] [Google Scholar]
- Zhan Y., Ou Y., Feldman M., Tomaszeweski J., and Davatzikos C., “Registering histologic and MR images of prostate for image-based cancer detection,” Acad. Radiol. 14, 1367–1381 (2007). 10.1016/j.acra.2007.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappelow J., Bloch B. N., Rofsky N., Genega E., Lenkinski R., DeWolf W., Viswanath S., and Madabhushi A., “COLLINARUS: Collection of image-derived non-linear attributes for registration using splines,” Proc. SPIE 7259, 72592N-1–72592N-12 (2009). 10.1117/12.812352 [DOI] [Google Scholar]
- Bhatia K. K., Hajnal J. V., Puri B. K., Edwards A. D., and Rueckert D., “Consistent groupwise non-rigid registration for atlas construction,” in Proceedings of the IEEE International Symposium on Biomedical Imaging, pp. 908–911, 2004.
- Studholme C., “Simultaneous population based image alignment for template free spatial normalisation of brain anatomy,” in Workshop on Biomedical Imaging Registration, pp. 81–90, 2003.
- Cootes T. F., Marsland S., Twining C., Smith K., and Taylor C., “Groupwise diffeomorphic non-rigid registration for automatic model building,” in European Conference on Computer Vision, pp. 316–327, 2004.
- Balci S., Golland P., Shenton M., and Wells W., “Free-form B-spline deformation model for groupwise registration,” in Proceedings of MICCAI, 2007.
- http://www.bic.mni.mcgill.ca/brainweb/
- Neemuchwala H., Hero A., and Carson P., “Image matching using alpha-entropy measures and entropic graphs,” Signal Process. 85, 277–296 (2005). 10.1016/j.sigpro.2004.10.002 [DOI] [Google Scholar]
- Staring M., van der Heide U. A., Klein S., Viergever M. A., and Pluim J., “Registration of cervical MRI using multifeature mutual information,” IEEE Trans. Med. Imaging 28, 1412–1421 (2009). 10.1109/TMI.2009.2016560 [DOI] [PubMed] [Google Scholar]
- Collins D. L., Zijdenbos A. P., Kollokian V., Sled J. G., Kabani N. J., Holmes C. J., and Evans A. C., “Design and construction of a realistic digital brain phantom,” IEEE Trans. Med. Imaging 17, 463–468 (1998). 10.1109/42.712135 [DOI] [PubMed] [Google Scholar]
- Lagarias J. C., Reeds J. A., Wright M. H., and Wright P. E., “Convergence properties of the Nelder–Mead simplex method in low dimensions,” SIAM J. Optim. 9, 112–147 (1998). 10.1137/S1052623496303470 [DOI] [Google Scholar]
- Xiao G., Bloch B. N., Chappelow J., Genega E. M., Rofsky N. M., Lenkinski R. E., Tomaszewski J., Feldman M. D., Rosen M., and Madabhushi A., “Determining histology-MRI slice correspondences for defining MRI-based disease signatures of prostate cancer,” Comput. Med. Imaging Graph. (in press). [DOI] [PubMed]
- Chappelow J., Tomaszewski J. E., Feldman M., Shih N., and Madabhushi A., “Histostitcher: An interactive program for accurate and rapid reconstruction of digitized whole histological sections from tissue fragments,” Comput. Med. Imaging Graph. (2011) (in press). [DOI] [PMC free article] [PubMed]
- Nicolas V., Beyersdorff D., Mueller-Lise U., Pennekamp W., and Heyer C., “The prostate and seminal vesicles,” in MR Imaging of the Abdomen and Pelvis (Thieme, 2002), pp. 220–240. [Google Scholar]
- Schiebler M. L., Tomaszewski J. E., Bezzi M., Pollack H. M., Kressel H. Y., Cohen E. K., Altman H. G., Gefter W. B., Wein A. J., and Axel L., “Prostatic carcinoma and benign prostatic hyperplasia: Correlation of high-resolution MR and histopathologic findings,” Radiology 172, 131–137 (1989). [DOI] [PubMed] [Google Scholar]