Abstract
Proteins that are expressed outside the cell must be synthesized, folded and assembled in a way that ensures they can function in their designate location. Accordingly these proteins are primarily synthesized in the endoplasmic reticulum (ER), which has developed a chemical environment more similar to that outside the cell. This organelle is equipped with a variety of molecular chaperones and folding enzymes that both assist the folding process, while at the same time exerting tight quality control measures that are largely absent outside the cell. A major post-translational modification of ER-synthesized proteins is disulfide bridge formation, which is catalyzed by the family of protein disulfide isomerases. As this covalent modification provides unique structural advantages to extracellular proteins, multiple pathways to their formation have evolved. However, the advantages that disulfide bonds impart to these proteins come at a high cost to the cell. Very recent reports have shed light on how the cell can deal with or even exploit the side reactions of disulfide bond formation to maintain homeostasis of the ER and its folding machinery.
Introduction
The extracellular environment where cell surface and secreted proteins must function is generally quite different than that of the cytosol, particularly in terms of redox potential and certain ion concentrations. Thus a major obstacle for the eukaryotic cell is to not only fold and assemble complex hetero-oligomeric proteins from extended polypeptide chains, but to do so under cellular conditions where the outcome can be monitored and the protein will function in its requisite location. The solution to this challenge was the development of the endoplasmic reticulum (ER), a vast reticular organelle present in all nucleated eukaryotic cells. The ER is similar to the extracellular environment in terms of its ion concentrations and oxidizing capabilities; but in contrast provides dedicated folding, oxidation, and quality control machineries. The biosynthesis of secretory proteins is closely aided and monitored by a vast array of resident ER proteins that comprise the quality control apparatus of ER, which allows only properly matured proteins to transit to the Golgi [1;2] Some quality control measures exist in post-ER compartments, but they become much more limited [3;4]. Thus, the ER must set and uphold high standards for protein quality control that can function under the complicating conditions of high concentrations of unfolded polypeptide chains, significantly differing clients, and changing metabolic needs.
Protein folding, post-translational modification and quality control in the ER
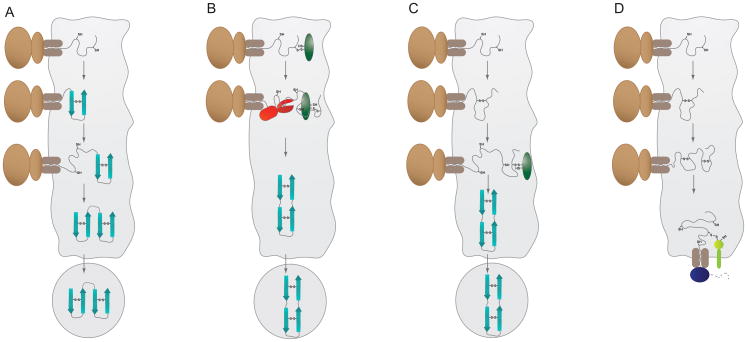
In addition to the free energy-driven conformational folding of the newly synthesized polypeptide chains (e.g., hydrogen bond formation and burying hydrophobic residues), a number of additional reactions take place in the ER that can influence the folding and the final state of a protein, including N-linked glycosylation (reviewed in Chapter XX of this issue), oligomerization, and formation of disulfide bonds (Figure 1). Their facilitation by resident ER chaperones and enzymes aids folding by restricting “off pathway” possibilities and accelerating slow folding reactions like disulfide bridge formation or peptidyl-prolyl isomerization [2]. While regulating steps on the pathway to the biologically active state, they also provide means to detect proteins that are not completely or properly matured (Figure 1).
Figure 1. Folding reactions occurring in the ER and features recognized by ER quality control components.
Folding and oligomerization take place in the ER and features that occur on non-native structures are identified by various chaperones and folding enzymes, which serve to prevent incompletely folded or assembled proteins from moving further along the secretory pathway. (A) Exposed hydrophobic regions (yellow) that will eventually be buried upon folding or oligomerization are often recognized by the Hsp70 chaperone BiP (red). (B) The lectins calreticulin (blue) and calnexin bind to sugar moieties possessing one terminal glucose residue (grey hexagon), which can be found on incompletely folded glycoproteins. (C) PDIs (green) form mixed disulfide bonds with free thiol groups to catalyze disulfide bond formation, reduction, or isomerization.
Disulfide bond formation in protein folding and oligomerization
Due to their covalent nature, disulfide bonds can have profound effects on the folding pathway and the stability of a protein, thus increasing its suitability for existence in the extracellular milieu. Disulfide bonds can stabilize a protein by reducing the entropy of the unfolded state [5]. Furthermore, they can facilitate the path to the native state if they link parts of a protein that must come into contact early during a folding reaction and can make unfolding less likely if they occur in particularly labile parts of a protein [5]. During the folding of secretory pathway proteins, native intra- and inter-molecular disulfide bonds can form co-translationally, well before more C-terminal regions of the protein have entered the ER [6]. This can occur when the bonds form between adjacent cysteines [7;8] or when the protein is composed of autonomously folded domains [9] (Figure 2). However, when long range disulfide bonds are part of the native structure and other cysteine residues occur between the correct cysteine pair the situation is more complex (Figure 2). Since the more N-terminal cysteines enter the ER lumen first, they must be shielded to prevent the formation of non-native bonds, which would need to be broken once the native partner cysteine was translocated. For the influenza hemagglutinin protein, UDP-glucose:glycoprotein glucosyltransferase 1 (UGT1) and calnexin/calreticulin binding shield regions of the N-terminus of the protein early in folding that will ultimately be involved in long range disulfide bonds with cysteines that occur nearer the C-terminus [10]. Whereas in the case of the immunoglobulin (Ig) heavy chain (HC), the binding of the BiP:ERdj3 chaperone complex prevents intradomain disulfide bonds from forming between linear cysteines in the CH1 domain until light chain (LC) assembly induces folding of the domain [11–13•]. Conversely, non-native disulfide bonds have been shown to occur as normal folding intermediates for a number of proteins, which then ultimately must be broken as folding proceeds (Figure 2). Cysteine knot proteins have a complex fold in which at least two non-successive disulfide bonds form prior to slipping a portion of the protein into this structure, which is then stabilized by a third non-successive bond [14]. This has been shown in some cases to involve several non-native bonds along the folding pathway [15]. Similarly, the LDL receptor, which has 30 native disulfide bonds, requires the formation of a number of non-native bonds that are reshuffled to achieve the proper final structure [16].
Figure 2. Different pathways to disulfide bridge formation in the ER.
(A) For some proteins, correct disulfide bonds can form co-translationally due to independent domain-like structures in the protein. (B) In other cases, the ER chaperone machinery, for simplicity only BiP (red) and PDI (green) are shown, inhibits premature folding and disulfide bridge formation of a protein with disulfide bonds that have to form between non-adjacent cysteines. Once the complete polypeptide chain has emerged from the translocon and is released from the chaperones, folding and disulfide bridge formation can proceed. (C) If non-native disulfide bonds between adjacent cysteines form co-translationally, these must be isomerized by a PDI (green) to allow the correct pair to form. Once folded, proteins can proceed to the Golgi and further along the secretory pathway (A–C). (D) In some cases, incorrect disulfide bonds are formed and terminal misfolding occurs, which renders the protein a substrate for ERAD. Retrotranslocation and proteolysis by the proteasome (blue) is likely preceded by reduction of the wrong disulfide bonds (reductase shown in light green) or even native ones in correctly folded regions of the protein [91].
Assembled subunits of secreted multimeric proteins are often covalently linked by disulfide bonds, which likely serve to stabilize their quaternary structure once outside the cell. The correct oligomerization and covalent linkage of multimeric proteins poses a particularly demanding task for the ER quality control system, which has guided the evolution of various mechanisms leading to correct folding, oligomerization and covalent assembly of multimeric proteins. While individual subunits or domains can in some cases fold autonomously and then assemble, a recent study demonstrated that the CH1 domain of isolated Ig HCs remains unstructured until it combines with LCs [13]. This allows it to remain associated with molecular chaperones, which, in addition to preventing aggregation, also provides a mechanism to identify it as unassembled. Only once folded correctly on the LC template, HC and LC become covalently attached [11–13]. In the case of assembling pentameric IgM, five monomers must combine with a J chain. This involves the binding of ERGIC-53 to a glycan near the C-terminus of the IgM heavy chain and covalent attachment of ERp44 to a C-terminal cysteine [17•], which paves the way for subsequent disulfide bond formation with a J chain. Even in the case of subunits that fold independent of assembly (e.g., some Ig LC [18] and β2 microglobulin [19]), the free subunit often shows characteristics of an incompletely folded protein [20;21]. Molecular chaperones that recognize unfolded or unassembled hydrophobic regions (e.g., the Hsp70 chaperone BiP) provide a solution by preventing aggregation and blocking transport of unassembled subunits [22;23]. However, it is still not clear how the ER quality control system ultimately distinguishes between nascent proteins that have not yet folded and those that are unable to fold and must be degraded [2;24].
Mechanisms and biological implications of disulfide bond formation, isomerization and reduction
The oxidizing extracellular environment has the potential to induce intra- or intermolecular disulfide bonds between random unpaired cysteines that find themselves even transiently juxtaposed. Accordingly, secreted proteins could have been selected to be cysteine free at the cost of the additional stability afforded by disulfide bonds as is the case for extracellular proteins produced by facultative anaerobic bacteria [25]. Instead, the cysteine content of mammalian extracellular proteins is actually higher than that of cytosolic proteins; arguing for an evolutionary advantage of a relatively high cysteine content in secreted mammalian proteins [26]. This is made possible by the evolution of dedicated chaperone and catalysis systems, which help ensure correct disulfide bridge formation prior to secretion. In part, the relatively high ratio of GSSG:GSH and other small molecule oxidants contribute to the oxidizing environment of the ER [27;28], and glutathione can be detected as mixed disulfides with nascent ER proteins [29]. However, more recent data argue that disulfide bond formation is significantly regulated by the endoplasmic reticulum oxidoreductin 1 (Ero1) protein, which can initiate a relay of disulfide bonds to various ER localized protein disulfide isomerases that in turn catalyze the oxidation of disulfide bonds in substrates [30;31].
Protein disulfide isomerases
PDI is the founding member of the ER protein disulfide isomerase family. It contains two enzymatically active (a and a′) and two inactive (b and b′) thioredoxin-like (Trx) domains, which are the structural and functional hallmark of the PDI family [32;33]. The catalytically active center of the Trx fold is a CXXC motif where the amino acids separating the two cysteines influence their redox potential [34]. Free PDI is present in different oxidation states in the ER [35], although there is apparently a slight preference for its reduced state in mammalian cells [36]. This is likely due to the slightly reducing GSH/GSSG system, which is required to ensure that reductive pathways are active allowing PDIs to reduce and isomerize non-native disulfides [35;37;38]. Much recent data argue that in order to catalyze disulfide bond formation in substrates, PDI is oxidized by Ero1 in yeast [30;39], or Ero1α or β in mammals [40;41]. Oxidized Ero1 first transfers its disulfide bond to PDI, which in turn oxidizes substrates, with the backward flow of electrons ultimately being transferred to oxygen [30]. The recently solved structure human Ero1α suggests a substrate-like recognition of Ero1α by a hydrophobic cleft on the b′ domain of PDI, making the displacement of Ero1α by substrates possible once Ero1α has oxidized PDI [42••]. The PDIs ERp57 and ERp72 bind to Ero1 with lower affinity, likely due to their less hydrophobic b′ domains [43;44], which also limits direct substrate binding [45;46•], and are thus oxidized to a lower extent than PDI [42;47•]. The different binding affinity of Ero1α for various PDI family members might influence redox reactions in the ER. For instance, ERp57, which is primarily reduced and partners with calnexin and calreticulin [36], could theoretically preferentially catalyze isomerization reactions required in later stages of a folding reaction [45], whereas reduction of the substrate, or even ERp57, could be performed by TMX4, which is also part of this complex [48]. It is noteworthy that PDI is also generally more reduced in mammalian cells [36;49] where more complex disulfide bond patterns are established than in yeast cells where PDI is mainly oxidized [39].
20 PDI family members have been described to date in human cells, most of which are ubiquitously expressed [33]. In fact, only a few PDIs show tissue specificity, like testis-localized PDILT [50] or PDIp which is synthesized in pancreatic beta cells [51]. Although the reason for having so many PDIs is not entirely clear, recent studies suggest some possibilities. First, some family members have extra domains (e.g., DnaJ-like domain in the PDI family member ERdj5 [52••], which can recruit BiP to terminally misfolded proteins and aid in their degradation). Second, the ability of certain PDIs to interact with chaperone subfamilies (e.g., ERp57 with calnexin/calreticulin [53], Sep15 with UGT1 [54;55], and PDI, ERp72, and P5 with BiP [12;56•]) may provide some substrate specificity. A recent study testing the ability of four different PDIs to interact with and oxidize five substrates in a human hepatoma line showed that PDI had the broadest substrate range and ERp57 was more important for glycoproteins; whereas ERp72 and P5 had little effect on the clients tested [57•]. Even in the case of PDI depletion, the effect on these substrates was modest, arguing for redundancy among family members in agreement with a previous study on ERp57 [46]. And third, the ER is comprised of functionally different regions (e.g., folding versus degradation) [58;59], which might require them to be populated with PDIs more suited to oxidation versus reduction reactions. Inspection of active site residues suggests some family members are more likely to act as reductases (e.g., ERdj5, which has CXPC in three sites), others as oxidases (e.g., PDI with CXHC in its two sites), and others form more stable mixed disulfides (e.g., ERp44, which is missing the C terminal cysteine of the active site, CXXS) [32;33;60•].
Unpaired cysteines and quality control
Although many surface or secreted proteins possess numerous cysteines, they do not usually leave the ER unpaired but form intra- and inter-chain disulfide bonds [61]. That a redox reaction itself is involved in this control step was demonstrated when the addition of mild reducing agents to cultured cells allowed the secretion of some incompletely oxidized Ig proteins [62;63]. ERp44, which possesses an incomplete active site and an ER retention sequence, can form fairly stable mixed disulfides with Ero1 [64], as well as with incompletely oxidized substrates like formylglycine-generating enzyme [65], IgM, κ LC, J chains [64], and adiponectin [66], serving to retain them. Interestingly, ERp44 possesses a hydrophobic cleft in its b′ domain, similar to the one of PDI, which might aid in Ero1 and substrate interactions [60;65]. Indeed, deletion of the auto-inhibitory C-terminus of ERp44, which normally binds its own b′ domain, increases the substrate binding range of ERp44 [60]. Several other members of the PDI family possess incomplete active sites and might play a more specialized role in retention or quality control of dedicated substrates [50;67]. Alternatively some PDI family members form transient mixed disulfide bonds with client through their CXXC motif [45] and either directly retain them via their ER-retention motif or indirectly through their involvement in multi-chaperone complexes [12;46;56] (Figure 3) until the client is folded or degraded [52]. Lastly, several PDIs (i.e., PDI and PDIp) not only recognize and react with unpaired cysteine residues but also bind exposed hydrophobic surfaces on substrates [68;69] thereby integrating different characteristic recognition features of incompletely matured proteins.
Figure 3. An overview over the oxidative folding and quality control mechanisms in the ER.
Proteins enter the ER co-translationally as unfolded polypeptide chains. (A) If they are non-glycosylated or the first N-linked glycan occurs fairly far into the sequence, they can bind BiP (red) along with PDI (green oval). Further components like the ERdj proteins are omitted for simplicity. (B) If the substrate is glycosylated, they interact with the calreticulin/calnexin system. Calreticulin (dark blue) interacts with the PDI family member ERp57 (dark green hexagon), which contributes to the oxidative folding of glycoproteins. If folding is successful, the proteins can leave the calreticulin/calnexin cycle, if not, UGT1 (light blue), which can cooperate with Sep15, another member of the PDI family (light green circle), re-glucosylate the substrate and reenter the protein into the calreticulin/calnexin cycle. (C) In both cases, once folding is nearly complete, any free thiols remaining can covalently engage ERp44 (yellow circle), which inhibits premature transport to the Golgi until these thiols are buried or part of a disulfide bond. (D) PDIs can be oxidized by Ero1 (dark brown), which generates H2O2. This can be used indirectly to oxidize Prx4 (one dimer of its decameric structure is shown in light brown), which in turn oxidizes PDI (green) generating two disulfide bonds per molecule of O2. PDIs are further coupled to the GSH/GSSG system, which could regulate Ero1 activity or even oxidize substrates that do not require other PDI-associated functions.
The costs of oxidative folding
While disulfide bonds clearly confer stability to proteins making them more suited to the extracellular environment, the cost to the cell appears to be even greater than the energy and machineries required to form them. The discovery of the Ero1-mediated transfer of disulfides, in which oxygen is the electron acceptor, suggests that for each disulfide bond formed one molecule of H2O2, a potent oxidant, will be produced [30]. While this has not been directly measured, for dedicated secretory cells like plasma cells or pancreatic beta islet cells, where it has been estimated that as many as 100,000 disulfide bonds can be formed a second, one might imagine that the oxidative stress encountered would be staggering. In support of the likely oxidative stress generated by the Ero1 cascade, reduced Ero1 activity has been implicated in increasing cell longevity and resistance against further ER stress [70–72]. If oxidative stress persists, CHOP-induced Ero1α seems to play an important role in inositol 1,4,5-triphosphate receptor mediated Ca2+ release from the ER and subsequent induction of apoptosis [73•]. Indeed, there is evidence that enzymes to counter oxidative stress are induced in response to high secretory loads [74;75]. Additionally, Ero1 function can be directly controlled by regulatory disulfide bonds, which form upon oxidative stress in the ER and diminish Ero1 activity [76;77••] in response to decreased levels of its substrate reduced PDI. Interestingly, whereas deletion of the pancreas-specific Ero1β has a significant negative effect on insulin production [78•], even Ero1α/β deleted mice are viable and show surprisingly little affect on IgM production [78]. As IgM molecules are highly disulfide bonded proteins produced at enormous rates, plasma cells may have evolved additional pathways to perform oxidative folding reactions [79•;80•;81] (and reviewed in Chapter XX of this issue).
Recently discovered alternative pathways to disulfide bridge formation in the ER [82••–84] might partially explain how cells reduce oxidative stress by re-using H2O2 directly [85•] or indirectly for the formation of disulfide bonds. In addition to converting H2O2 to H2O, which would help alleviate oxidative stress generated in the ER and other organelles [82;86•;87], recent studies suggest that Prx4 can be reduced by several different PDI family members following its oxidation by H2O2 [82;88••]. This would allow two disulfide bonds to be formed in substrate proteins per molecule of O2, providing a direct link between H2O2 generation and PDI-catalyzed disulfide bond formation and may explain the mild effect of Ero1α/β absence on disulfide bond formation observed in higher eukaryotes [78;82] compared to that observed in yeast [39], which lack a Prx4 homologue. Finally, some oxidation may be GSSG mediated, as GSSG overproduction was observed immediately after treatment of cells with the reducing agent DTT [47]. In this case, some PDIs could act as a conduit in GSSG production [88], which might be sufficient to form disulfide bonds in proteins that do not depend on the additional functions of the various PDIs or alternatively, GSSG could regulate Ero1α activity, thereby indirectly contributing to disulfide formation.
Summary and outlook
The ER has evolved specific posttranslational modifications and quality control mechanisms to prepare proteins for the extracellular environment. Although these modifications come at a significant cost to the cell, they dramatically enhance the stability of secreted proteins. Indeed, mammalian cells do not show an increase in ER chaperone expression or induction of the unfolded protein response upon heat shock [89]; a treatment which readily disturbs protein folding homeostasis in the cytosol [90;91]. Efforts to monitor completeness of folding and assembly are also centered in the ER, and in many cases utilize the same modifications, highlighting the integrative character of the ER folding and quality control machinery. Much has been discovered about the cellular components and pathways that guide oxidative folding in the ER and cellular mechanisms for dealing with the potentially toxic by products to maintain homeostasis in this essential organelle. However, there is still much to be learned, as our current methods for monitoring protein folding in a cell are largely restricted to the formation of disulfide bonds and changes in sugar processing, both of which are only an indication of the folded state. Our ability to potentially correct the ever-growing group of protein folding diseases will depend on a more complete understanding of oxidative folding in the ER.
Acknowledgments
MJF gratefully acknowledges funding by a Paul Barrett endowed fellowship of St. Jude Children’s Research Hospital and LMH from NIH GM54068. We are grateful to Neil Bulleid, Roberto Sitia, Roger Müller and Moritz Marcinowski for constructive comments on the manuscript. Our apologies go to all our colleagues whose work in the rapidly evolving field of oxidative protein folding in the ER could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Ng DT. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell. 2010;21:1153–1165. doi: 10.1091/mbc.E09-10-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feige MJ, Buchner J. The role of disulfide bonds in protein folding and stability. In: Moroder Luis, Buchner Johannes., editors. Oxidative Folding of Peptides and Proteins. 2010. [Google Scholar]
- 6.Chen W, Helenius J, Braakman I, Helenius A. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters T, Jr, Davidson LK. The biosynthesis of rat serum albumin. In vivo studies on the formation of the disulfide bonds. J Biol Chem. 1982;257:8847–8853. [PubMed] [Google Scholar]
- 8.Braakman I, Hoover-Litty H, Wagner KR, Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergman LW, Kuehl WM. Formation of an intrachain disulfide bond on nascent immunoglobulin light chains. J Biol Chem. 1979;254:8869–8876. [PubMed] [Google Scholar]
- 10.Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 11.Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •13.Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. The CH1 domain of IgG antibodies is shown to be unfolded in isolation and to fold only upon association with the light chain CL domain in a proline-isomerization limited reaction; recognition of the unfolded CH1 domain by BiP provides a mechanism for Ig quality control in the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly NL, Clark RJ, Craik DJ. Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J Biol Chem. 2003;278:6314–6322. doi: 10.1074/jbc.M210492200. [DOI] [PubMed] [Google Scholar]
- 15.Wilken JA, Bedows E. Disulfide bond rearrangement during formation of the chorionic gonadotropin beta-subunit cystine knot in vivo. Biochemistry. 2004;43:5109–5118. doi: 10.1021/bi049856x. [DOI] [PubMed] [Google Scholar]
- 16.Jansens A, van Duijn E, Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- •17.Cortini M, Sitia R. ERp44 and ERGIC-53 synergize in coupling efficiency and fidelity of IgM polymerization and secretion. Traffic. 2010;11:651–659. doi: 10.1111/j.1600-0854.2010.01043.x. This study reveals a mechanism for limiting the size of IgM polymers and provides insights into how unoxidized thiols are monitored and used to retain incompletely assembled molecules. [DOI] [PubMed] [Google Scholar]
- 18.Dul JL, Aviel S, Melnick J, Argon Y. Ig light chains are secreted predominantly as monomers. J Immunol. 1996;157:2969–2975. [PubMed] [Google Scholar]
- 19.Donaldson JG, Williams DB. Intracellular assembly and trafficking of MHC class I molecules. Traffic. 2009;10:1745–1752. doi: 10.1111/j.1600-0854.2009.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitzgen K, Knittler MR, Haas IG. Assembly of immunoglobulin light chains as a prerequisite for secretion. A model for oligomerization-dependent subunit folding. J Biol Chem. 1997;272:3117–3123. doi: 10.1074/jbc.272.5.3117. [DOI] [PubMed] [Google Scholar]
- 21.Hodkinson JP, Jahn TR, Radford SE, Ashcroft AE. HDX-ESI-MS reveals enhanced conformational dynamics of the amyloidogenic protein β2-microglobulin upon release from the MHC-1. J Am Soc Mass Spectrom. 2009;20:278–286. doi: 10.1016/j.jasms.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendershot L, Bole D, Kohler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol. 1987;104:761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng DT, Watowich SS, Lamb RA. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol Biol Cell. 1992;3:143–155. doi: 10.1091/mbc.3.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollocl MR, Richmond MH. Low cyst(e)ine content of bacterial extracellular proteins: its possible physiological significance. Nature. 1962;194:446–449. doi: 10.1038/194446a0. [DOI] [PubMed] [Google Scholar]
- 26.Fahey RC, Hunt JS, Windham GC. On the cysteine and cystine content of proteins. Differences between intracellular and extracellular proteins. J Mol Evol. 1977;10:155–160. doi: 10.1007/BF01751808. [DOI] [PubMed] [Google Scholar]
- 27.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 28.Margittai E, Banhegyi G. Oxidative folding in the endoplasmic reticulum: towards a multiple oxidant hypothesis? FEBS Lett. 2010;584:2995–2998. doi: 10.1016/j.febslet.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 29.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J Biol Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 30.Tu BP, Weissman JS. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 31.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Kozlov G, Maattanen P, Thomas DY, Gehring K. A structural overview of the PDI family of proteins. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- 34.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 36.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 39.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 40.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 41.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- ••42.Inaba K, Masui S, Iida H, Vavassori S, Sitia R, Suzuki M. Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 2010 doi: 10.1038/emboj.2010.222. Comparison of structures for the hyperactive and inactive forms of Ero1α revealed that its activity is regulated by proper positioning of regulatory cysteines within a flexible loop and by fine-tuning the electron shuffle ability of this protein through disulfide rearrangements and how Ero1 can interact with PDI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozlov G, Maattanen P, Schrag JD, Pollock S, Cygler M, Nagar B, Thomas DY, Gehring K. Crystal structure of the bb′ domains of the protein disulfide isomerase ERp57. Structure. 2006;14:1331–1339. doi: 10.1016/j.str.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov G, Maattanen P, Schrag JD, Hura GL, Gabrielli L, Cygler M, Thomas DY, Gehring K. Structure of the noncatalytic domains and global fold of the protein disulfide isomerase ERp72. Structure. 2009;17:651–659. doi: 10.1016/j.str.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Jessop CE, Chakravarthi S, Garbi N, Hammerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••46.Jessop CE, Tavender TJ, Watkins RH, Chambers JE, Bulleid NJ. Substrate specificity of the oxidoreductase ERp57 is determined primarily by its interaction with calnexin and calreticulin. J Biol Chem. 2009;284:2194–2202. doi: 10.1074/jbc.M808054200. This paper provided evidence that substrate specificity of some PDI family members is determined by their ability to form complexes with particular chaperone systems and not by their direct ability to interact with substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47.Appenzeller-Herzog C, Riemer J, Zito E, Chin KT, Ron D, Spiess M, Ellgaard L. Disulphide production by Ero1α-PDI relay is rapid and effectively regulated. EMBO J. 2010 doi: 10.1038/emboj.2010.203. The study shows that re-establishment of redox homeostasis in the ER is a rapid process after reductive stress of the cells and is partly, but not completely, mediated by Ero1 and PDI suggesting alternative pathways involved in ER redox reactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiura Y, Araki K, Iemura S, Natsume T, Hoseki J, Nagata K. Novel thioredoxin-related transmembrane protein TMX4 has reductase activity. J Biol Chem. 2010;285:7135–7142. doi: 10.1074/jbc.M109.082545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers JE, Tavender TJ, Oka OB, Warwood S, Knight D, Bulleid NJ. The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1α. J Biol Chem. 2010;285:29200–29207. doi: 10.1074/jbc.M110.156596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Lith M, Hartigan N, Hatch J, Benham AM. PDILT, a divergent testis-specific protein disulfide isomerase with a non-classical SXXC motif that engages in disulfide-dependent interactions in the endoplasmic reticulum. J Biol Chem. 2005;280:1376–1383. doi: 10.1074/jbc.M408651200. [DOI] [PubMed] [Google Scholar]
- 51.Desilva MG, Lu J, Donadel G, Modi WS, Xie H, Notkins AL, Lan MS. Characterization and chromosomal localization of a new protein disulfide isomerase, PDIp, highly expressed in human pancreas. DNA Cell Biol. 1996;15:9–16. doi: 10.1089/dna.1996.15.9. [DOI] [PubMed] [Google Scholar]
- ••52.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. The identification of this PDI family member with both BiP binding and reductase activity provided a mechanism for reducing partially oxidized, incompletely folded proteins so they could be retrotranslocated to the cytosol for degradation. [DOI] [PubMed] [Google Scholar]
- 53.Oliver JD, Roderick HL, Llewellyn DH, High S. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol Biol Cell. 1999;10:2573–2582. doi: 10.1091/mbc.10.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- 55.Labunskyy VM, Yoo MH, Hatfield DL, Gladyshev VN. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry. 2009;48:8458–8465. doi: 10.1021/bi900717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •56.Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. This study demonstrates that one way to target PDI family members to substrates is through their direct interaction with specific molecular chaperones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •57.Rutkevich LA, Cohen-Doyle MF, Brockmeier U, Williams DB. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell. 2010;21:3093–3105. doi: 10.1091/mbc.E10-04-0356. The impact of the depletion of different ER PDIs on a variety of proteins is tested; PDI is found to have the broadest substrate range and its depletion to have the most significant effect on oxidative folding in the ER yet overlapping substrate range and compensatory finctions are reported for the different PDIs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Hebert DN, Bernasconi R, Molinari M. ERAD substrates: which way out? Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- •60.Wang L, Wang L, Vavassori S, Li S, Ke H, Anelli T, Degano M, Ronzoni R, Sitia R, Sun F, Wang CC. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 2008;9:642–647. doi: 10.1038/embor.2008.88. The crystal structure of ERp44 provides insight into its function and regulation by its C-terminal tail, which folds back onto ERp44 to prevent nonspecific binding of hydrophobic substrates to ERp44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sitia R, Neuberger M, Alberini C, Bet P, Fra A, Valetti C, Williams G, Milstein C. Developmental regulation of IgM secretion: the role of the carboxy-terminal cysteine. Cell. 1990;60:781–790. doi: 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 62.Alberini CM, Bet P, Milstein C, Sitia R. Secretion of immunoglobulin M assembly intermediates in the presence of reducing agents. Nature. 1990;347:485–487. doi: 10.1038/347485a0. [DOI] [PubMed] [Google Scholar]
- 63.Valetti C, Sitia R. The differential effects of dithiothreitol and 2-mercaptoethanol on the secretion of partially and completely assembled immunoglobulins suggest that thiol-mediated retention does not take place in or beyond the Golgi. Mol Biol Cell. 1994;5:1311–1324. doi: 10.1091/mbc.5.12.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mariappan M, Radhakrishnan K, Dierks T, Schmidt B, von Figura K. ERp44 mediates a thiol-independent retention of formylglycine-generating enzyme in the endoplasmic reticulum. J Biol Chem. 2008;283:6375–6383. doi: 10.1074/jbc.M709171200. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Lam KS, Yau MH, Xu A. Post-translational modifications of adiponectin: mechanisms and functional implications. Biochem J. 2008;409:623–633. doi: 10.1042/BJ20071492. [DOI] [PubMed] [Google Scholar]
- 67.Park SW, Zhen G, Verhaeghe C, Nakagami Y, Nguyenvu LT, Barczak AJ, Killeen N, Erle DJ. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci USA. 2009;106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freedman RB, Klappa P, Ruddock LW. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klappa P, Ruddock LW, Darby NJ, Freedman RB. The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 71.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blais JD, Chin KT, Zito E, Zhang Y, Heldman N, Harding HP, Fass D, Thorpe C, Ron D. A small molecule inhibitor of endoplasmic reticulum oxidation 1 (ERO1) with selectively reversible thiol reactivity. J Biol Chem. 2010;285:20993–21003. doi: 10.1074/jbc.M110.126599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •73.Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, Tabas I. Role of ERO1-α-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186:783–792. doi: 10.1083/jcb.200904060. CHOP-induced Ero1α is shown to play a role in apoptosis upon ER-stress via its effect on IP3R regulated calcium release from the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimizu Y, Hendershot LM. Oxidative folding: cellular strategies for dealing with the resultant equimolar production of reactive oxygen species. Antioxid Redox Signal. 2009;11:2317–2331. doi: 10.1089/ars.2009.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertolotti M, Yim SH, Garcia-Manteiga JM, Masciarelli S, Kim YJ, Kang MH, Iuchi Y, Fujii J, Vene R, Rubartelli A, Rhee SG, Sitia R. B- to plasma-cell terminal differentiation entails oxidative stress and profound reshaping of the antioxidant responses. Antioxid Redox Signal. 2010;13:1133–1144. doi: 10.1089/ars.2009.3079. [DOI] [PubMed] [Google Scholar]
- 76.Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- ••77.Appenzeller-Herzog C, Riemer J, Christensen B, Sorensen ES, Ellgaard L. A novel disulphide switch mechanism in Ero1α balances ER oxidation in human cells. EMBO J. 2008;27:2977–2987. doi: 10.1038/emboj.2008.202. Mutational analysis revealed a feedback loop allowing human Ero1 activity to be linked to the availability of its substrate reduced PDI, providing a mechanism to prevent hyperoxidation of the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •78.Zito E, Chin KT, Blais J, Harding HP, Ron D. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol. 2010;188:821–832. doi: 10.1083/jcb.200911086. Ero1β deletion is shown to compromise the oxidative folding of proinsulin whereas even double knockouts of Ero1α and β are able to produce immunoglobulins suggesting novel pathways to oxidative folding in the ER and a role for Ero1 in insulin biogenesis and toxicity of insulin mutants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •79.Shimizu Y, Meunier L, Hendershot LM. pERp1 is significantly up-regulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. Proc Natl Acad Sci USA. 2009;106:17013–17018. doi: 10.1073/pnas.0811591106. This study describes a lymphoid specific protein with an unusual structure that plays a role in the oxidative folding of individual immunoglobulin heavy chain domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •80.van Anken E, Pena F, Hafkemeijer N, Christis C, Romijn EP, Grauschopf U, Oorschot VM, Pertel T, Engels S, Ora A, Lastun V, Glockshuber R, Klumperman J, Heck AJ, Luban J, Braakman I. Efficient IgM assembly and secretion require the plasma cell induced endoplasmic reticulum protein pERp1. Proc Natl Acad Sci USA. 2009;106:17019–17024. doi: 10.1073/pnas.0903036106. In this study, the same protein described above was shown to contribute to the assembly of immunogloblin heavy and light chains, which is required for their assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- ••82.Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum localized peroxiredoxin. Mol Cell. in press. The demonstration that Prx4 can catalyze disulfide bond formation, in addition to reducing H2O2 to H2O, provides both an explanation for the rather mild phenotype observed in Ero1α/β mice compared to that seen in Ero1 deficient yeast as well as an additional pathway to disulfide bond formation in the ER. [Google Scholar]
- 83.Kodali VK, Thorpe C. Oxidative protein folding and the quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid Redox Signal. 2010;13:1217–1230. doi: 10.1089/ars.2010.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •85.Karala AR, Lappi AK, Saaranen MJ, Ruddock LW. Efficient peroxide-mediated oxidative refolding of a protein at physiological pH and implications for oxidative folding in the endoplasmic reticulum. Antioxid Redox Signal. 2009;11:963–970. doi: 10.1089/ars.2008.2326. Hydrogen peroxide in modest amounts is shown to productively contribute to the oxidative folding of proteins. [DOI] [PubMed] [Google Scholar]
- 86•.Tavender TJ, Bulleid NJ. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J Cell Sci. 2010 doi: 10.1242/jcs.067843. Mammalian Peroxiredoxin IV is demonstrated to metabolize Ero1-derived hydrogen peroxide and protect cells from oxidative stress. Its oxidation status can serve as a marker for ER-stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••88.Tavender TJ, Springate JF, Bulleid NJ. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. doi: 10.1038/emboj.2010.273. in press. Prx4 is shown to productively re-use H2O2 to oxidize different PDIs to various extents thus contributing indirectly to disulfide bond formation in substrate proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finka A, Mattoo RU, Goloubinoff P. Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperones. 2010 doi: 10.1007/s12192-010-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.OkudaShimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]