Abstract
Anthropogenic climate change may threaten many species with extinction. However, species at risk today survived global climate change in recent geological history. Describing how habitat tracking and adaptation allowed species to survive warming since the end of the Pleistocene can indicate the relative importance of dispersal and natural selection during climate change. By taking this historical perspective, we can identify how contemporary climate change could interfere with these mechanisms and threaten the most vulnerable species. We focused on a group of closely related plant species in the genus Dodecatheon (Primulaceae) in eastern North America. Two rare species (Dodecatheon amethystinum and Dodecatheon frenchii) that are endemic to patchy cool cliffs may be glacial relicts whose ranges constricted following the last glacial maximum. Alternatively, these species may be extreme ecotypes of a single widespread species (Dodecatheon meadia) that quickly adapted to microclimatic differences among habitats. We test support for these alternative scenarios by combining ecophysiological and population genetic data at a regional scale. An important ecophysiological trait distinguishes rare species from D. meadia, but only a few northern populations of D. amethystinum are genetically distinctive. These relict populations indicate that habitat tracking did occur with historical climate change. However, relatively stronger evidence for isolation by distance and admixture suggests that local adaptation and genetic introgression have been at least as important. The complex response of Dodecatheon to historical climate change suggests that contemporary conservation efforts should accommodate evolutionary processes, in some cases by restoring genetic connectivity between ecologically differentiated populations.
Keywords: phylogeography, specific leaf area
By many estimates, life on earth today faces an extinction event as catastrophic as the Big Five documented in the fossil record (1). Each previous mass extinction involved sudden global environmental changes (2). Anthropogenic global climate change (AGCC) may pose such a threat to biodiversity today. The effects of AGCC on biodiversity are ubiquitous. Around the world, species have shifted their ranges, changed their behavior, and some have gone extinct (3). The scope of these changes is alarming, but it is far from unprecedented. During the Pleistocene-to-Holocene transition, global temperature increased by 7 °C (4). The fossil record from this period documents diverse responses (5) but relatively few extinctions (6). If species survived rapid climate change in the past, can they survive AGCC, or does AGCC threaten them differently? Answering this question first requires describing how species responded to historical climate change. Then, by contrasting historical climate change with contemporary climate change, we can identify why some species that survived in the past may face extinction in the near future.
In general, climate change threatens species by disrupting the match between their adaptive traits and their environments. Three kinds of responses can restore this match: habitat tracking, behavioral shifts/phenotypic plasticity, and adaptation. Inferring the relative roles of these responses during climate change is complicated by variation in data quality. Evidence for habitat tracking is strongest. Fossil pollen assemblages clearly show latitudinal shifts in plant species ranges since the last glacial maximum (5), and contemporary resurveys of species’ distributions often find evidence for rapid altitudinal habitat tracking (3). Indonesian moths (7) and Californian plants (8) have moved more than 60 m upslope during the last half-century. The data used to infer habitat tracking are relatively precise and straightforward to collect. However, physiological traits that mediate adaptation to climate are seldom preserved in fossils and can be difficult to measure through time. Changes in these traits due to behavior and plasticity can occur relatively quickly, within single generations. Several recent examples illustrate roles for behavior and plasticity during responses to contemporary climate change. As northeastern North America has warmed during the last 150 y, some groups of plants flower earlier, and those that do not have declined in abundance (9). In very different systems, complex links between climate, phenology, and growth rates have resulted in smaller sheep in the North Atlantic (10) and larger populations of marmots in the Rocky Mountains (11). In contrast to plasticity, adaptation occurs across generations and is the least well-documented response. Some have argued that adaptation occurs too slowly to contribute (3, 12). However, adaptation may be the only possible response for many species at geographic or ecological boundaries. Without clear data on historical adaptation to climate, the assumption that adaptation plays a relatively insignificant role during responses to rapid climate change has yet to be adequately tested.
As an alternative to other data sources, the contemporary distribution of genetic variation may indicate the relative roles of different responses to historical climate change among closely related species. Many species inhabit regions that experienced historical climate change and exhibit contemporary microclimate heterogeneity. If microclimate variation along local gradients exceeds the fitness-buffering capacity of plasticity and behavioral shifts for a particular group of species, then the contemporary match between adaptive traits and local habitats must reflect historical habitat tracking or ongoing adaptation. These two responses are not mutually exclusive (12). However, they involve biological mechanisms that operate at different spatial scales and on different levels of biological organization. With habitat tracking, the match between traits and environments results from the regional process of dispersal and ecological sorting of species (13). With adaptation, the match results from local natural selection among alternative alleles (14). Just as these mechanisms differ, so do their consequences for the evolution and biogeography of specific lineages. If habitat tracking predominates, species’ geographic distributions change more quickly than do their traits. If adaptation predominates, traits may change in situ, without range shifts. These complementary landscape and lineage perspectives highlight how alternative responses to climate change may generate different relationships between genetic variation, trait variation, and geographic distributions. In this study, we integrate population genetic and ecophysiological data at a regional scale to test the relative roles of different responses to historical climate change in a temperate species complex.
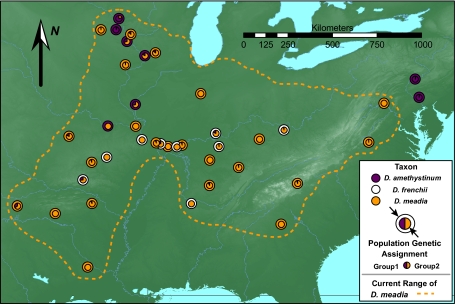
Shooting Stars (Primulaceae, Dodecatheon) are perennial herbaceous plants that occupy diverse habitats across North America. Their vegetative morphology is relatively simple, consisting of a basal rosette of leaves (15). Though the taxonomy of this group is notoriously complex, the most recent revision (16) recognizes three species in eastern North America. Two species, Dodecatheon frenchii (Vasey) Rydb. and Dodecatheon amethystinum (Fassett) Fassett, are rare moist-cliff endemics. Like their habitat, these species have patchy distributions. The third species, Dodecatheon meadia L., is widespread and occupies diverse habitats from woodlands to rock outcrops. The range of D. meadia almost completely encompasses the ranges of both rare species (Fig. 1), and where they cooccur in the upper midwestern and southcentral United States, populations often grow within the foraging range of shared pollinators (17). All three species produce small seeds in woody capsules on short stalks with no obvious adaptation for long-distance dispersal by wind, water, or animals (18).
Fig. 1.
Distribution of samples, taxa, and inferred genetic groups. Outer circles at sampling localities represent taxonomic determinations. Inner circles at sampling localities show the average proportion within each population assigned to two genetic groups across 20 independent Bayesian clustering analyses of 1,182 AFLPs. Samples of rare taxa represent most of their distributions. Dashed line represents the approximate extent of the distribution of D. meadia.
The rare and widespread species differ with respect to an ecologically important morphological trait. D. meadia has thick leaves, whereas both moist-cliff endemics have much thinner leaves (15). Leaf thickness mediates a tradeoff between light capture and water loss that is important for performance among habitats that differ in water stress (19). Reciprocal transplant studies between D. frenchii and D. meadia indicated that their leaf thickness difference may have a genetic basis (20). Ultimately, the colony of D. meadia that had been transplanted into the habitat of D. frenchii went extinct, suggesting divergent local adaptation (21). Another reciprocal transplant experiment between glade and forest subspecies of D. meadia also demonstrated local adaptation to microclimate (22). These findings suggest that leaf thickness may contribute to ecological sorting among species and local adaptation within species to contemporary microclimate heterogeneity.
Two scenarios have been proposed to explain how traits match patchy microclimates in a group with no obvious long-distance dispersal mechanism. First, the rare species may be glacial relicts that were widespread during cooler conditions in the Pleistocene (23). When recent climate warming increased water stress, the species did not adapt and became restricted to isolated microclimate refuges, whereas D. meadia colonized intervening habitats. As relicts with limited adaptive potential, rare species would be conservation priorities under AGCC. Alternatively, the rare species may be convergent ecotypes of one highly polymorphic, quickly adapting lineage (15). As ecotypes, rare species may not merit species recognition, and the entire group may show considerable potential to adapt to future climate change.
These two scenarios highlight different historical roles for habitat tracking and adaptation, and make distinct predictions for the contemporary distribution of genetic variation within lineages and across the landscape. If rare species are relicts and their ecological differences evolved before historical climate change, then much of the genetic variation in the entire group should occur between these long-isolated species. Conversely, if rare species are recently evolved ecotypes, then this lineage should show little hierarchical genetic structure. Likewise, if ecological sorting and migration predominated during historical warming, then ecophysiologically similar populations should share genetic variation regardless of their geographic proximity. However, if divergent natural selection on causative genes maintains the match between traits and environments despite introgression between different ecotypes, then nearby populations could share more neutral genetic variation regardless of their ecophysiological traits. In this study, we evaluate support for these alternative scenarios by testing (i) the degree of hierarchical structure in genetic variation and (ii) the relative strength of the correlation between population genetic distance and ecophysiological difference vs. geographic distance.
Results and Discussion
Evidence for Historical Responses to Climate Change.
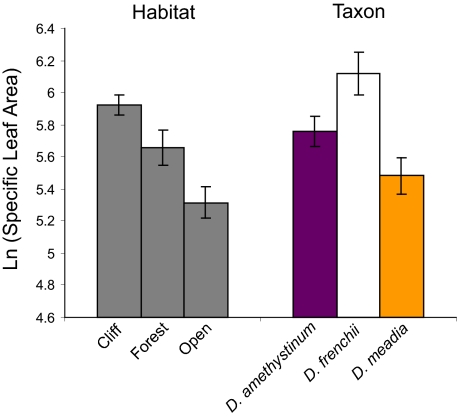
At a regional scale, variation in an important ecophysiological trait-matched microclimatic variation among habitats in eastern North American Dodecatheon (Fig. 2). Specific leaf area (SLA), which is strongly correlated with leaf thickness across plants (24), was higher among populations in more sheltered environments (mixed-model ANOVA, numDF = 2, denDF = 37, F = 19.38, P < 0.001). Plants growing near cliffs had the highest SLA, followed by plants growing in forests and those growing in open habitats. Correspondingly, both cliff endemic taxa had leaves with higher SLA than D. meadia (mixed-model ANOVA, numDF = 2, denDF = 37, F = 18.30, P < 0.001).
Fig. 2.
Specific leaf area (log transformed) among eastern North American Dodecatheon by habitat type and by taxon. Error bars represent SEs as inferred from mixed-model ANOVA.
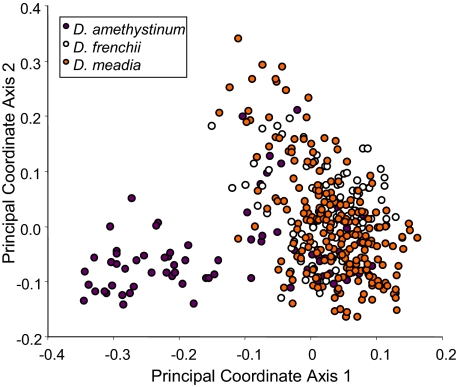
Although rare taxa have similar SLA and occur in similar habitats, the hierarchical structure of genetic variation within the lineage suggests that different historical scenarios explain their origins. The overall amount of cpDNA variation in the sample was low, with only 10 haplotypes occurring among all 400 samples (Table S1). Dodecatheon amethystinum showed weak genetic divergence from D. meadia (Fig. S1, clade 1-1, χ2 = 0.67, simulated P < 0.01), whereas D. frenchii showed no significant genetic differentiation. Low polymorphism and weak differentiation are often encountered in phylogeographic studies based on cpDNA due to slow substitution rates for this marker (25). Genome-wide dominant markers [amplified fragment-length polymorphism markers (AFLPs)], which are generally more sensitive to evolutionary divergence in closely related groups, provided better resolution. Among 383 plants scored at all 1,182 AFLP loci, 1,110 of these loci were polymorphic, and the estimated error rate was only 4.6%. An AMOVA detected significant AFLP variation among taxa (Table 1). However, these differences accounted for only 2.64% of the variation. Based on a Bayesian clustering analysis, the best description of genetic structure was provided by two groups rather than three or more groups (ΔK2 = 53.01, ΔK3 = 2.16, ΔK4 = 0.81, ΔK5 = 0.61). One of these groups consisted of four disjunct populations of D. amethystinum, including the population with distinct cpDNA haplotypes; the other group included individuals from all other populations (Fig. 1). This characterization of genetic structure was congruent with phenetic clustering. The first principal coordinate axis distinguished individuals from these D. amethystinum populations from all others, whereas individuals from D. frenchii and D. meadia broadly overlapped in genetic space (Fig. 3). Differences between the two groups identified in the clustering analyses accounted for 10.7% of variation in the dataset (AMOVA, df = 1, sum of squares = 920.9, Va = 9.44, P < 0.001). These analyses indicated that widely disjunct populations of D. amethystinum are characterized by distinctive genetic variation, supporting the hypothesis of a glacial relict origin. In contrast, D. frenchii appears to be an ecotype of widespread D. meadia. These two species show no range-wide genetic distinction at neutral loci.
Table 1.
Analysis of molecular variance among eastern Dodecatheon taxa
| Source | df | Sum of squares | % variation |
| Among taxa | 2 | 947.2 | 2.64 |
| Among populations in taxa | 37 | 9,000.6 | 23.56 |
| Within populations | 347 | 20,575.8 | 73.81 |
All sources of variation are significant at P < 0.0001.
Fig. 3.
Principal coordinate analysis of pairwise Jaccard distances between AFLP profiles for 383 plants.
The spatial distribution of genetic variation across the entire group suggests how different biological mechanisms interacted during the response to historical climate change. Pairs of populations that were nearby tended to be more genetically similar, but pairs of populations more ecologically similar were not [multiple matrix regression, R2 = 0.174, mean pairwise Jaccard distance = 4.16 × 10−8 geographic distance (P < 0.001) − 3.36 × 10−3 mean pairwise difference in ln(SLA) (P = 0.35)]. The observed pattern of isolation by distance indicates spatially restricted gene flow and is consistent with the lack of long-distance dispersal mechanisms in these species. The relative strength of the isolation by distance pattern has an important implication—it suggests that local introgression and adaptation contributed more to the match between traits and habitats than habitat tracking by distinct species.
Roles of Habitat Tracking and Adaptation.
The relative roles of habitat tracking and adaptation appear to have differed between the two rare species. In the case of D. frenchii, its ecophysiological distinction from D. meadia appears to reflect divergent local adaptation. Considering the very low level of genetic differentiation across more than 1,000 polymorphic AFLPs, the loci responsible for adaptation may represent a small portion of the genome. In this respect, these two species are similar to other plant groups that show local adaptation to microclimate despite extensive introgression of neutral polymorphisms (26, 27). Collectively, these two species show strong evidence for local adaptation to microclimate since the last glacial maximum.
In the case of D. amethystinum, habitat tracking may explain why some disjunct populations of this species are distinct from other Dodecatheon, but ongoing interactions with D. meadia may explain why these populations occur where they do. If range dynamics and ecological sorting were solely responsible for the distribution of D. amethystinum, then all populations should be genetically distinct from other Dodecatheon. However, only a northern subset of D. amethystinum populations was different. Other populations that resemble D. amethystinum further south were genetically indistinct from D. meadia, much like populations of the putative ecotype D. frenchii. The latitudinal replacement of the D. amethystinum gene pool is consistent with a hybrid zone that is shifting north as current conditions favor plants from the D. meadia/D. frenchii gene pool. Shifting hybrid zones can produce idiosyncratic patterns and may explain why few other relict species occur in the same two areas where distinct D. amethystinum occurs (28). Though habitat tracking appears to have played some role in the response of D. amethystinum to historical warming, genetic introgression from D. meadia and divergent natural selection appear to have contributed as well, much as they have for D. frenchii.
Conservation Strategies for Dodecatheon.
Our findings provide a historical reference point for evaluating how AGCC could affect eastern North American Dodecatheon. The data support a strong role for ongoing adaptation to microclimate mediated by both natural selection and genetic introgression. Adaptive genes appear to have dispersed among Dodecatheon populations since historical warming. In this regard, the inferred response of Dodecatheon is similar to the predicted response of some other plants. A recent study of an alpine grass suggested that upslope gene flow could promote adaptation to warming (27). For groups such as these, AGCC may pose less of a threat than it does to other groups with less genetic variation and connectivity. However, AGCC differs from historical climate change in two specific ways that could preclude a sustained evolutionary response. First, models predict that the rate of warming during AGCC may exceed that which occurred at the end of the Pleistocene (29). The associated increase in the strength of selection may exceed the ability of perennial plants to evolve fitness traits, especially if these traits are negatively correlated (30). Second, AGCC is occurring across a highly fragmented landscape. Empirical and simulation studies suggest that habitat fragmentation can greatly slow migration in response to climate change (26, 31). For Dodecatheon, any decrease in connectivity could slow the northward spread of warm-adapted alleles. The general pattern of isolation by distance and the latitudinal replacement of gene pools suggest that the spread of adaptive genes may lag behind a change in climate even before habitats became extensively fragmented.
Our findings also provide concrete conservation recommendations for these taxa under climate change. First, we found little evidence that D. frenchii is a distinct evolutionary lineage. Conservation managers may apply limited resources more efficiently by concentrating on taxa that are more likely to merit species recognition. Second, we found strong evidence that introgression has played an important role during these species’ responses to historical climate change. This result suggests that assisting movement of warm-adapted alleles may sustain the potential of this group to adapt to AGCC.
Implications of Evolutionary Responses for Contemporary Conservation.
Research into the effects of climate change on biodiversity often assumes that habitat tracking will predominate (3, 32). However, the complex response to historical climate change reported here joins a growing body of evidence that evolutionary processes are relevant for predicting and mitigating the effects of AGCC (12, 26).
A popular group of methods to predict habitat tracking with AGCC are species distribution models. These models infer species’ climatic tolerance and project future suitable habitats under the assumption that all populations are ecologically exchangeable (33). All three species of Dodecatheon violate this assumption. Both D. frenchii and D. meadia belong to a single lineage but are adapted to different microclimates. Different populations of D. amethystinum have different genetic backgrounds. Other studies have found evidence that genetic structure influences climatic tolerance. Genetically differentiated subgroups of a Mediterranean wasp have different climate niches (34). In groups such as these, species distribution models may be inaccurate because different populations have evolved different tolerances. Emerging phylogenetic comparative methods could describe evolutionary shifts in climate tolerances relative to shared ancestors (9, 35, 36). However, these methods require bifurcating population trees, and they would poorly accommodate the complex processes of introgression and local adaptation evident in Dodecatheon.
A controversial conservation strategy that also assumes habitat tracking is assisted migration. This strategy faces a suite of technical challenges, including predicting suitable habitats and establishing populations in intact communities (37). Despite these challenges, some advocates have suggested that assisted migration may be the only way to preserve slowly dispersing, long-lived species such as Dodecatheon (38). Our findings suggest a complementary strategy: assisted introgression. Introducing individuals or gametes from warm-adapted source areas into genetically isolated populations could restore connectivity and enhance adaptive potential (26). As with other conservation interventions, this approach bears risks, especially outbreeding depression (39) and pathogen spread. Whether assisted introgression or some other approach is the best conservation strategy for a particular species in a particular area, managers and policy makers should not assume that habitat tracking is the only response to climate change. Adaptation played a role in the past that is likely to continue into the future.
Materials and Methods
During Spring 2007 and 2008, populations were sampled from across the ranges of three species (Fig. 1). If the majority of individuals in a population occurred within 2 m of a vertical rock face, the habitat was categorized as cliff. For populations not near cliffs, the habitat was categorized as forest if plants occurred under a continuous tree canopy or open if otherwise. Ten reproductive plants were sampled per population at 3-m intervals along linear transects through population centers. The dataset included 400 plants from 40 populations, with nine populations of D. frenchii, eight populations of D. amethystinum, and 23 populations of D. meadia. All populations of each rare taxon occurred in cliff habitats. Populations of D. meadia occurred in open (11 populations), forest (10 populations), and cliff (two populations) habitats (Table S1). Vouchers are deposited at the Missouri Botanical Garden herbarium.
To quantify ecophysiological trait variation, SLA of the largest undamaged leaf from each individual was measured. SLA is the ratio of fresh leaf area to dry leaf mass and is closely related to leaf thickness (23). Area was measured from a digital photograph of leaves pressed inside a modified picture frame in the field. Leaves were then dried in a plant press and weighed to a precision of 0.1 mg. Differences in log-transformed SLA among taxa and habitats were tested as fixed effects, treating populations as nested random effects with mixed-model ANOVAs, using the package nlme (40) in R version 9.0 (41).
To quantify genetic variation, additional leaf material from each plant was preserved in silica gel and DNA extracted using Viogene plant DNA miniprep kits. Genetic variation was quantified for both cpDNA sequences and genome-wide dominant markers, AFLPs. Preliminary analyses of several cpDNA regions identified sequence polymorphism at psbA–trnHGUG. This intergenic spacer was amplified using the protocol described by Shaw et al. (42) and sequenced at the Washington University Genome Sequencing Center. A haplotype network was reconstructed under statistical parsimony (43). The original reconstruction produced two loops (haplotypes A-B-C-G and haplotypes G-J-C; Fig. S1), indicating homoplasy. Each loop involved both a substitution and length variation at a polynucleotide repeat. Because polynucleotide repeats are prone to homoplasy, loops were broken by allowing multiple changes in those characters (GxE, JxG) (44).
AFLPs were generated using the protocol described by Trybush et al. (45) with four different primer combinations that had previously been used to detect genetic structure among closely related Primula species (46). The primers began with the preselective sequences EcoRI 5′-GAC TGC GTA CCA ATT C XXX and MseI 5′-GAT GAG TCC TGA GTA A XXX, and involved the following 5′ fluorescent dyes: (i) Mse CTC, Eco ACT, 6-Fam; (ii) Mse CTC, Eco AAG HEX; (iii) Mse CAG, Eco ACT, 6-Fam; and (iv) Mse CAG, Eco ACT HEX. Selective amplifications were conducted for each Mse primer in multiplex PCR with both dye-labeled Eco primers. AFLP profiles were generated on an ABI 3130xl Genetic Analyzer. Loci were scored using GeneMapper 3.7 (Applied Biosystems) with the following peak-detection parameters: peak height threshold = 160, bin width = 1.0 bp, peak half-width = 4 points, polynomial degree = 5, window size = 9. To estimate error, one individual was selected from every other population, and a second series of AFLP profiles was generated from a second DNA extraction. All individuals that failed for one or more AFLP primer combinations were excluded from all subsequent analyses.
To quantify the hierarchical structure of genetic variation, taxon-centered and taxon-free approaches were used. For the cpDNA sequence variation, the haplotype network was nested following the rules of Templeton and Sing (47). The association between the frequency of haplotype clades in each population and taxonomy was tested by comparing the observed χ2 statistic against a reference distribution generated through 105 replicates of Monte Carlo simulation as implemented in R version 9.0 (package stats) (48). For the AFLP data, the proportion of variation within populations, among populations, and among taxa was tested with an analysis of molecular variance (AMOVA) implemented in Arlequin version 3.1 (49, 50). To identify genetic groups without reference to taxonomy, two genetic clustering procedures were applied to the AFLP data. The first was a Bayesian approach that assigns individuals to a specified number of groups (=K) based on inferred allele frequency differences (51, 52). The optimal number of genetic groups was identified using the ad hoc statistic ΔK (53) based on 20 independent runs at K = 1–6. All runs were iterated for 104 Markov Chain Monte Carlo generations following a 104 generation burn-in using the admixture model with correlated allele frequencies and population location information as implemented in Structure version 2.3.3. A phenetic clustering approach based on principal coordinates analysis of the pairwise Jaccard distances followed by modal clustering (PCO-MC) was also used. The PCO was conducted in R version 9.0 (package stats). Modal clustering was implemented with PROC MODECLUS (54) in SAS version 9.1 (SAS Institute) using the following parameters: STANDARD, METHOD = 6, CASCADE = 1, and MAXCLUSTERS = 3 (55).
The relative strength of the correlation between population genetic distance and ecophysiological differences vs. geographic distance was tested with multiple matrix regression. Population genetic distances were estimated as the mean pairwise Jaccard distance among individuals. Ecophysiological differences were quantified as the pairwise Euclidean distance in mean ln(SLA) among populations. Geographic distance was calculated in meters using ArcGIS version 9.0 (ESRI). The significance of regression coefficients was tested against 104 permutations of the response matrix as a one-tailed test following the procedure of Legendre et al. (56) as implemented in the R package ecodist (57).
Supplementary Material
Acknowledgments
We gratefully acknowledge the Nature Conservancy, the US Department of Agriculture Forest Service, the Illinois Department of Natural Resources (DNR) and Nature Preserves Commission, the Missouri DNR, the Arkansas Division of Nature Preserves, the Tennessee Division of Nature Preserves, the Kentucky Department of Parks and State Nature Preserves Commission, the Indiana DNR Division of Nature Preserves, the Virginia Department of Conservation of Resources Division of Nature Preserves, the Lancaster County Pennsylvania Department of Parks and Recreation, the Iowa DNR, the Wisconsin DNR, and private landowners for logistical support and permits. T. Lacefield, T. Witsell, T. Draude, L. Klotz, C. Loeffler, B. Wender, and K. Oberle assisted with fieldwork. K. Vijayakumar and K. Oberle contributed to data collection. P. Raven, A. Templeton, T. Knight, P. Hoch, K. Olsen, and two anonymous reviewers provided valuable comments on earlier drafts of the manuscript. Funding was provided by National Science Foundation Grant DEB-0608317, the Division of Biology and Biomedical Sciences at Washington University in St. Louis, and the Graduate Program at the Missouri Botanical Garden.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences reported in this paper have been deposited in the GenBank database (accession nos. HM778141–HM778150).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012302108/-/DCSupplemental.
References
- 1.Wake DB, Vredenburg VT. Colloquium paper: Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Twitchett R. The palaeoclimatology, palaeoecology and palaeoenvironmental analysis of mass extinction events. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;232:190–213. [Google Scholar]
- 3.Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Syst. 2006;37:637–669. [Google Scholar]
- 4.Kim S, et al. High-resolution climate simulation of the last glacial maximum. Clim Dyn. 2007;31:1–16. [Google Scholar]
- 5.Davis MB, Shaw RG. Range shifts and adaptive responses to Quaternary climate change. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- 6.Roy K, Valentine JW, Jablonski D, Kidwell SM. Scales of climatic variability and time averaging in Pleistocene biotas: Implications for ecology and evolution. Trends Ecol Evol. 1996;11:458–463. doi: 10.1016/0169-5347(96)10054-9. [DOI] [PubMed] [Google Scholar]
- 7.Chen IC, et al. Elevation increases in moth assemblages over 42 years on a tropical mountain. Proc Natl Acad Sci USA. 2009;106:1479–1483. doi: 10.1073/pnas.0809320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly AE, Goulden ML. Rapid shifts in plant distribution with recent climate change. Proc Natl Acad Sci USA. 2008;105:11823–11826. doi: 10.1073/pnas.0802891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis CG, Ruhfel B, Primack RB, Miller-Rushing AJ, Davis CC. Phylogenetic patterns of species loss in Thoreau's woods are driven by climate change. Proc Natl Acad Sci USA. 2008;105:17029–17033. doi: 10.1073/pnas.0806446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozgul A, et al. The dynamics of phenotypic change and the shrinking sheep of St. Kilda. Science. 2009;325:464–467. doi: 10.1126/science.1173668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozgul A, et al. Coupled dynamics of body mass and population growth in response to environmental change. Nature. 2010;466:482–485. doi: 10.1038/nature09210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis MB, Shaw RG, Etterson JR. Evolutionary responses to changing climate. Ecology. 2005;86:1704–1714. [Google Scholar]
- 13.Ackerly D. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int J Plant Sci. 2003;164:S165–S184. [Google Scholar]
- 14.Endler JA. Natural Selection in the Wild. Princeton, NJ: Princeton Univ Press; 1986. [Google Scholar]
- 15.Fassett N. Dodecatheon in eastern North America. Am Midl Nat. 1944;31:455–486. [Google Scholar]
- 16.Reveal JL. Flora of North America: North of Mexico, eds Flora of North America Editorial Committee. Vol. 8. New York: Oxford Univ Press; 2009. pp. 268–286. [Google Scholar]
- 17.Macior L. Pollination ecology of Dodecatheon amethystinum (Primulaceae) Bull Torrey Bot Club. 1970;97:150–153. [Google Scholar]
- 18.Oberle B, Esselman EJ. Fruit and seed characters help distinguish southern Illinois Dodecatheon (Primulaceae) species and highlight unusual intergrading populations. Rhodora. 2011 in press. [Google Scholar]
- 19.Westoby M, Falster DS, Moles AT, Vesk PA, Wright IJ. Plant ecological strategies: Some leading dimensions of variation between species. Annu Rev Ecol Syst. 2002;33:125–159. [Google Scholar]
- 20.Voigt J, Swayne J. French's shooting star in southern Illinois. Rhodora. 1955;57:325–332. [Google Scholar]
- 21.Mohlenbrock RH. Alum Cove, Arkansas. Nat Hist. 1987;96:60–62. [Google Scholar]
- 22.Turner B, Quarterman E. Ecology of Dodecatheon meadia L.(Primulaceae) in Tennessee glades and woodland. Ecology. 1968;49:909–915. [Google Scholar]
- 23.Ugent D, Verdun M, Mibb M. The jeweled shooting star (Dodecatheon amethystinum): A postglacial migrant in the Mississippi Valley. Phytologia. 1982;51:323–329. [Google Scholar]
- 24.Vile D, et al. Specific leaf area and dry matter content estimate thickness in laminar leaves. Ann Bot (Lond) 2005;96:1129–1136. doi: 10.1093/aob/mci264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaal B, Hayworth D, Olsen K, Rauscher J, Smith W. Phylogeographic studies in plants: Problems and prospects. Mol Ecol. 1998;7:465–474. [Google Scholar]
- 26.Jump AS, Penuelas J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol Lett. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalo-Turpin H, Hazard L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J Ecol. 2009;97:742–751. [Google Scholar]
- 28.Barton NH. The dynamics of hybrid zones. Heredity. 1979;43:341–359. [Google Scholar]
- 29.Loarie SR, et al. The velocity of climate change. Nature. 2009;462:1052–1055. doi: 10.1038/nature08649. [DOI] [PubMed] [Google Scholar]
- 30.Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- 31.Honnay O, et al. Possible effects of habitat fragmentation and climate change on the range of forest plant species. Ecol Lett. 2002;5:525–530. [Google Scholar]
- 32.Thomas CD, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- 33.Wiens JA, Stralberg D, Jongsomjit D, Howell CA, Snyder MA. Niches, models, and climate change: Assessing the assumptions and uncertainties. Proc Natl Acad Sci USA. 2009;106(Suppl 2):19729–19736. doi: 10.1073/pnas.0901639106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozier JD, Mills NJ. Ecological niche models and coalescent analysis of gene flow support recent allopatric isolation of parasitoid wasp populations in the Mediterranean. PLoS ONE. 2009;4:e5901. doi: 10.1371/journal.pone.0005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans ME, Smith SA, Flynn RS, Donoghue MJ. Climate, niche evolution, and diversification of the “bird-cage” evening primroses (Oenothera, sections Anogra and Kleinia) Am Nat. 2009;173:225–240. doi: 10.1086/595757. [DOI] [PubMed] [Google Scholar]
- 36.Carstens BC, Knowles LL. Shifting distributions and speciation: Species divergence during rapid climate change. Mol Ecol. 2007;16:619–627. doi: 10.1111/j.1365-294X.2006.03167.x. [DOI] [PubMed] [Google Scholar]
- 37.Richardson DM, et al. Multidimensional evaluation of managed relocation. Proc Natl Acad Sci USA. 2009;106:9721–9724. doi: 10.1073/pnas.0902327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLachlan JS, Hellmann JJ, Schwartz MW. A framework for debate of assisted migration in an era of climate change. Conserv Biol. 2007;21:297–302. doi: 10.1111/j.1523-1739.2007.00676.x. [DOI] [PubMed] [Google Scholar]
- 39.Fenster CB, Galloway LF. Inbreeding and outbreeding depression in natural populations of Chamaecrista fasciculata (Fabaceae) Conserv Biol. 2000;14:1406–1412. [Google Scholar]
- 40.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-92. 2009. Available at http://cran.r-project.org/web/packages/nlme/index.html.
- 41.R Development Core Team. Vienna: R Foundation for Statistical Computing; 2010. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 42.Shaw J, et al. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 43.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Templeton AR, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF. Cladistic structure within the human Lipoprotein lipase gene and its implications for phenotypic association studies. Genetics. 2000;156:1259–1275. doi: 10.1093/genetics/156.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trybush S, et al. Getting the most out of fluorescent amplified fragment length polymorphism. Can J Bot. 2006;84:1347–1354. [Google Scholar]
- 46.Kelso S, Beardsley P, Weitemier K. Phylogeny and biogeography of Primula sect. Parryi (Primulaceae) Int J Plant Sci. 2009;170:93–106. [Google Scholar]
- 47.Templeton AR, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics. 1993;134:659–669. doi: 10.1093/genetics/134.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matos JA, Schaal BA. Chloroplast evolution in the Pinus montezumae complex: A coalescent approach to hybridization. Evolution. 2000;54:1218–1233. doi: 10.1111/j.0014-3820.2000.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 49.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 51.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 54.Sarle W, Kuo A. SAS Technical Report P-256. Cary, NC: SAS Inst; 1993. The MODECLUS procedure. [Google Scholar]
- 55.Reeves PA, Richards CM. Distinguishing terminal monophyletic groups from reticulate taxa: Performance of phenetic, tree-based, and network procedures. Syst Biol. 2007;56:302–320. doi: 10.1080/10635150701324225. [DOI] [PubMed] [Google Scholar]
- 56.Legendre P, Lapointe F, Casgrain P. Modeling brain evolution from behavior: A permutational regression approach. Evolution. 1994;48:1487–1499. doi: 10.1111/j.1558-5646.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 57.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22:1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.