Abstract
We show here that the promoters for many of the Escherichia coli ribosomal protein operons are regulated directly by two transcription factors, the small RNA polymerase-binding protein DksA and the nutritional stress-induced nucleotide ppGpp. ppGpp and DksA work together to inhibit transcription initiation from ribosomal protein promoters in vitro and in vivo. The degree of promoter regulation by ppGpp/DksA varies among the r-protein promoters, but some are inhibited almost as much as rRNA promoters. Thus, many r-protein operons are regulated at the level of transcription in addition to their control by the classic translational feedback systems discovered ~30 y ago. We conclude that direct control of r-protein promoters and rRNA promoters by the same signal, ppGpp/DksA, makes a major contribution to the balanced and coordinated synthesis rates of all of the ribosomal components.
Keywords: ribosome synthesis, transcriptional control, stringent response, translational control
Protein synthesis is the major consumer of cellular energy in bacteria. Because the number of ribosomes is the primary determinant of the level of translation, and ribosome synthesis itself is an energy-intensive process, there are mechanisms that prevent over- or underinvestment of cellular resources in ribosome synthesis (1). Both ribosomal RNA (rRNA) and ribosomal protein (r-protein) synthesis rates are thereby tightly regulated in Escherichia coli (for reviews, see refs. 2, 3).
One of the earliest reported examples of regulation of bacterial ribosome synthesis is a stress response referred to as “the stringent response,” in which rRNA transcription is inhibited in cells starved for amino acids or some other nutrients (4). In this response, uncharged tRNAs induce the ribosome-associated RelA and/or SpoT proteins to synthesize ppGpp (5–7). [The term “ppGpp” is used here to describe both the unusual nucleotide guanosine-3′,5′-(bis)pyrophosphate and its pentaphosphate precursor.] ppGpp concentrations change not only after complete starvations but also after less severe shifts in nutritional conditions, coordinating rRNA synthesis with the need for protein synthesis. Shifts to a more favorable nutritional condition result in a decrease in the concentration of ppGpp and a corresponding increase in rRNA promoter activity, whereas shifts to a less favorable condition result in an increase in ppGpp and a corresponding decrease in rRNA transcription (8).
ppGpp binds directly to E. coli RNA polymerase (RNAP) and inhibits transcription from rRNA promoters (9), although the identity of the ppGpp binding site on RNAP remains unclear (10). However, for ppGpp to exert its full effect on transcription, RNAP has to be modified by the small protein, DksA (11, 12). Unlike ppGpp, DksA is present at high concentrations in cells under all conditions that have been examined (11, 13). rRNA transcription initiation is also regulated by the concentration of the first nucleotide in the transcript (8, 14) and by at least one DNA binding factor, the 11.2-kDa Fis protein (15). Together, ppGpp, DksA, the concentration of the first NTP (iNTP), and Fis match ribosome synthesis rates to the availability of nutrients (8, 11, 14–19).
The 55 different r-protein genes are spread throughout the E. coli genome, some in units encoding only one or two proteins, but others in long operons encoding 10 or more proteins (Table S1). Early studies demonstrated that most (or all) r-protein synthesis is regulated by the stringent control system but could not distinguish between direct effects of ppGpp on transcription of r-protein mRNAs and indirect effects of ppGpp on r-protein mRNA levels through its effects on rRNA synthesis (20, 21). A series of elegant studies performed primarily in the 1970s and 1980s demonstrated convincingly that most (or all) ribosomal protein operons are regulated by translational feedback mechanisms (reviewed in ref. 2) in which each operon encodes a single bifunctional r-protein that can bind not only to a high-affinity site on 16S or 23S rRNA but also to a single lower-affinity site on its own mRNA. By inhibiting translation of r-protein operons only when the r-protein concentration exceeds that of rRNA, the translational feedback mechanisms couple r-protein synthesis to rRNA synthesis and could be sufficient to account for the coordination of the synthesis rates of all of the ribosomal components with changes in nutritional conditions (2).
Recent genome-wide expression studies have reported that at least five and as many as 40 r-protein transcripts decrease following amino acid limitation (22, 23). However, as in the earlier studies, conclusions about the mechanistic basis for the decrease in r-protein mRNA levels were constrained by reports that r-protein transcripts had lifetimes of only a few seconds when they were translationally repressed (24, 25).
Each cistron in an r-protein operon has its own potential ribosome binding site, but there is only one binding site on the mRNA for the translational repressor. Thus, an explanation was needed to account for regulation of r-protein synthesis from the entire operon. Previous studies had shown that translation of downstream cistrons in the trp operon was coupled to translation of upstream cistrons (26, 27). Although the mechanism of translational coupling in the trp operon did not involve mRNA binding by a translational repressor, Nomura et al. reasoned that translational coupling might also occur in r-protein operons. They demonstrated that a single repressor r-protein binding event could inhibit expression of an entire r-protein operon, because translation of each cistron in the mRNA was coupled to translation of the target cistron at which the translational repressor acted (28, 29). A general mechanism was envisioned in which inhibition of translation of upstream cistrons in r-protein operons resulted in the formation of mRNA secondary structures that precluded translation of the downstream cistrons.
Translational coupling mechanisms have been demonstrated for a limited number of r-protein operons. In the L11-L1 operon, the translational repressor L1 binds to a site on the mRNA upstream of the L11 cistron. The L1 binding site on the mRNA resembles the L1 binding site on 23S rRNA, and L1 inhibits translation of both proteins by binding to its mRNA target (29). In the L35-L20 operon, which is transcribed from several promoters (thrS, infC P1, infC P2, and rpmI), L20 (encoded by rplT) binds upstream of the L35 coding region, blocks the L35 ribosome binding site, and allows a secondary structure to form downstream that occludes the L20 ribosome binding site, blocking both L35 and L20 translation (30).
In the resulting model for r-protein synthesis regulation, the translational repressor r-proteins bind to their mRNA targets when free rRNA is unavailable, and in conjunction with translational coupling, coordinate the synthesis rates of most (or all) r-proteins with each other and with rRNA (2). An additional mechanism, transcription attenuation, helps coordinate expression of the S10 operon. This operon is transcribed by the rpsJ promoter and encodes 11 r-proteins, but attenuation is still dependent on the action of L4, the same bifunctional r-protein that serves as the operon's translational repressor (31). The elegance of the translational feedback mechanism, its widespread occurrence in bacteria, and its place in the history of molecular biology as one of our earliest and best-understood examples of translational regulation, have resulted in an assumption that this is the primary mechanism controlling r-protein synthesis in E. coli.
Because inactivation of the dksA gene increases the activities of numerous promoters in vivo in addition to those for rRNA, and ppGpp/DksA directly inhibits some of these promoters in vitro (11, 32–34), we decided to examine whether control of transcription initiation by ppGpp/DksA might also play a direct role in the regulation of r-protein synthesis. Here we report that many of the major r-protein promoters are specifically inhibited by ppGpp/DksA in vitro, that the activities of the same r-protein promoters increase in strains lacking dksA, and that ppGpp/DksA-regulated r-protein promoters are not stringently controlled in strains lacking dksA. Thus, many E. coli r-protein operons are regulated by a mechanism that acts at transcription initiation in addition to by the well-established translational control paradigm.
Results
Some r-Protein Promoters Are Inhibited by ppGpp/DksA.
To investigate whether DksA and ppGpp play a direct role in regulating transcription initiation in r-protein operons, we studied 17 promoters responsible for expression of 39 of the 55 r-proteins (as well as some other proteins; e.g., the set also includes the promoters for the α, β, β′, σ70, and ω subunits of RNAP). The rrnB P1 and lacUV5 promoters were included as positive and negative controls, respectively, for inhibition by DksA and ppGpp (Tables S1–S3 for operon structures, promoter fragment endpoints, and promoter sequences).
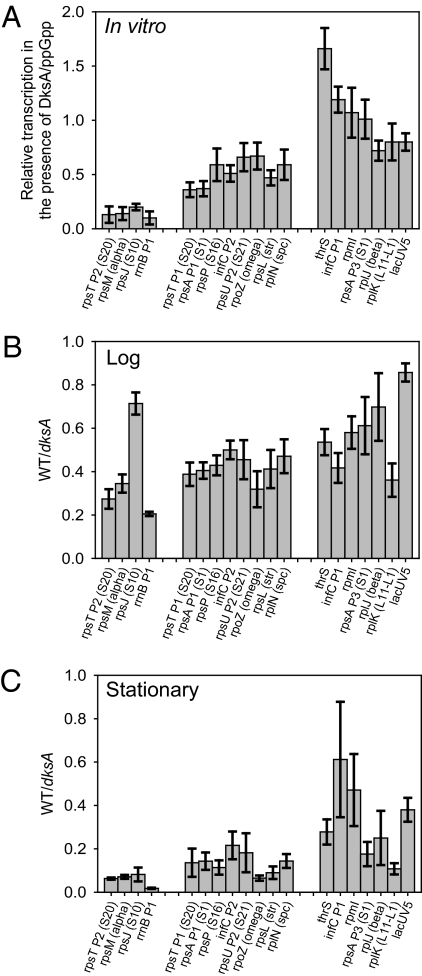
As an initial screen, we performed single-round in vitro transcription reactions in the presence and absence of DksA and ppGpp (Fig. 1A). Of the 17 promoters examined, three promoters, rpsM (which transcribes a five-cistron mRNA encoding four r-proteins and the RNAP α subunit), rpsT P2 (r-protein S20), and rpsJ (S10 operon) were inhibited strongly by ppGpp/DksA (more than fourfold under these reaction conditions), almost as much as the rRNA promoter rrnB P1. Transcription from eight other promoters was inhibited ~twofold, and six promoters were inhibited to the same extent as (or even less than) the promoter used as a control, lacUV5 (Fig. 1A). Based on these results in vitro, the promoters were sorted into three groups for display purposes, with arbitrary boundaries between them: group I for promoters inhibited by ppGpp/DksA at least fourfold in vitro; group II for promoters inhibited 1.5- to fourfold in vitro; and group III for promoters inhibited less than 1.5-fold in vitro.
Fig. 1.
Effects of ppGpp/DksA on transcription from r-protein promoters in vitro and in vivo. (A) Single-round transcription in vitro. Bars represent transcription in the presence of DksA and ppGpp relative to that in the absence of either factor. Error bars represent the SD from at least six reactions. Promoters are generally identified by the first gene in the operon; alternative names are given in parentheses. (B) Expression from promoter-lacZ fusions, log phase (OD600 ~0.3). Bars represent β-galactosidase activity in the WT strain relative to that in the ΔdksA strain. Absolute activities varied slightly from day to day, but the WT:ΔdksA ratio remained constant. Reported error bars represent means and SDs from WT: ΔdksA ratios in multiple experiments. (C) Expression from promoter-lacZ fusions, stationary phase. β-Galactosidase assays were performed on aliquots from the same cultures used in B but 22–24 h after inoculation.
Expression from r-Protein Promoters Inhibited by DksA/ppGpp in Vitro Increases in a ΔdksA Mutant in Vivo.
Next we tested the effects of DksA on r-protein promoters by measuring the β-galactosidase activities of ΔdksA and WT strains containing promoter-lacZ fusions carried on bacteriophage λ prophages in single copy (Figs. 1 B and C). In log phase, effects of the dksA mutation on a control promoter, lacUV5, were minimal (~10%). In contrast, 16 of the 17 test promoters were affected by the dksA mutation more than the lacUV5 promoter [the statistical significance of the ΔdksA effect on the rplJ (β) promoter was uncertain]. In stationary phase (22–24 h after inoculation), there was a ~twofold effect on lacUV5. The loss of dksA had a much larger effect on most of the test promoters. The significance of the effect of the dksA mutation on four of the promoters (thrS, infC P1, rpmI, rplJ) was uncertain (the effect was within error of that on lacUV5). With one exception, the rpsJ (S10) promoter, there was a strong correlation between the promoters affected most by dksA in vitro and those affected in log and stationary phase (Fig. 1 A–C). For reasons that remain unclear, the rpsJ (S10) promoter was strongly affected by the ΔdksA mutation in vitro and in stationary phase, but not in log phase.
Because β-galactosidase is stable, the measurements made in stationary phase reflected the accumulation of this enzyme and not necessarily the promoter activity at the time of assay. In fact, ppGpp concentrations return to basal levels after a few hours in stationary phase (8). However, the effects of the ΔdksA mutation were larger in stationary phase in almost all cases than the effects in log phase, even when considering the general effects on transcription (~twofold) reflected by the activity of the lacUV5 construct in stationary phase. We conclude that the larger effects of the ΔdksA mutation at this time derive from the inability of the cell to turn down transcription at early times in stationary phase, when ppGpp concentrations rise and NTP concentrations fall (8).
Because the promoter-lacZ fusions used in this study do not contain sequences corresponding to the mRNA targets of the translational repressor r-proteins, and because the promoters most strongly affected by the ΔdksA mutation in stationary phase correlate with those inhibited by ppGpp/DksA in vitro [rpsM (α), rpsT P2 (S20), rpsJ (S10)], we conclude that many r-protein promoters are regulated directly by ppGpp/DksA.
Our collection of promoters included those responsible for expression of all of the subunits of the major form of RNAP, α (expressed from the rpsM promoter), β and β′ (rplJ promoter), ω (rpoZ promoter), and σ70 (rpsU P2 promoter). These promoters were affected to different extents by ppGpp/DksA in the three assays (in vitro, log phase, stationary phase). Thus, the data do not support a model in which coordination of expression of the RNAP subunits results from regulation by ppGpp/DksA.
Stringent Control of Synthesis of Some r-Proteins in Response to Amino Acid Starvation Occurs at the Level of Transcription.
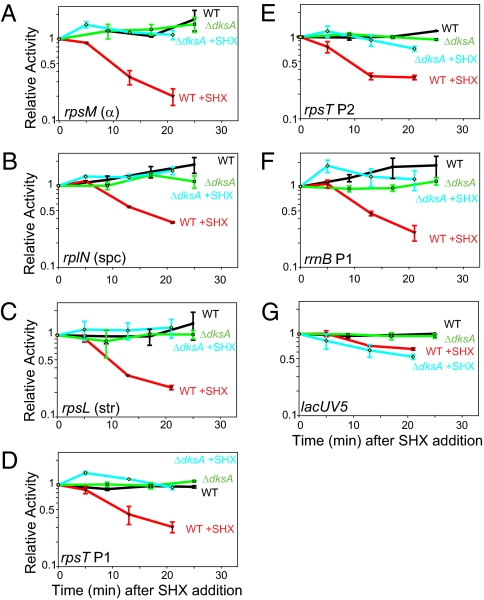
To determine RNA synthesis directly in response to ppGpp induction, we examined transcription by primer extension from seven representative promoters (rpsM, rpsT P2, and rrnB P1 from group I; rplN, rpsL, and rpsT P1 from group II; and lacUV5 from group III) after amino acid starvation in vivo. These measurements detect RNA directly after exposure to high levels of ppGpp (compared with indirectly by measuring β-galactosidase accumulation in the presence of the lower levels of ppGpp present in steady-state growth). After addition of serine hydroxamate (SHX), a serine analog that prevents charging of seryl-tRNAs and thus causes partial serine starvation and an increase in ppGpp levels, transcription from the rrnB P1 promoter and each of the 5 r-protein promoters decreased three- to fivefold in the WT strain (Fig. 2 A–F) but not in the ΔdksA strain. Much less inhibition of the lacUV5 promoter was observed (Fig. 2G), even though growth of that culture, as well as the others, was arrested by SHX. These results are consistent with the model that transcription inhibition of r-protein promoters by ppGpp/DksA is specific and direct.
Fig. 2.
Stringent control of r-protein promoters in WT and ΔdksA mutant strains. The promoters tested in A–G are listed in the lower left of each panel. Transcription was from the same promoter–lacZ fusion constructs used in Fig. 1 B and C, but quantified from direct measurement of RNA by primer extension at different times after addition of serine hydroxamate (SHX) to starve for amino acids (Materials and Methods). Activities are plotted relative to that from the same culture before addition of SHX. Error bars represent range from two independent experiments.
DksA and ppGpp Work Together to Inhibit Transcription.
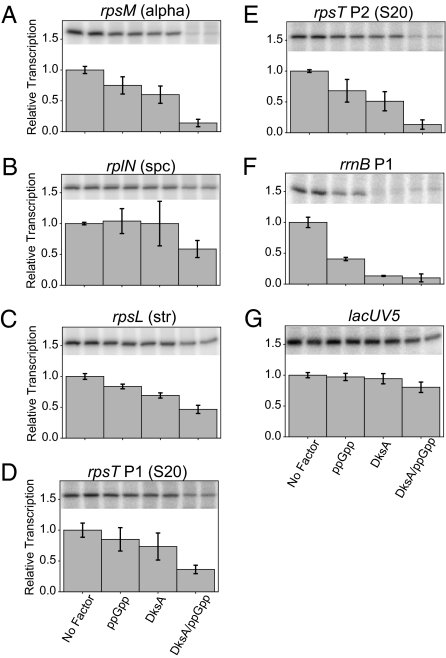
We showed previously that subsaturating concentrations of ppGpp and DksA together inhibit transcription in vitro from rrnB P1 much more strongly than either factor alone (11). We next examined effects of ppGpp and DksA individually and together on r-protein promoters (Fig. 3). DksA (2 μM) and ppGpp (100 μM) individually had relatively small effects on each of the r-protein promoters examined (Fig. 3 A–E), although the effects of ppGpp or DksA alone were significantly greater on each of the r-protein promoters except the rplN (spc) promoter than on the control promoter, lacUV5 (Fig. 3G). At these same concentrations, ppGpp and DksA together more strongly inhibited the r-protein promoters. The promoters most inhibited by dksA in vivo, rpsM (α) and rpsT P2 (S20), as measured by the promoter-lacZ fusions in Figs. 1 B and C, were the most strongly inhibited by ppGpp/DksA in vitro, approaching the degree of inhibition observed with rrnB P1.
Fig. 3.
Independent and combined effects of ppGpp and DksA on r-protein promoters in vitro. The promoters tested in A–G are listed at the top of each panel. Promoter activities were measured in the presence of DksA, ppGpp, both, or neither as indicated under the bottom panels using single-round in vitro transcription assays (Materials and Methods). Representative duplicate lanes of relevant bands from a single gel are shown above the bars in the histograms, but error bars indicate the SD from at least six reactions. The amount of transcription was normalized to that in the absence of either factor.
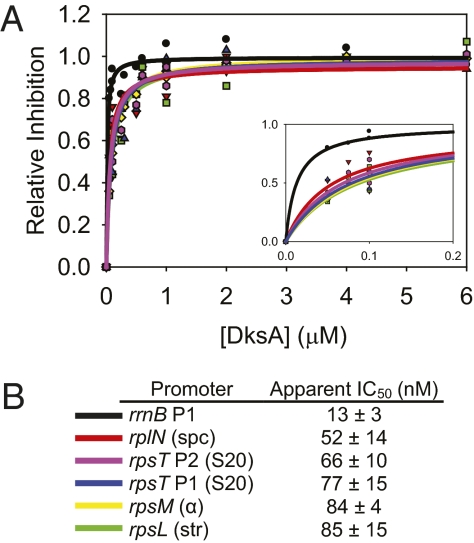
In contrast to its relatively small effects on the r-protein promoters, 2 μM DksA by itself almost eliminated transcription from rrnB P1 (Fig. 3F), masking detection of synergistic effects of the two factors together. These results suggested that the identity of the promoter might influence the concentration of DksA needed for inhibition in the presence of ppGpp. Therefore, as depicted in Fig. 4, we measured the DksA concentration-dependence (low nM to low μM DksA with 100 μM ppGpp) for inhibition of transcription from each of the same promoters examined and depicted in Fig. 3. The IC50 (DksA concentration needed for half-maximal inhibition) ranged from 52 to 85 nM for the five r-protein promoters, values that were within experimental error (Fig. 4). As a result, we cannot conclude whether the heterogeneity in the IC50 values among the different r-protein promoters accounts for the quantitative differences in the degree of inhibition by DksA. However, the difference in IC50 between the r-protein promoters and rrnB P1 (13 nM) is significant and could account for the larger effects of 2 μM DksA on rrnB P1 than on the r-protein promoters in the absence of ppGpp (Fig. 3). Further studies will be needed to understand the relationship between promoter DNA sequence and IC50 by DksA in vitro and whether this contributes to the differential effects of ppGpp/DksA on different promoters in vivo.
Fig. 4.
Ribosomal protein promoter complexes require similar DksA concentrations for transcription inhibition. Half-maximal concentration of DksA required for transcription inhibition in vitro (IC50) was measured at a range of DksA concentrations in the presence of 100 μM ppGpp. The r-protein promoter corresponding to the colored titration curves in A are identified in B, along with the IC50 for each promoter and the SE.
Discussion
The Same Factors Regulate rRNA and r-Protein Synthesis.
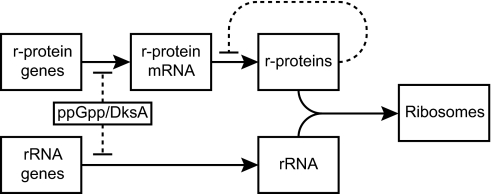
Figure 5 schematically illustrates that multiple r-protein promoters in E. coli are regulated not only by the previously described translational feedback mechanism but also by the same transcription factors that regulate rRNA promoters, ppGpp, and DksA. Using the same factors to regulate the synthesis of multiple components of the ribosome, most of which are needed in equimolar amounts, would seem logical. Regulating expression at the transcription initiation step might also seem logical from the perspective of energy efficiency. However, translation is far more costly than transcription in terms of energy expenditure, and it has been proposed that the production of equimolar quantities of each ribosomal component likely evolved to compensate for the lack of complete cooperativity in ribosome assembly (i.e., to prevent formation of incomplete ribosome particles that could potentially bind to mRNAs and block translation) rather than for efficiency reasons (35).
Fig. 5.
Schematic diagram illustrating transcriptional and translational contributions to regulation of ribosome synthesis in E. coli. Many r-protein promoters and all rRNA promoters are directly regulated by ppGpp/DksA. In addition, r-protein expression is translationally regulated by binding of excess repressor r-proteins to r-protein mRNAs.
It has long been recognized that a translational coupling mechanism would be necessary to account for translational control of all of the members of an r-protein operon, as each cistron appears to have its own ribosome binding site (2). Control of r-protein promoters by ppGpp/DksA would reduce reliance on translational coupling for regulation of all of the members of an r-protein operon [as does the transcriptional attenuation mechanism that contributes to regulation of the rpsJ (S10) operon]. However, the degree of inhibition of r-protein promoters by ppGpp/DksA does not appear to correlate with the number of proteins encoded by the operon. For example, the rpsT P2 promoter transcribes only the mRNA encoding S20, yet this promoter is one of those most strongly regulated by ppGpp/DksA. Conversely, the rplN (spc) promoter transcribes one of the longest of the r-protein operons, but it is not one of those most strongly affected by ppGpp/DksA. Therefore, relief of reliance on translational coupling cannot be the only reason for regulation at the transcription initiation step.
We noticed that some r-protein promoters were affected more by a ΔdksA deletion in vivo than by ppGpp/DksA in vitro or vice versa (for example, compare rplK, rpoZ, and rpsJ in Fig. 1 A and C). Although the mRNA sequence signals needed for translational repression are not present in our constructs, and thus translational feedback could not be responsible for the observed in vivo–in vitro discrepancy, perhaps additional factors affected by the ΔdksA mutation contribute to regulation of some r-protein promoters in vivo.
Sequence Features Responsible for Regulation of r-Protein Operons by ppGpp/DksA.
DksA modifies the conformation of RNAP, interfering with open complex formation and exacerbating the intrinsic instability of the rRNA promoter open complex. It thereby enhances the inhibitory effects of ppGpp in vitro and sensitizes rRNA promoter complexes to changing concentrations of ppGpp in vivo (11, 36). The promoter sequences that contribute to the susceptibility of rRNA promoter complexes to inhibition by ppGpp/DksA are complex. They include (but are not limited to) a G+C-rich “discriminator” sequence just upstream from the transcription start site (especially a C residue on the nontemplate strand two positions downstream from the −10 hexamer) and the absence of an extended −10 sequence. The discriminator sequence in rRNA promoters disfavors a specific interaction with region 1.2 of the σ subunit, and the absence of an extended −10 sequence disfavors an interaction with region 3.0 of σ (37). Although the sequences of the ppGpp/DksA-regulated r-protein promoters (Table S3) do not suggest an absolute requirement for the absence of promoter interactions with σ regions 1.2 and 3.0, most conform to this pattern. No other simple sequence pattern correlates with the degree of r-protein promoter regulation. A better understanding of the mechanism of ppGpp/DksA function will be needed before predictions about regulation can be made from promoter sequence alone.
Comparison with Other Results.
Although regulation of many r-protein operons was examined previously, in some cases it was not determined whether it resulted from effects on transcription, translation, or both. Furthermore, as indicated above, the observation that r-protein mRNAs can be extraordinarily short-lived when they are translationally repressed (25) limited conclusions that could be made about the mechanism(s) responsible for regulation.
There were previous suggestions that the same factors might be capable of regulating both rRNA and r-protein synthesis. For example, an increase in r-protein synthesis was reported when ribosome assembly was inhibited, and this correlated with an increase in rRNA synthesis (38). However, it was not possible to indentify the molecular mechanism responsible.
Some effects on specific promoters reported here are consistent with observations made previously. For example, Lindahl et al. (39) reported that addition of ppGpp to a coupled transcription–translation system reduced incorporation of radioactive nucleotide into r-protein mRNAs generated from a DNA template containing the rpsL (str), rpsJ (S10), rplN (spc), and rpsM (α) operons. However, only very weak, promoter-nonspecific inhibition was observed from the same template after ppGpp addition to transcription reactions containing only purified components. In contrast, we found that two of the four promoters on this template were strongly inhibited by ppGpp when DksA was present. Perhaps the extract used to make the coupled transcription–translation system contained DksA, whereas it was absent in the purified system.
Cole and Nomura (24) concluded that stringent control of L11–L1 synthesis occurred at the translational level, consistent with our observation that the rplK (L11–L1) promoter was affected only very slightly by ppGpp/DksA in vitro. The potential for different r-protein operons to be regulated at different (or multiple) steps in gene expression has also been noted (24, 39), a prediction borne out here.
Finally, it was reported that ppGpp alone directly inhibited the rpsA P1 (S1) promoter in vitro (40) (Fig. 1). However, not all promoters inhibited by ppGpp in that report were regulated by ppGpp in another report (41) or by ppGpp/DksA in our study.
RelA/SpoT homologs are found throughout the bacterial kingdom (5), although the mechanism of ppGpp action on rRNA transcription appears to be indirect in some cases (10, 42). DksA has been studied far less than ppGpp in organisms other than E. coli. Sequence homologs of DksA are present throughout proteobacteria, but they are not obvious in many other species, including some Gram-negative or some Gram-positive thermophiles (10, 42). However, structurally similar proteins could be present but not recognized in sequence comparisons. For example, Gre factors are structurally very similar to DksA in their coiled-coil domain and bind in the RNAP secondary channel like DksA. E. coli GreB can even complement some activities of DksA when at high concentration (13), but it bears little or no sequence resemblance to DksA. Thus, sequence comparisons are insufficient to determine whether regulation of r-protein synthesis by ppGpp/DksA is widespread in nature.
Prospect.
We have demonstrated specific regulation of a large number of r-protein promoters by ppGpp/DksA using purified components in vitro, with reporter constructs at two stages of growth in vivo, and by direct measurement of RNA made from r-protein promoters in cells starved for amino acids. Although the transcriptional and translational mechanisms for control of r-protein synthesis could have evolved to be redundant to ensure that this crucial regulation persists in case one of the systems fails, it would not be surprising if the relative contributions of the two mechanisms to regulation varied with growth stage or nutritional conditions. In this way, the two mechanisms could complement each other like the multiple mechanisms that regulate rRNA promoters (8). For example, one could be more important for rapid responses during severe upshifts and downshifts whereas the other could be more important for fine adjustments during exponential growth or during changes in growth phase. Thus, nuances of regulation of E. coli r-protein synthesis remain to be explored.
Materials and Methods
Strains, Plasmids, and Proteins.
Strains and plasmids used in this work are listed in Table S2. Endpoints of the promoter fragments used to make lacZ fusions are numbered relative to the transcription start site. Promoter fragments were amplified from chromosomal DNA by PCR from VH1000 (14) and cloned into plasmid pMSB1 (43). All except one of the r-protein promoter-lacZ fusions (see below) were then inserted into phage λRS468 using an in vivo recombination method (43, 44), the fusions were introduced by infection into VH1000 (14), an MG1655-derivative lacking lacZ, lysogens were identified on LB plates containing 40 μg/mL X-gal, and single-copy λ lysogens were chosen for analysis. The r-protein promoter-lacZ fusions lack the sequences needed for translational control. Lysogens containing the ΔdksA::tetR allele were obtained by transduction with phage P1vir grown on donor strain RLG8124 (11).
We were unable to clone the rpsJ (S10) promoter fragment into pMSB1. Therefore, a promoter-lacZ fusion was created by ligation of the promoter fragment with phage λ arms followed by packaging in vitro (43, 45). Difficulties in cloning strong promoters into pMSB1 have been noted previously (43).
Plasmids used for in vitro transcription were constructed by amplifying a promoter fragment from the bacterial chromosome by PCR using primers containing EcoRI or HindIII sites. The promoter fragment was then inserted into the HindIII and EcoRI sites of plasmid pRLG770 (15), which contains rrnB T1T2 transcription termination sequences ~150 nt (for T1) or ~300 nt (for T2) downstream of the HindIII site. The previously annotated rpsU P1 promoter was inactive in vitro, and a promoter directly upstream of rplT (46) was inactive in vivo. Therefore, these promoters were not examined further. Promoter fragments used for transcription in vitro generally had endpoints slightly further downstream than those used in vivo to create transcripts that migrated to a clear position in the gel to facilitate quantitation (Table S2).
RNA polymerase holoenzyme and His-tagged DksA were purified as described elsewhere (13).
In Vitro Transcription.
Single-round in vitro transcription assays were performed at 30 °C as described (11). The reactions contained 40 mM Tris·acetate, pH 7.9, 60 mM NaCl, 10mM MgCl2, 1 mM DTT, 1 nM supercoiled plasmid DNA, 10 nM RNAP, 200 μM ATP, CTP ,and GTP, 10 μM UTP, and ~1 μCi [α-32P]-UTP. Heparin (10 μg/mL) was used as a competitor to prevent reinitiation and was added together with the NTPs to start the reactions. DksA (2 μM or the concentration indicated) and/or ppGpp (100 μM; Trilink) were preincubated with RNAP and buffer components for 10 min before addition of NTPs and heparin (where indicated).
β-Galactosidase Assays.
Cells were grown in Mops medium supplemented with glycerol and all 20 amino acids at 37 °C, except the rpsJ-lacZ fusion strain was grown at 30 °C because it contains a phage with a temperature-sensitive λ repressor. β-Galactosidase activity was determined as previously described (29). Error bars in Fig. 1 B and C represent the SD from at least six independent cultures inoculated from colonies and grown for at least three generations to an OD600 of ~0.3. Growth was continued overnight (a total of 22–24 h) for the stationary-phase measurements.
RNA Extraction and Primer Extension.
WT and ΔdksA strains containing the same promoter-lacZ fusions used for measuring r-protein promoter activity in the experiments described in Fig. 1 B and C were grown at 37 °C in Mops medium supplemented with glycerol and all 20 amino acids. At OD600 ~0.3, WT or ΔdksA mutant cultures were each divided into 25-mL samples in prewarmed flasks containing 1 mL either 25 mg/mL serine hydroxamate to induce amino acid starvation (SHX-treated) or water (untreated). Aliquots from the SHX-treated or untreated cultures were removed at the indicated times. RNA extraction (by boiling lysis followed by phenol-chloroform extraction) and primer extension were performed as previously described (47), except for using SuperScript III (Invitrogen) in the buffer supplied by the manufacturer. A primer that annealed near the 5′ end of the lacZ gene (34) was used for all strains.
Supplementary Material
Acknowledgments
We thank Masayasu Nomura for comments on the manuscript. This work was supported by National Institutes of Health Grant R37 GM37048 (to R.L.G.). P.S.-V. and H.L.B. were participants in a program sponsored by the National Science Foundation (Research Experience for Undergraduates) during the course of this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019383108/-/DCSupplemental.
References
- 1.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 2.Nomura M, Gourse RL, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 3.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 4.Sands MK, Roberts RB. The effects of a tryptophan-histidine deficiency in a mutant of Escherichia coli. J Bacteriol. 1952;63:505–511. doi: 10.1128/jb.63.4.505-511.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 6.Xiao H, et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 7.Jiang M, Sullivan SM, Wout PK, Maddock JR. G-protein control of the ribosome-associated stress response protein SpoT. J Bacteriol. 2007;189:6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 9.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 10.Vrentas CE, et al. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004a;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Perederina A, et al. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 13.Rutherford ST, et al. Effects of DksA, GreA, and GreB on transcription initiation: Insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr., Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 15.Ross W, Thompson JF, Newlands JT, Gourse RL. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appleman JA, Ross W, Salomon J, Gourse RL. Activation of Escherichia coli rRNA transcription by FIS during a growth cycle. J Bacteriol. 1998;180:1525–1532. doi: 10.1128/jb.180.6.1525-1532.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gralla JD. Escherichia coli ribosomal RNA transcription: Regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55:973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- 18.Magnusson LU, Farewell A, Nyström T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Dennis PP, Nomura M. Stringent control of ribosomal protein gene expression in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:3819–3823. doi: 10.1073/pnas.71.10.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis PP, Nomura M. Stringent control of the transcriptional activities of ribosomal protein genes in E. coli. Nature. 1975;255:460–465. doi: 10.1038/255460a0. [DOI] [PubMed] [Google Scholar]
- 22.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole JR, Nomura M. Translational regulation is responsible for growth-rate-dependent and stringent control of the synthesis of ribosomal proteins L11 and L1 in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4129–4133. doi: 10.1073/pnas.83.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR, Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986;188:383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- 26.Oppenheim DS, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das A, Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 1984;12:4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yates JL, Nomura M. Feedback regulation of ribosomal protein synthesis in E. coli: Localization of the mRNA target sites for repressor action of ribosomal protein L1. Cell. 1981;24:243–249. doi: 10.1016/0092-8674(81)90520-1. [DOI] [PubMed] [Google Scholar]
- 29.Baughman G, Nomura M. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell. 1983;34:979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- 30.Lesage P, et al. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J Mol Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl L, Archer R, Zengel JM. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983;33:241–248. doi: 10.1016/0092-8674(83)90353-7. [DOI] [PubMed] [Google Scholar]
- 32.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol. 2006;188:5775–5782. doi: 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aberg A, Shingler V, Balsalobre C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol Microbiol. 2006;60:1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- 34.Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodd J, Kolb JM, Nomura M. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie. 1991;73:757–767. doi: 10.1016/0300-9084(91)90055-6. [DOI] [PubMed] [Google Scholar]
- 36.Rutherford ST, Villers CL, Lee J-H, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haugen SP, et al. rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 38.Takebe Y, Miura A, Bedwell DM, Tam M, Nomura M. Increased expression of ribosomal genes during inhibition of ribosome assembly in Escherichia coli. J Mol Biol. 1985;184:23–30. doi: 10.1016/0022-2836(85)90040-3. [DOI] [PubMed] [Google Scholar]
- 39.Lindahl L, Post L, Nomura M. DNA-dependent in vitro synthesis of fibosomal proteins, protein elongation factors, and RNA polymerase subunit α: Inhibition by ppGpp. Cell. 1976;9:439–448. doi: 10.1016/0092-8674(76)90089-1. [DOI] [PubMed] [Google Scholar]
- 40.Kajitani M, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J Biol Chem. 1984;259:1951–1957. [PubMed] [Google Scholar]
- 41.Raghavan A, Kameshwari DB, Chatterji D. The differential effects of guanosine tetraphosphate on open complex formation at the Escherichia coli ribosomal protein promoters rplJ and rpsA P1. Biophys Chem. 1998;75:7–19. doi: 10.1016/s0301-4622(98)00185-9. [DOI] [PubMed] [Google Scholar]
- 42.Krásný L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao L, et al. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 44.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 45.Gourse RL, de Boer HA, Nomura M. DNA determinants of rRNA synthesis in E. coli: Growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 46.Fayat G, et al. Escherichia coli phenylalanyl-tRNA synthetase operon region. Evidence for an attenuation mechanism. Identification of the gene for the ribosomal protein L20. J Mol Biol. 1983;171:239–261. doi: 10.1016/0022-2836(83)90092-x. [DOI] [PubMed] [Google Scholar]
- 47.Ross W, Gourse RL. Analysis of RNA polymerase-promoter complex formation. Methods. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.