Abstract
Deletion of the late cornified envelope (LCE) genes LCE3B and LCE3C has recently been identified as a risk factor for psoriasis. Expression of 16 LCE genes of LCE groups 1, 2, 3, 5, and 6 was examined in vivo and in vitro. Quantitative PCR demonstrated that moderate to high LCE expression was largely confined to skin and a few oropharyngeal tissues. Genes of the LCE3 group demonstrated increased expression in lesional psoriatic epidermis and were induced after superficial injury of normal skin, whereas expression of members of other LCE groups was down-regulated under these conditions. Immunohistochemistry and immunoelectron microscopy demonstrated that LCE2 protein expression was restricted to the uppermost granular layer and the stratum corneum. Stimulation of in vitro reconstructed skin by several psoriasis-associated cytokines resulted in induction of LCE3 members. The data suggest that LCE proteins of groups 1, 2, 5, and 6 are involved in normal skin barrier function, whereas LCE3 genes encode proteins involved in barrier repair after injury or inflammation. These findings may provide clues to the mechanistic role of LCE3B/C deletion in psoriasis.
Psoriasis is a common inflammatory skin disease caused by both genetic and environmental factors.1–3 A substantial number of psoriasis susceptibility loci has been identified that harbor genes involved in adaptive and innate immunity, such as HLA-Cw6, IL12B, IL23R, TNFAIP3, TNIP1, and the β-defensins.2 Recently, a deletion of two members of the late cornified envelope genes, LCE3B and LCE3C, was identified as the first psoriasis risk factor with a putative role in skin barrier function.4 In support of this finding, an independent genome-wide association study of a large Chinese cohort found that single nucleotide polymorphisms in strong linkage disequilibrium with the LCE3B/C deletion are associated with psoriasis.5 This association has been widely replicated in cohorts of European and Asian origin.6,7
The LCE genes are located on chromosome 1q21 in a region called the epidermal differentiation complex.8 This region is enriched for genes expressed during epidermal differentiation including loricrin, involucrin, filaggrin, the small proline-rich protein genes, and the LCE genes.9,10 With 18 members, the LCE gene family is divided into six groups, LCE1 to LCE6, based on similarities of amino acid sequences, genomic organization, and patterns of expression.9
The role of the LCE proteins in epidermal biology has not been extensively studied. In mice, it was observed that members of the Lce1 group are expressed rather late in epithelial development, where they are incorporated into the cornified envelope via cross-linking by transglutaminases.11 Other investigators used quantitative PCR (qPCR) to demonstrate that the human LCE1 and LCE2 genes were primarily expressed in skin, whereas LCE4 and LCE5 gene expression was largely undetectable in the human tissues examined.9 LCE3 gene expression was absent or variable at low levels in internal and external epithelia, although at that time the LCE3B/C deletion was not known, which may have affected these results. Studies in cultured keratinocytes demonstrated that LCE2 expression is induced by Ca++, whereas UVB irradiation induced expression of LCE3E and members of the LCE1 and LCE2 groups.9
In a previous study based on the observed LCE3C induction in lesional psoriatic skin and injured skin, a role for the LCE3 genes in skin barrier repair was hypothesized.4 The present study sought to extend these studies, to find a possible clue to the mechanistic relationship between the LCE3B/C deletion and psoriasis. Expression of 16 LCE genes of the LCE groups 1, 2, 3, 5, and 6 was investigated in vivo and in vitro at the mRNA and the protein level. The results demonstrate that, compared with other LCE groups, members of the LCE3 group are differently expressed, which suggests nonredundant functions that may be relevant for psoriasis.
Material and Methods
Human Tissues
Patients and healthy volunteers (N = 6 for all groups) were recruited by the Dermatology Clinic of Radboud University Nijmegen Medical Centre (Nijmegen, The Netherlands). All diagnoses were made by a dermatologist. The study was approved by the local medical ethical committee, “Commissie Mensgebonden Onderzoek Arnhem-Nijmegen,” and conducted according to the Declaration of Helsinki principles. Written informed consent was obtained. Patients had moderate to severe plaque-type psoriasis. Biopsy specimens were taken from lesional and nonlesional skin (N = 4). For stratum corneum removal by tape stripping in healthy control individuals (N = 5), two areas on the lower back measuring 1 × 2 cm each were tape stripped until the surface glistened after repeated application and removal of adhesive tape (20 to 40 times). At 24 and 48 hours after tape stripping, 3-mm biopsy specimens were taken from the tape-stripped area and from healthy skin for RNA isolation and histologic analysis, respectively. Archival autopsy material from a large number of normal human tissues was obtained from the Department of Pathology (Radboud University Nijmegen Medical Centre).
Three-Dimensional Reconstructed Skin
Reconstructed skin was generated as described previously.12 In brief, de-epidermized human dermis, 0.8 mm thick and 8 mm in diameter, was used as a scaffold for keratinocytes. Constructs were cultured submerged for 3 days in tissue culture inserts in a 24-well plate, and subsequently the medium level was lowered to enable air exposure to induce terminal differentiation. For analysis of LCE expression during normal skin development, the constructs were cultured air-exposed for the indicated times. This cultured epidermis is negative for psoriasis-associated proteins such as cytokeratin 16, elafin, and human β-defensin.12 At this point, a combination of proinflammatory cytokines [10 ng/mL of IL-1α, 5 ng/mL of tumor necrosis factor-α (TNF-α), and 5 ng/mL of IL-6] or Th17 cytokines (100 ng/mL of IL-17 or 100 ng/mL of IL-22) was added for 72 hours (N = 5 per group) to induce cytokine-specific gene expression. Nine healthy donors of human keratinocytes were used.
Real-Time qPCR
Epidermal sheets from skin biopsy tissue or skin constructs were separated from the underlying dermis as described previously.13 The extracted and DNase I–treated RNA from these epidermal sheets was used for first-strand cDNA synthesis using a kit (iScript cDNA synthesis kit; Bio-Rad Laboratories, Inc., Hercules, CA) using 1 μg of RNA as input.14 The reverse transcriptase reaction products were used for qPCR amplification of genes of interest using a quantification system (MyiQ Single-Color Real-Time Detection System; Bio-Rad Laboratories, Inc.) and melting curve analysis as described previously.15 Expression of target genes was normalized to expression of the housekeeping gene human ribosomal phosphoprotein P0 (RPLP0) in the same sample. For analysis of LCE1A, LCE1C, LCE1D and LCE1E, and the complete LCE2 group, primers were used as described previously.9 Primers for LCE1B, LCE1F, LCE3A, LCE3C, LCE3D, LCE3E, LCE5A, and LCE6A were designed, and PCR products were sequenced to check primer specificity. Primers (Biolegio BV, Nijmegen, The Netherlands) for qPCR were accepted only if their efficiency was between 90% and 110%. For LCE3B and LCE4A, none of the designed primer pairs met this criterion, and, therefore, these genes were not analyzed in this study. Primer sequences and efficiency are given in Table 1. Because the LCE primer pairs were not intron-spanning, each primer pair was also tested with input in which during cDNA synthesis no reverse transcriptase had been used. No specific product was found in any of these reactions. Relative mRNA expression levels were calculated using the ΔΔ-Ct method.16 For graphic representation of mRNA data, all in vivo data are plotted relative to LCE1A expression in normal skin (Figures 1, 2A, and 3A) or uninvolved skin (Figure 2B). The in vitro data are plotted relative to expression of the same gene in the control condition (Figure 4).
Table 1.
Primers for qPCR
| Gene | Forward primer | Reverse primer | E⁎ |
|---|---|---|---|
| LCE1A† | 5′-TGCAAGAGTGGCTGAGATGC-3′ | 5′-AGACAACACAGTTGGTGTCAGG-3′ | 2.18 |
| LCE1B | 5′-TGAGCCTAGAAGAACACAA-3′ | 5′-GAAGGAGAAGGAAGACGG-3′ | 1.94 |
| LCE1C† | 5′-GAATCCAGGACCGCAAACTG-3′ | 5′-TGGACCTGTGAGCCTCTCAG-3′ | 2.03 |
| LCE1D&E† | 5′-TGAATAGCTGAGAGGTTCCAGC-3′ | 5′-CAGCCATGGATCTGCAGAAG-3′ | 2.06 |
| LCE1F | 5′-CCACACCGAAGTGCC-3′ | 5′-GCTGCAGCAGGAAGAG-3′ | 1.83 |
| LCE2A† | 5′-GGACCTGTCCCAGAGTGATG-3′ | 5′-GATCCAGGATGGGCTCTTG-3′ | 2.10 |
| LCE2B† | 5′-GGTTGACTAAACTCTGCCAGG-3′ | 5′-CACTGGGGCAGGCATTTA-3′ | 2.04 |
| LCE2C† | 5′-CTTGGGACTGAATGGCCAAG-3′ | 5′-GACTTGCAATTGGGGTGTTAC-3′ | 1.91 |
| LCE2D† | 5′-CTGCAGAAGAGCTCTGGTACTG-3′ | 5′-CTCCATCAAGCACAAAGTTCTG-3′ | 2.15 |
| LCE3A | 5′-GAGTCACCACAGATGCC-3′ | 5′-CTTGCTGACCACTTCCC-3′ | 2.02 |
| LCE3C | 5′-CTGAGCCACCACAGGCACTT-3′ | 5′-TGTTTCCTCCAAAGATCACTTGTC-3′ | 2.03 |
| LCE3D | 5′-CCCCATCTTGATGCATGAG-3′ | 5′-TGTGACATCCTGGACATCAG-3′ | 2.07 |
| LCE3E | 5′-CTGATGCTGAGACAAGCGATCTT-3′ | 5′-GATCCCCCACAGGAAAACCT-3′ | 2.20 |
| LCE5A | 5′-CCCTCTTTATCCTGCCC-3′ | 5′-ACAACACCTAGTTCTCCAA-3′ | 1.87 |
| LCE6A | 5′-AGAAGCAGCAATCTTGGA-3′ | 5′-CCTTTGGGAACTGGAATG-3′ | 2.05 |
Efficiency as fold increase in fluorescence per PCR cycle.
Primer was previously described by Jackson et al.9
Figure 1.
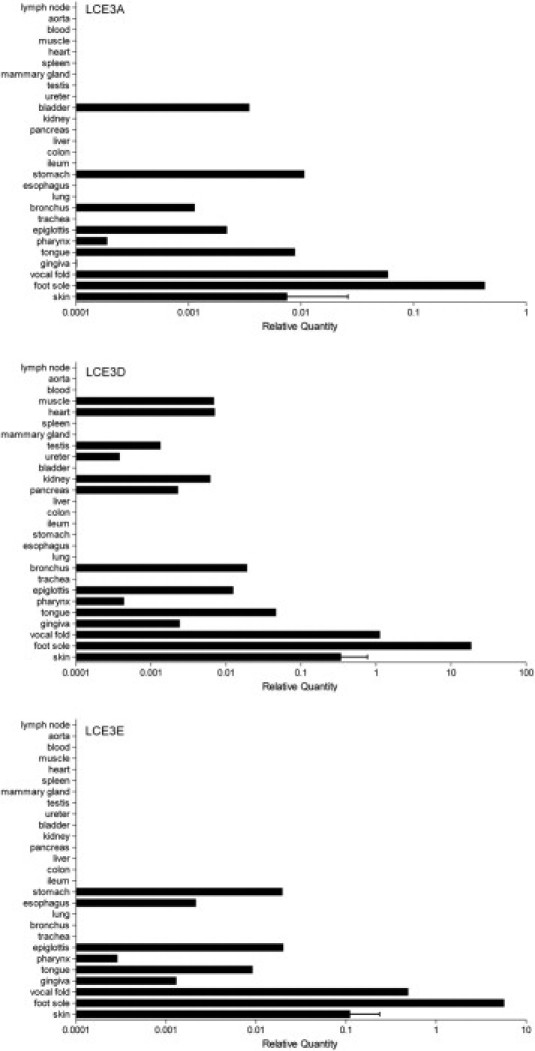
Relative mRNA expression levels of LCE3 genes in normal human tissues. Expression of target genes was normalized to that of RPLP0. For graphic representation, all values are expressed relative to LCE1A in skin, which was set at unity, to enable comparison of expression levels between tissues and genes (see Livak and Schmittgen16). Primer sequences and efficiency of amplification are given in Table 1. Expression of other LCE family members is shown in Supplemental Figure S1 (available at http://ajp.amjpathol.org). Normal human tissues are from a single individual, and data from normal skin are from 12 individuals. Data are given as mean (SD).
Figure 2.
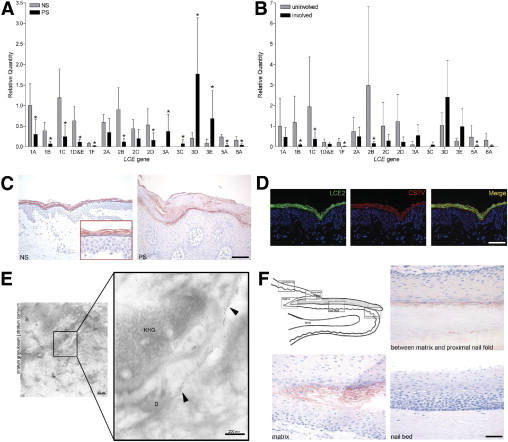
LCE expression in normal and psoriatic skin. Relative LCE gene mRNA expression levels in epidermal sheets from healthy control individuals (NS) and patients with psoriasis (PS) (N = 6) (A) and uninvolved and involved skin from patients with psoriasis (N = 4) (B). All values are expressed relative to LCE1A in normal or uninvolved skin. *P < 0.05 compared with normal or uninvolved skin [mean (SD)]. C: Immunohistochemical staining with the pan-LCE2 antibody on normal skin (NS) and lesional psoriatic skin (PS). Scale bar = 100 μm. D: Co-localization (merged yellow signal) of LCE2 (green) and cathepsin V (red) in the upper granular layer of normal skin. Scale bar = 50 μm. E: Immunoelectron microscopy. LCE2 labels (arrowheads) are associated with the cornified envelope in the stratum granulosum. D, desmosome; KHG, keratohyalin granule. Scale bar = 50 μm. F: Immunohistochemical staining for the LCE2 group in nail. Scale bar = 50 μm.
Figure 3.
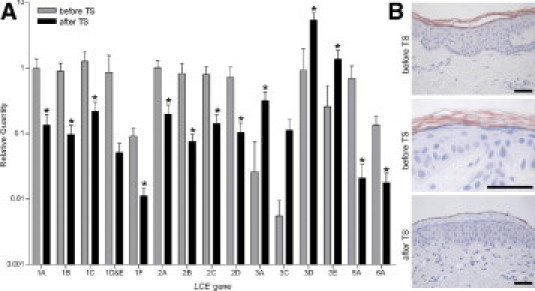
LCE expression before and after tape stripping in normal skin. A: Relative mRNA expression levels of LCE genes in epidermal sheets of healthy control individuals before and after tape stripping (TS) (N = 5). All values were expressed relative to LCE1A in skin to enable comparison of expression levels between genes. *P < 0.05 compared with before tape stripping [mean (SD)]. B: Immunohistochemical staining with the pan-LCE2 antibody on sections of normal skin before tape stripping (upper and middle panels), before enlarged (middle panel), and 48 hours after tape stripping (lower panel). Each illustration is representative of data for five different individuals. Scale bar = 50 μm.
Figure 4.
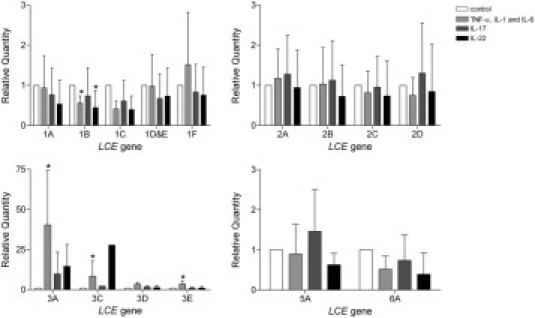
Cytokine-induced LCE mRNA expression in reconstructed skin. Relative mRNA expression levels of LCE genes in epidermal sheets from skin constructs after stimulation with different cytokine combinations (N = 5 per group): 10 ng/mL of IL-1α, 5 ng/mL of TNF-α, and 5 ng/mL of IL-6 or 100 ng/mL of IL-17 or 100 ng/mL of IL-22. Values for each gene were expressed relative to the control culture condition, which was set at unity. It is not possible to compare expression levels between genes. *P < 0.05 compared with control condition [mean (SD)].
LCE2 Antibody Production
A human pan-LCE2 antibody was raised in rabbits against the synthetic peptide RPRLFHRRRHQSPD. This peptide perfectly matches LCE2B, LCE2C, and LCE2D, and differs by one amino acid for LCE2A (fourth R→H in LCE2A). The antiserum was positively tested on recombinant human LCE2A from bacterial lysates at Western blot analysis (see Supplemental Figure S1 at http://ajp.amjpathol.org).
Immunohistochemistry
Tissues or skin constructs were fixed in a buffered 4% formalin solution (Mallinckrodt Baker, Inc., Deventer, The Netherlands) for 4 hours and subsequently embedded in paraffin. Material was cut in 7-μm sections and subsequently processed for staining using a kit (Vectastain ABC kit; Vector Laboratories, Inc., Burlingame, CA) as previously described.17 LCE2 staining was performed using the primary rabbit anti-human pan-LCE2 antibody and a goat anti-rabbit antibody as a secondary antibody (Vector Laboratories, Inc.).
Immunofluorescence Analysis
Skin biopsy specimens were fixed and cut as described (see “Immunohistochemistry”). Immunofluorescence analysis was performed as previously described18 using the rabbit anti-human pan-LCE2 antibody and a mouse anti-cathepsin V antibody (R&D Systems, Inc., Minneapolis, MN) as primary antibodies. The secondary reagents were Alexa Fluor 488 goat anti-rabbit IgG highly cross-absorbed and Alexa Fluor 594 goat anti-mouse IgG highly cross-absorbed (Molecular Probes, Inc., Eugene, OR). Nuclei were stained with DAPI (Dako-Cytomation A/S, Copenhagen, Denmark).
Immunoelectron Microscopy
Postembedding immunoelectron microscopy using resin (Lowicryl K11M; Chemische Werke Lowi GmbH, Waldkraiburg, Germany) was performed as described previously.19 The secondary antibody used for immunoelectron microscopy was 5 nm of gold-conjugated goat anti-rabbit IgG (BBinternational, Cardiff, UK).
Statistical Analysis
Data are given as mean (SD). Statistical analysis of qPCR data was performed on ΔCt values using commercially available software (SPSS release 16.0; SPSS, Inc., Chicago, IL). One-way analysis of variance, followed by Dunnett post hoc testing, and paired t-tests were performed. P < 0.05 was considered statistically significant.
Results
LCE Expression in Human Tissues
Because quantitative information about relative expression levels of the LCE gene family in human beings is limited,9,11 qPCR analysis was performed on a large panel of tissues including many epithelia. Expression was analyzed of all LCE genes except LCE3B and LCE4A, for which validation of specific primers failed. Because of high sequence similarities between LCE1D and LCE1E, these genes were analyzed together (Figure 1, D and E). Figure 1 shows the expression of the LCE3 genes in a wide range of normal human tissues using the expression of LCE1A in skin as a reference (set at unity). The tissues originate from individuals homozygous for the LCE3B/C deletion; therefore, no LCE3C data are available. Gene expression data from the other LCE genes is shown in Supplemental Figure S2 (at http://ajp.amjpathol.org). Most samples demonstrated very low to undetectable LCE expression. Moderate expression was observed in trunk skin and some oral epithelia. High LCE gene expression was observed only in plantar skin.
Distinct Expression Patterns of LCE3 Genes in Normal and Psoriatic Skin
Figure 2A shows the mRNA expression of the various LCE genes as determined in epidermal sheets of normal skin and of lesional skin of patients with psoriasis (see Supplemental Table S1 at http://ajp.amjpathol.org for ΔCt values and exact P values). Expression levels of the various LCE groups seemed to be groupwise regulated. All members of the LCE groups 1, 2, 5, and 6 were significantly down-regulated in psoriatic skin, whereas all genes of the LCE3 group were barely present in normal skin and were significantly induced in psoriatic skin (for LCE3C only when present in the genome), as reported by others.20 In four patients with psoriasis, LCE gene expression levels in uninvolved psoriatic skin were also investigated, and exhibited a similar expression pattern as observed in normal skin (Figure 2B and Supplemental Table S2 at http://ajp.amjpathol.org). To confirm the LCE mRNA expression data at the protein level, an antiserum directed against all LCE2 members was used to stain paraffin sections of psoriatic and normal skin. Figure 2C shows that the pan-LCE2 antibody stained the upper granular layer and stratum corneum of normal and lesional psoriatic epidermis. No gross difference in staining intensity between normal and diseased skin was observed despite the decrease of mRNA expression observed for LCE2 members (notably LCE2B) in psoriasis. Co-localization studies using immunofluorescence microscopy on normal skin sections demonstrated overlap between LCE2 and cathepsin V expression (Figure 2D). Previous studies have shown that cathepsin V is present in the superficial granular layer and stratum corneum, where it is associated with corneodesmosomes.18 LCE2 co-localized completely with cathepsin V in the granular layer, whereas expression in the stratum corneum partly co-localized. In the stratum corneum, LCE2 is more broadly expressed. Immunoelectron microscopy analysis was performed to study the subcellular localization of the LCE2 proteins, which were present in a diffuse cytoplasmic distribution in the cells of the granular layer. In the stratum corneum and upper granular layer, LCE2 proteins were localized along the cornified envelopes (Figure 2E). LCE2 expression in skin appendages was limited to nail, where expression was observed in the matrix, between the matrix and the proximal nail fold, in the proximal nail fold, and in the hyponychium (Figure 2F). No expression was observed in hair follicles or sweat glands (data not shown).
Skin Injury Induces Specific Expression of LCE3 Genes
It has previously been demonstrated that expression of the LCE3C and LCE3E genes is induced after tape stripping of normal skin.4 Superficial skin injury in healthy volunteers leads to disruption of the skin barrier and induces epidermal activation as assessed by morphologic features, cell proliferation, and gene expression.21 qPCR analysis was used to investigate expression of all LCE genes before and after tape stripping of normal skin (Figure 3, and Supplemental Table S3 at http://ajp.amjpathol.org). The LCE genes of groups 1, 2, 5, and 6 were significantly down-regulated after tape stripping, whereas the LCE3 genes were significantly up-regulated. This pattern of groupwise induction and down-regulation was similar to that in normal versus psoriatic skin (Figure 2A). mRNA data were confirmed at the protein level for LCE2. Figure 3B shows that before tape stripping, LCE2 was detected in the uppermost layer of the stratum granulosum and in the entire orthokeratotic stratum corneum. Forty-eight hours after tape stripping, LCE2 protein was completely absent in the granular layer and in the parakeratotic stratum corneum.
LCE Gene Expression in Stratifying Epidermis in a Reconstructed Skin Model
To examine the dynamics of LCE gene expression during the process of epidermal stratification, reconstructed skin on de-epidermized dermis was used, as described previously.22 The mRNA data (see Supplemental Figure S3A and Supplemental Table S4 at http://ajp.amjpathol.org) demonstrate that expression of the LCE genes was very low at day 0 (mean Ct-value, 31). At this time, the cells are still present as an undifferentiated layer of a few cells. After 3 days of air exposure, expression of all LCE genes except LCE3A and LCE5A was highly induced. The LCE expression levels continued to rise until day 6, then stabilized until day 15 of the air-exposure phase. The qPCR results for LCE2 were confirmed at the protein level (Figure S3B). LCE2 proteins were not expressed in the skin construct at day 0; however, from day 3 onward, expression gradually emerged in the stratum granulosum, and later LCE2 proteins were also present in the stratum corneum. The expression data for reconstructed skin, at both the mRNA and protein levels (for LCE2), were similar to those observed in normal human skin (Figure 2B), which indicates that this is a suitable model for study of LCE biology in vitro. In this model, higher expression of LCE3D and LCE3E and lower expression of LCE5A was observed compared with expression levels of these genes in normal skin.
Cytokine Induction of LCE Gene Expression in a Reconstructed Skin Model
A recently developed in vitro psoriatic skin model was used to examine disease-specific keratinocyte gene expression levels.12 This model was used to study the effect of proinflammatory cytokines associated with psoriasis on expression of LCE genes. A combination of psoriasis-associated cytokines TNF-α, IL-1α, and IL-6 significantly induced expression of the LCE3 genes, whereas this combination and the Th17 cytokine IL-22 significantly down-regulated expression of LCE1B (Figure 4 and Supplemental Table S5 at http://ajp.amjpathol.org). IL-17 and IL-22 demonstrated nonsignificant up-regulation of the LCE3 genes. The investigated cytokine combinations did not significantly affect expression of other LCE members.
Discussion
Herein is reported a comprehensive study on the expression of human LCE genes in vivo and in vitro. The highest expression of all LCE genes was observed in skin, and moderate expression in some oral epithelia. Very low to undetectable expression was observed for all LCE genes in most samples from a broad panel of tissues including many epithelia. Of 16 investigated LCE genes, only expression of the LCE3 group members was significantly induced in psoriatic skin, whereas genes from the other LCE groups were significantly down-regulated. Similar groupwise expression patterns were observed after superficial injury of normal skin by tape stripping. Localization studies demonstrated that the LCE2 proteins are expressed in the uppermost layer of the stratum granulosum on cornified envelopes. Stimulation of in vitro reconstructed skin with psoriasis-associated cytokines demonstrated induction of LCE3 gene expression. This in vitro model also demonstrated that during the process of epidermal stratification, LCE gene and protein expression begins at the moment of stratum corneum formation.
Analysis of tissue expression suggests that all LCE genes are likely involved in skin-specific processes. The data presented herein confirmed the results of previous studies of LCE1, LCE2, and LCE5 expression. One study, however, demonstrated that LCE3 genes were primarily expressed in esophagus and tongue, and were barely present in skin.11 This may be because only normal skin was studied, in which LCE3 expression is low. The LCE2 proteins are localized in the superficial cells of the stratum granulosum and in premature cornified envelopes, resembling involucrin staining in normal skin.23 In mouse embryos, similar localizations were observed for Lce1.11 LCE2 is also present in the nail unit, at similar locations as involucrin, filaggrin, and transglutaminase 3,24 which suggests that LCE2 is not only involved in terminal differentiation in the epidermis but also in the nail unit. From in vitro data in stratifying epidermis, it was observed that LCE expression begins at the moment of stratum corneum formation, which is similar to loricrin expression in a previous study.22 Together these results strengthen previous observations that LCE proteins are expressed late and are finally localized in the stratum corneum as part of the cornified envelope.11
The present study demonstrated that induction of LCE3 gene expression observed in lesional psoriatic epidermis can be mimicked by adding a combination of TNF-α, IL-1α, and IL-6 to the reconstructed skin model. In addition, the Th17 cytokines seem to have an effect on LCE3 expression; IL-22 demonstrated a trend toward induction, especially of LCE3A. The effects on LCE3D and LCE3E gene expression are smaller. It could be that the basal expression levels of these genes in the reconstructed skin model are higher in the control situation than in normal skin; thus, induction of gene expression is more difficult to demonstrate. Previous in vitro studies used submerged keratinocyte cultures in which LCE2 gene expression was induced by adding Ca++-rich medium.9 UVB irradiation caused induction of the LCE1 and LCE2 groups and of LCE3E. Expression of other LCE genes remained low in both situations. These results do not show the same groupwise expression as observed in the present study. This might be caused by the use of submerged cultures in which the basal expression levels of the LCE genes are very low (data not shown).
Deletion of LCE3B and LCE3C is a widely replicated risk factor for psoriasis.4–7 Although the associated odds ratio is modest (1.21 in a recent meta-analysis7) compared with that of HLA-Cw6 (2.8 according to another meta-analysis25), the high prevalence of the deletion in patients with psoriasis results in a considerable population-associated risk (21%4). Its reported epistasis with HLA-Cw6 in at least some white populations makes it a major risk factor for psoriasis.7 Inasmuch as all LCE genes are similar and would be predicted to exert similar functions, the question arises as to why deletion of 2 of 18 quite similar LCE genes could have a role in the etiology of psoriasis. Herein it was demonstrated that four of the five LCE3 genes are differently expressed compared with other LCE groups. It was interpreted that LCE3 members may function as barrier repair proteins whose expression is driven by proinflammatory cytokines, whereas other LCE proteins have a constitutive role in barrier function of normal skin. Heterozygous or homozygous deletion of LCE3B and LCE3C, which is observed in greater than 90% of patients with psoriasis, could potentially affect skin barrier properties (compared with the homozygous ancestral status of two intact chromosomes), in particular under conditions of stress.7 Thus, concerning the relevance for psoriasis, there is deletion of two of five LCE3 genes rather than two of 18 LCE genes. Because LCE3B and LCE3C are expressed at low levels, if at all, in normal skin, the situation is different from the filaggrin (FLG) gene and atopic dermatitis.26 In atopic dermatitis, it has been suggested that a slightly leaky skin barrier could enable penetration of environmental allergens and would favor development of atopy.27 For LCE3B and LCE3C, expression is induced only after barrier disruption. Although 18 LCE genes are present on the genome, only five genes from this family seem to have a function after skin barrier disruption or inflammation. It was hypothesized that because of deletion of LCE3B and LCE3C, in combination with other unknown factors, an incomplete skin barrier repair response could occur after minor skin injury. It is speculated that the presence of only the LCE3A, LCE3D, and LCE3E proteins is not sufficient for a complete barrier repair response. A consequence could be increased permeability for environmental stimuli (eg, microbial components), leading to activation of innate immune mechanisms and subsequent recognition by the adaptive immune system in predisposed (eg, HLA-Cw6+) individuals (Figure 5). This is a testable hypothesis that merits further investigation.
Figure 5.
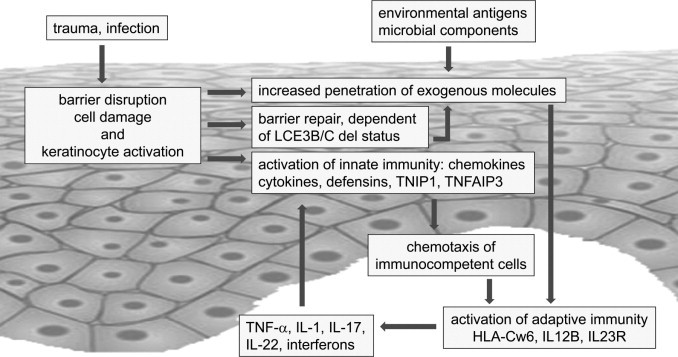
Tentative model of psoriasis based on genetic and cell biological data. It was proposed that minor trauma such as by mechanical stimuli or subclinical infection will breach the skin barrier and induce cell damage, which subsequently causes activation of keratinocytes. Keratinocytes will respond to superficial injury by innate immune responses such as activation of nuclear factor κB signaling and secretion of chemokines, cytokines, and β-defensins. This has been well documented in in vivo (tape stripping) and in vitro models. Skin barrier disruption will activate repair mechanisms such as up-regulation of normal constituents (involucrin and transglutaminase-1) and induce expression of LCE3 genes (present study). The speed and quality of the temporary skin barrier may depend on the genotype for the LCE3B/C deletion. Individuals harboring a deleted allele may be subject to increased penetration of environmental or microbial antigens. The low-grade inflammation caused by keratinocyte activation will attract immunocompetent cells (T cells and dendritic cells) to the skin and could lead to activation of adaptive immunity in genetically predisposed individuals that responds to unidentified antigens that have penetrated the skin. The ensuing immune response characterized by secretion of Th1 and Th17 cytokines will in turn cause activation of keratinocytes, resulting in a vicious cycle and chronicity of inflammation.
Acknowledgments
We thank the volunteers who participated in this study and Ms. Noriko Takashita for assisting with the immunoelectron microscopy experiments.
Footnotes
Supported in part by DPTE grant bgt.6739 and grant 114000084 from the Alternatives to Animal Experiments program of ZonMW; by the University of Cambridge, Cancer Research UK, Hutchison Whampoa, Ltd. (M.N.); and by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (A.I.-Y.).
P.L.J.M.Z and J.S contributed equally to this work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2010.12.017.
Current address of J.-Y.T.: CEA, iBiTec-S, Gif-sur-Yvette, France.
Supplementary data
Validation anti-LCE2 antibody on bacterial lysates expressing recombinant human flag-LCE2A. The anti-LCE2 antibody was raised in a rabbit against a synthetic peptide. This peptide perfectly matches LCE2B, C, and D and differs one amino acid from LCE2A. Validation of 2 different bleeds (Nos. 73 and 74) on bacterial lysates expressing human flag-LCE2A showed a band at about 20 to 25 kDa, which was specific. This band was detected by both anti-Flag and anti-LCE2A antibodies. Antibodies 74 give higher specific signal than antibodies 73, but also higher background. Both bleeds were used in immunohistochemistry and yielded similar results.
Relative mRNA expression levels of LCE genes in normal human tissues. Expression of target genes was normalized to that of RPLP0. For graphic representation, all values are expressed relative to LCE1A in skin, which was set at unity, to enable comparison of the expression levels between tissues and genes (see Livak and Schmittgen16). For primer sequences and efficiency of amplification see Table 1. Normal human tissues are from a single individual. Skin data was obtained from 12 normal skin biopsy specimens (mean [SD]).
LCE expression in stratifying epidermis in reconstructed skin. A: Relative mRNA expression levels of LCE genes in epidermal sheets from skin constructs. Adult human keratinocytes were seeded on a de-epidermized dermis and cultured for 3 days submerged, followed by 0, 3, 6, 9, 12, or 15 days of culturing at the air-liquid interphase. All values are expressed relative to LCE1A at day 0, which was set at unity, to enable comparison of the expression levels between genes. Values are given as mean (SD). B: Immunohistochemical staining with an anti-LCE2 antibody on sections of reconstructed skin at different times during the process of epidermal stratification. Scale bar = 50 μm.
References
- 1.Bowcock A.M., Krueger J.G. Getting under the skin: the immunogenetics of psoriasis. Nat Rev Immunol. 2005;5:699–711. doi: 10.1038/nri1689. [DOI] [PubMed] [Google Scholar]
- 2.Nestle F.O., Kaplan D.H., Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 3.Schon M.P., Boehncke W.H. Psoriasis. N Engl J Med. 2005;352:1899–1912. doi: 10.1056/NEJMra041320. [DOI] [PubMed] [Google Scholar]
- 4.de Cid R., Riveira-Munoz E., Zeeuwen P.L., Robarge J., Liao W., Dannhauser E.N., Giardina E., Stuart P.E., Nair R., Helms C., Escaramis G., Ballana E., Martin-Ezquerra G., den Heijer M.D., Kamsteeg M., Joosten I., Eichler E.E., Lazaro C., Pujol R.M., Armengol L., Abecasis G., Elder J.T., Novelli G., Armour J.A., Kwok P.Y., Bowcock A., Schalkwijk J., Estivill X. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X.J., Huang W., Yang S., Sun L.D., Zhang F.Y., Zhu Q.X., Zhang F.R., Zhang C., Du W.H., Pu X.M., Li H., Xiao F.L., Wang Z.X., Cui Y., Hao F., Zheng J., Yang X.Q., Cheng H., He C.D., Liu X.M., Xu L.M., Zheng H.F., Zhang S.M., Zhang J.Z., Wang H.Y., Cheng Y.L., Ji B.H., Fang Q.Y., Li Y.Z., Zhou F.S., Han J.W., Quan C., Chen B., Liu J.L., Lin D., Fan L., Zhang A.P., Liu S.X., Yang C.J., Wang P.G., Zhou W.M., Lin G.S., Wu W.D., Fan X., Gao M., Yang B.Q., Lu W.S., Zhang Z., Zhu K.J., Shen S.K., Li M., Zhang X.Y., Cao T.T., Ren W., Zhang X., He J., Tang X.F., Lu S., Yang J.Q., Zhang L., Wang D.N., Yuan F., Yin X.Y., Huang H.J., Wang H.F., Lin X.Y., Liu J.J. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 6.Huffmeier U., Bergboer J.G., Becker T., Armour J.A., Traupe H., Estivill X., Riveira-Munoz E., Mossner R., Reich K., Kurrat W., Wienker T.F., Schalkwijk J., Zeeuwen P.L., Reis A. Replication of LCE3C-LCE3B CNV as a risk factor for psoriasis and analysis of interaction with other genetic risk factors. J Invest Dermatol. 2010;130:979–984. doi: 10.1038/jid.2009.385. [DOI] [PubMed] [Google Scholar]
- 7.Riveira-Munoz E, He SM, Escaramis G, Stuart PE, Huffmeier U, Lee C, Kirby B, Oka A, Giardina E, Liao W, Bergboer J, Kainu K, de CR, Munkhbat B, Zeeuwen PL, Armour JA, Poon A, Mabuchi T, Ozawa A, Zawirska A, Burden AD, Barker JN, Capon F, Traupe H, Sun LD, Cui Y, Yin XY, Chen G, Lim HW, Nair RP, Voorhees JJ, Tejasvi T, Pujol R, Munkhtuvshin N, Fischer J, Kere J, Schalkwijk J, Bowcock A, Kwok PY, Novelli G, Inoko H, Ryan AW, Trembath RC, Reis A, Zhang XJ, Elder JT, Estivill X: Meta-analysis confirms the LCE3C_LCE3B deletion as a risk factor for psoriasis in several ethnic groups and finds interaction with HLA-Cw6. J Invest Dermatol, doi: 10.1038/jid.2010.350. [DOI] [PMC free article] [PubMed]
- 8.Mischke D., Korge B.P., Marenholz I., Volz A., Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 9.Jackson B., Tilli C.M., Hardman M.J., Avilion A.A., MacLeod M.C., Ashcroft G.S., Byrne C. Late cornified envelope family in differentiating epithelia-response to calcium and ultraviolet irradiation. J Invest Dermatol. 2005;124:1062–1070. doi: 10.1111/j.0022-202X.2005.23699.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X.P., Elder J.T. Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics. 1997;45:250–258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- 11.Marshall D., Hardman M.J., Nield K.M., Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc Natl Acad Sci USA. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tjabringa G., Bergers M., van Rens D., de Boer R., Lamme E., Schalkwijk J. Development and validation of human psoriatic skin equivalents. Am J Pathol. 2008;173:815–823. doi: 10.2353/ajpath.2008.080173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Ruissen F., Jansen B.J., de Jongh G.J., Zeeuwen P.L., Schalkwijk J. A partial transcriptome of human epidermis. Genomics. 2002;79:671–678. doi: 10.1006/geno.2002.6756. [DOI] [PubMed] [Google Scholar]
- 14.de Jongh G.J., Zeeuwen P.L., Kucharekova M., Pfundt R., van der Valk P.G., Blokx W., Dogan A., Hiemstra P.S., van de Kerkhof P.C., Schalkwijk J. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J Invest Dermatol. 2005;125:1163–1173. doi: 10.1111/j.0022-202X.2005.23935.x. [DOI] [PubMed] [Google Scholar]
- 15.Franssen M.E., Zeeuwen P.L., Vierwinden G., van de Kerkhof P.C., Schalkwijk J., van Erp P.E. Phenotypical and functional differences in germinative subpopulations derived from normal and psoriatic epidermis. J Invest Dermatol. 2005;124:373–383. doi: 10.1111/j.0022-202X.2004.23612.x. [DOI] [PubMed] [Google Scholar]
- 16.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Le M., Schalkwijk J., Siegenthaler G., van de Kerkhof P.C., Veerkamp J.H., van der Valk P.G. Changes in keratinocyte differentiation following mild irritation by sodium dodecylsulphate. Arch Dermatol Res. 1996;288:684–690. doi: 10.1007/BF02505278. [DOI] [PubMed] [Google Scholar]
- 18.Zeeuwen P.L., Ishida-Yamamoto A., van Vlijmen-Willems I.M., Cheng T., Bergers M., Iizuka H., Schalkwijk J. Colocalization of cystatin M/E and cathepsin V in lamellar granules and corneodesmosomes suggests a functional role in epidermal differentiation. J Invest Dermatol. 2007;127:120–128. doi: 10.1038/sj.jid.5700480. [DOI] [PubMed] [Google Scholar]
- 19.Ishida-Yamamoto A., Simon M., Kishibe M., Miyauchi Y., Takahashi H., Yoshida S., O'Brien T.J., Serre G., Iizuka H. Epidermal lamellar granules transport different cargoes as distinct aggregates. J Invest Dermatol. 2004;122:1137–1144. doi: 10.1111/j.0022-202X.2004.22515.x. [DOI] [PubMed] [Google Scholar]
- 20.Guttman-Yassky E., Suarez-Farinas M., Chiricozzi A., Nograles K.E., Shemer A., Fuentes-Duculan J., Cardinale I., Lin P., Bergman R., Bowcock A.M., Krueger J.G. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124:1235–1244. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Van Duijnhoven-Avontuur W.M., Alkemade J.A., Schalkwijk J., Mier P.D., van der Valk P.G. The inflammatory and proliferative response of normal skin in a model for acute chemical injury: ornithine decarboxylase induction as a common feature in various models for acute skin injury. Br J Dermatol. 1994;130:725–730. doi: 10.1111/j.1365-2133.1994.tb03409.x. [DOI] [PubMed] [Google Scholar]
- 22.Cheng T., Tjabringa G.S., van Vlijmen-Willems I.M., Hitomi K., van Erp P.E., Schalkwijk J., Zeeuwen P.L. The cystatin M/E-controlled pathway of skin barrier formation: expression of its key components in psoriasis and atopic dermatitis. Br J Dermatol. 2009;161:253–264. doi: 10.1111/j.1365-2133.2009.09156.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishida-Yamamoto A., Eady R.A., Watt F.M., Roop D.R., Hohl D., Iizuka H. Immunoelectron microscopic analysis of cornified cell envelope formation in normal and psoriatic epidermis. J Histochem Cytochem. 1996;44:167–175. doi: 10.1177/44.2.8609373. [DOI] [PubMed] [Google Scholar]
- 24.Cheng T., van Vlijmen-Willems I.M., Hitomi K., Pasch M.C., van Erp P.E., Schalkwijk J., Zeeuwen P.L. Colocalization of cystatin M/E and its target proteases suggests a role in terminal differentiation of human hair follicle and nail. J Invest Dermatol. 2009;129:1232–1242. doi: 10.1038/jid.2008.353. [DOI] [PubMed] [Google Scholar]
- 25.Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., Ruether A., Schreiber S., Weichenthal M., Gladman D., Rahman P., Schrodi S.J., Prahalad S., Guthery S.L., Fischer J., Liao W., Kwok P.Y., Menter A., Lathrop G.M., Wise C.A., Begovich A.B., Voorhees J.J., Elder J.T., Krueger G.G., Bowcock A.M., Abecasis G.R. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., Goudie D.R., Sandilands A., Campbell L.E., Smith F.J., O'Regan G.M., Watson R.M., Cecil J.E., Bale S.J., Compton J.G., DiGiovanna J.J., Fleckman P., Lewis-Jones S., Arseculeratne G., Sergeant A., Munro C.S., El H.B., McElreavey K., Halkjaer L.B., Bisgaard H., Mukhopadhyay S., McLean W.H. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 27.Cork M.J., Danby S.G., Vasilopoulos Y., Hadgraft J., Lane M.E., Moustafa M., Guy R.H., Macgowan A.L., Tazi-Ahnini R., Ward S.J. Epidermal barrier dysfunction in atopic dermatitis. J Invest Dermatol. 2009;129:1892–1908. doi: 10.1038/jid.2009.133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation anti-LCE2 antibody on bacterial lysates expressing recombinant human flag-LCE2A. The anti-LCE2 antibody was raised in a rabbit against a synthetic peptide. This peptide perfectly matches LCE2B, C, and D and differs one amino acid from LCE2A. Validation of 2 different bleeds (Nos. 73 and 74) on bacterial lysates expressing human flag-LCE2A showed a band at about 20 to 25 kDa, which was specific. This band was detected by both anti-Flag and anti-LCE2A antibodies. Antibodies 74 give higher specific signal than antibodies 73, but also higher background. Both bleeds were used in immunohistochemistry and yielded similar results.
Relative mRNA expression levels of LCE genes in normal human tissues. Expression of target genes was normalized to that of RPLP0. For graphic representation, all values are expressed relative to LCE1A in skin, which was set at unity, to enable comparison of the expression levels between tissues and genes (see Livak and Schmittgen16). For primer sequences and efficiency of amplification see Table 1. Normal human tissues are from a single individual. Skin data was obtained from 12 normal skin biopsy specimens (mean [SD]).
LCE expression in stratifying epidermis in reconstructed skin. A: Relative mRNA expression levels of LCE genes in epidermal sheets from skin constructs. Adult human keratinocytes were seeded on a de-epidermized dermis and cultured for 3 days submerged, followed by 0, 3, 6, 9, 12, or 15 days of culturing at the air-liquid interphase. All values are expressed relative to LCE1A at day 0, which was set at unity, to enable comparison of the expression levels between genes. Values are given as mean (SD). B: Immunohistochemical staining with an anti-LCE2 antibody on sections of reconstructed skin at different times during the process of epidermal stratification. Scale bar = 50 μm.


