Abstract
Plasminogen activator inhibitor-1 (PAI-1) is a key physiological inhibitor of fibrinolysis. Previously, we reported placenta growth factor (PlGF) mediated transcriptional upregulation of PAI-1 mRNAexpression via activation of hypoxia-inducible factor-1α and activator protein-1 in human pulmonary microvascular endothelial cells (HPMVEC); which resulted in elevated PAI-1 in humans with sickle cell anemia (SCA). Herein, we identified the role of post-transcriptional mechanism(s) of PlGF-mediated accumulation of PAI-1 mRNA in HPMVEC by examining the role of microRNAs in PlGF-induced PAI-1 mRNA stability. Our results show reduced expression of miR-30c and miR-301a, but not of miR-99a in response to PlGF, which have evolutionarily conserved binding sites in the 3′-untranslated region (3′UTR) of PAI-1 mRNA. Transfection of anti-miR-30c or anti-miR-301a oligonucleotides resulted in increased PAI-1 mRNA levels, which were further increased with PlGF stimulation. Conversely, overexpression of pre-miR-30c or pre-miR-301a resulted in attenuation of PlGF-induced PAI-1 mRNA and protein levels. Luciferase reporter assays using wild-type and mutant 3′UTR constructs confirmed that PAI-1-3′UTR is indeed a direct target of miR-30c and miR-301a. Finally, plasma levels of miR-30c and miR-301a were significantly downregulated in patients with SCA, compared to normal controls. These data provide a post-transcriptional regulatory mechanism of PlGF-inducedPAI-1elevation.
Keywords: 3′ untranslated region, adenosine- and uridine-rich elements, microRNAs, Plasminogen activator inhibitor-1, Placenta growth factor, Sickle cell anemia
INTRODUCTION
Vascular injury plays a significant role in the pathogenesis of a number of acute and chronic disorders, wherein initial injury triggers a cascade of events such as inflammation, activation of coagulation and repair processes so as to maintain vascular integrity. However, a loss in homeostasis leads to overt pathological disease. Although many factors contribute to vascular dysfunction, the plasminogen activator inhibitor-1 (PAI-1) has been shown to play a key role in the pathogenesis of cardiovascular, renal, hepatic and pulmonary disorders [1, 2], and hypercoagulability in sickle cell anemia (SCA) [3, 4]. Elevated plasma levels of circulating PAI-1 and tissue factor have been observed in SCA patients under steady state, which further increase during vaso-occlusive crises [5]. The prothrombotic state predisposes patients to both stroke and pulmonary hypertension, two distressing complications of SCA [6].
PAI-1, a secreted 50 kDa protein and a member of the serpin family of serine protease inhibitors, is the main circulating inhibitor of plasminogen activation and hence fibrinolysis [7]. X-ray crystal structure has revealed that serpins, including PAI-1, are globular proteins, which have an exposed peptide loop and the central reactive loop, the latter is essential for inhibitory mechanisms [7]. PAI-1 is synthesized by a variety of cell types, such as endothelial cells, smooth muscle cells, mononuclear cells, hepatocytes and platelets [8, 9]. The expression of PAI-1 is regulated by hypoxia [8, 10], insulin [11] and transforming growth factor-β (TGF-β) [12] in a variety of tissue and cell types. The hypoxia-induced expression of murine PAI-1 in monononuclear cells involves cis elements for early growth response-1 (Egr-1), hypoxia-inducible factor-1α (HIF-1α) and CCAAT enhancer binding protein α (C/EBPα) in the promoter region [10]. Oxidative stress induced PAI-1 expression requires AP-1 binding to cis-elements within the promoter [13]. In our recent studies [9] we showed placenta growth factor (PlGF), released by sickle erythroblasts at a higher level compared to normal erythroblasts, increased PAI-1 expression in primary human pulmonary microvascular endothelial cells (HPMVEC) and monocytes. PlGF-induced PAI-1 mRNA expression occurred through activation and binding of transcription factors, HIF-1α and activator protein-1 (AP-1) to their cognate elements in the promoter region of PAI-1 at 4–6 hr post PlGF stimulation [9]. In addition, we observed that PlGF also increased PAI-1 mRNA at an early time point of 1 hr by an unidentified mechanism. Therefore, we studied the role of microRNAs (miRNAs) in PlGF-mediated post transcriptional PAI-1 mRNA stabilization in the present study.
miRNAs are a class of endogenous, small noncoding RNAs of approximately 22–25 nucleotides, which repress gene expression by binding to imperfect complementary sequences in the 3′-untranslated regions (UTRs) of target mRNA, leading to mRNA degradation and translational repression [1–17]. In mammals, miRNAs have been shown to be associated with diverse biological processes including cell proliferation, differentiation, apoptosis, metabolism, and morphogenesis [18], innate immunity [19–22], kidney dysfunction in diabetes [23, 24], tumorigenesis [25–27] and leukotriene formation [28]. The 3′UTR of PAI-1 mRNA contains several putative adenosine- and uridine-rich elements (AREs), and several studies have identified the interaction of different mRNA binding proteins with these AREs for regulating turnover and stability. However, relatively less is known about the role of miRNAs in PAI-1 mRNA regulation.
In the present study, we show PlGF treatment of HPMVEC at early time points increased stabilization of PAI-1 mRNA through concomitant decrease in the expression of miR-30c and miR-301a. We further show that inhibitor oligonucleotides of miR-30c and miR-301a augmented PAI-1 mRNA expression. Conversely, miR-30c and miR-301 precursor oligonucleotides reduced PAI-1 mRNA expression. We confirmed that these miRNAs directly target PAI-1-3′UTR in a sequence specific manner from reporter studies. We also show that plasma concentrations of miR-30c and miR-301a were significantly reduced in patients with SCA. Our results demonstrate, for the first time, to the best of our knowledge, that PlGF-induced PAI-1 expression at early time-points is post-transcriptionally regulated by two miRNAs, -30c and -301a.
MATERIALS AND METHODS
Cells and reagents
HPMVEC were obtained and cultured as described earlier [9]. These cells were confirmed to express CD31 and von Willebrand factor (vWF) as previously described [9]. HPMVEC were incubated overnight in endothelial basal medium-2 (EBM-2) containing 2% serum followed by serum deprivation for 3 hr prior to treatment with either PlGF (250 ng/mL) or other indicated experimental conditions.
Reagents were obtained as follows: human recombinant PlGF (R&D Systems, Minneapolis, MN); HIF-1α siRNA, PlGF siRNA and corresponding control scrambled (sc) RNA, primary antibodies against PAI-1 and β-actin, and secondary antibodies conjugated to HRP (Santa Cruz Biotechnology, Santa Cruz, CA); c-Jun TranSilent siRNA vector (Panomics Inc., Fremont, CA); actinomycin D (Enzo Life Sciences, Plymouth Meeting, PA). The primers used for PCR amplification of PAI-1-3′UTR and mutagenesis were purchased from Integrated DNA Technologies, Inc. (San Diego, CA). Unless otherwise specified, all other reagents were purchased from Sigma Chemical Company (St. Louis, MO).
Human Subjects
All blood samples were obtained from children with homozygous SCA at steady state at their elective clinical appointment with routine clinical draws using Institutional Review Board approved protocols through the Hematology Repository at Cincinnati Children’s Hospital Medical Center. The plasma samples were obtained from the same SCA and healthy subjects as described previously [29].
Analysis of miRNA expression
Total RNA was extracted from HPMVEC using the mirVana miRNA Isolation kit (Applied Biosystems, Foster City, CA). For extraction of total RNA (including small RNA molecules) from plasma of human subjects, 400 μL of plasma was mixed completely with TRIzol reagent at a volume ratio of 1.0:0.8. After chloroform extraction, the aqueous layer was removed and mixed with absolute ethanol (1.25 volumes) and loaded onto the cartridge supplied in the mirVana isolation kit. Total small RNA was eluted according to the manufacturer’s instructions. The miRNA expression was determined by using the TaqMan MicroRNA Assay kits for indicated miRNA (Applied Biosystems) according to the manufacturer’s protocol. Briefly, 100 ng of total RNA was reverse transcribed at 42°C/15min, 95°C/5min and 4°C/5min. qRT-PCR (20 μl total reaction) was performed in a 384-well plate at 95°C for 10 min, followed by 40 cycles of 95°C×15s and 60°C×60s. All reactions were run in triplicate. Gene expression was normalized to reference gene, 5S rRNA for cell culture samples and U6 small nuclear RNA (RNU6B) for plasma samples. Relative quantitative (RQ) levels for miRNA expression were calculated as 2 −ΔΔCt by the comparative Ct method [30], where ΔΔCt= (Ct target miRNA of treated sample- Ct reference gene of treated sample) − (Ct target miRNA of control sample-Ct reference gene of control sample).
Transient transfection
HPMVEC (1×106) were resuspended in 100 μL of serum free RPMI medium containing appropriate siRNA constructs (50 nM) or luciferase reporter plasmids (0.5 μg) or 60 pmol anti-miRNA inhibitor (Life Technologies, Carlsbad, CA) or 1 μg pre-miRNA precursor constructs (Life Technologies) and transfected by nucleofection with the S-05 program in the nucleofector apparatus (Lonza, Basel, Switzerland) as previously described [31]. The β-galactosidase plasmid (0.5 μg) was cotransfected with luciferase reporter constructs to monitor transfection efficiency. Following nucleofection, the cells were incubated in EBM-2 complete medium overnight, kept in serum free EBM-2 for 3 hr and treated with PlGF for the indicated time periods. The cell lysates were analyzed for luciferase and β-galactosidase activity using appropriate kits (Promega, Madison, WI) [32]. Luciferase values were normalized to β-galactosidase values and expressed relative to the activity of the promoter-less pGL3 basic vector. For PAI-1 mRNA and miRNA analysis, cells were lysed with TriZOL and total RNA extracted. The expression was determined as described above.
Western blot analysis
The cytosolic extracts were isolated from untreated and PlGF treated cells. Briefly, 2×106 cells were resuspended in 400 μL of cell-lysis buffer for 20 min and cytosolic supernatants were collected after centrifugation at 10,000g for 5 min [32]. The cytosolic extracts were subjected to SDS-PAGE analysis for PAI-1 protein expression. The protein loading was monitored by stripping and reprobing the membrane with β-actin antibody. The protein bands were visualized using Immunobilon western reagents (Millipore Corporation, Billerica, MA).
PAI-1-3′UTR plasmid and its mutagenesis
The 455bp fragment spanning the region between +292 to +746 bp relative to the translation stop codon of PAI-1 mRNA was PCR-amplified using the forward primer containing HindIII restriction enzyme site and reverse primer containing SpeI site, as listed in Supplemental Table S1, and a Phusion high fidelity DNA polymerase (New England Biolabs, Ipswich, MA) according to the standard procedure. Human cDNA clone for serpin-1 (Genbank accession, NM_000602) was used as a template (Origene, Rockville, MD). The PCR product was cloned downstream of the firefly luciferase reporter gene in pMIR-REPORT plasmid (Ambion, Austin, TX). The orientation of the insert relative to the luciferase gene was confirmed by DNA sequencing and the plasmid was purified using EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA). The resulting plasmid is designated as pMIR-PAI-1-3′UTR. The mutant constructs of PAI-1-3′UTR in the binding sites for miR-30c, miR-301a or miR-99a were generated using pMIR-PAI-1-3′UTR as the template with primers listed in Supplemental Table S1 in accordance with the QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA). The double mutant construct for miR-30c and miR-301a binding sites was generated using miR-30c mutant construct as a template. The mutations were confirmed by DNA sequencing (Microchemical Core Facility, Norris Comprehensive Cancer Center, USC).
Statistical Analysis
Data are presented as mean ± SEM. The significance of difference in mean values between multiple groups was analyzed with parametric one way ANOVA followed by a Tukey-Kramer test using Instat 2 software suite (Graphpad, San Diego, CA). Student’s t test was used to evaluate the significance of difference between two groups of experiments. *** P< 0.001; ** P<0.01; ns, P> 0.05.
RESULTS
Post-transcriptional regulation of PlGF-mediated PAI-1 mRNA expression at early time period (1 and 2 hr)
We previously reported that PlGF-induces PAI-1 mRNA expression after 6 hr in HPMVEC via transcriptional activation of PAI-1 promoter by HIF-1α and AP-1 [9]. In the same study, we observed that PlGF could elevate PAI-1 mRNA levels as early as 1 hr post-induction [33]. In the present study, we confirmed the earlier results and observed increased PlGF-mediated PAI-1 mRNA expression at earlier time intervals (1, 2 hrs post-induction) using qRT-PCR (Supplementary Figure S1A). Moreover, PlGF-mediated PAI-1 expression at 1 and 2 hr occurred in the absence of transcriptional activation of the PAI-1 promoter, since PlGF activation of the PAI-1 promoter was delayed and observed only at later time points (4 and -6 hr post-treatment) (Supplementary Figure S1B).
Next, we assessed the role of transcription factors, HIF-1α and c-Jun in PlGF-induced PAI-1 mRNA expression at early (2 hr) and late (4 and 6 hr) time points. As shown in Supplementary Figure S1C, PlGF-mediated PAI-1 mRNA expression at 2 hr was not affected by transfection with either HIF-1α siRNA or HIF-1α scrambled (sc) RNA. However, PlGF-induced PAI-1 mRNA expression at later time points (4 and 6 hr) was significantly attenuated by HIF-1α siRNA, but not with HIF-1α scRNA, as previously observed [9]. Moreover, transfection with c-Jun siRNA, but not c-Jun scRNA, reduced PlGF-induced PAI-1 mRNA expression at 4 and 6 hr, as earlier reported (Supplementary Figure S1D) [9]. In contrast, PlGF-mediated PAI-1 mRNA expression was not affected by transfection of either c-Jun siRNA or c-Jun scRNA at 2 hr (Supplementary Figure S1D). Taken together, these data indicate that PlGF-induced PAI-1 mRNA expression at earlier time points (1 and 2 hr), most likely occurs by mRNA stabilization and at later time points (4 and 6 hr) via transcriptional activation of PAI-1 promoter by HIF-1α and AP-1.
Next, we performed experiments to determine whether the increased PAI-1 mRNA levels at the early time period (2 hr) in PlGF-treated HPMVEC was a result of increased mRNA stability. The cells were first treated with recombinant PlGF (250 ng/ml) for 2 hr, followed by treatment with actinomycin D to inhibit ongoing transcription and total RNA was isolated at indicated times following induction (10–120 min) for PAI-1 mRNA analysis. A comparison of PAI-1 turnover in the absence and presence of PlGF treatment showed a significant stabilization in mRNA from t1/2 of 12 ± 1.5 min to 28 ± 3 min (Supplementary Figure S1E). The kinetics show a biphasic nature of PAI-1 mRNA decline in untreated cells indicating a second order degradation process, consistent with an miRNA-dependent mechanism. In contrast, the linear or first order nature of PAI-1 mRNA decay in PlGF-treated cells implicated a process that was dependent only on mRNA concentration.
Identification of miRNAs involved in PlGF-mediated PAI-1 mRNA stabilization
We examined miRNAs that may be involved in stabilization and degradation of PAI-1 mRNA. Bioinformatics approaches were used to identify potential miRNA target sites harbored in 3′UTR of the PAI-1 mRNA. [17, 34, 35]. We used a web-based miRNA target prediction program (Microcosm, http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl) that considers complementarity, target site accessibility, and the extent of evolutionary conservation. This program predicted ~20 putative miRNA target sites in the PAI-1 3′UTR. In the present study, we selected five candidates (i.e. miR-30c, miR-301a, miR-99a, miR-299-5p and miR-609) based on good complementarity (ΔG°~−17–27 kcal/mol) and a high degree of site conservation among different mammalian species (Supplementary Figure S2). There is only a single predicted target site for each of these miRNAs in the 3′UTR of PAI-1 mRNA.
Next, we examined whether PlGF affected the expression of these five candidate miRNAs in HPMVEC after 2 hr of PlGF treatment. As shown in Figure 1A, there was approximately a 10-fold and 4-fold downregulation in the expression of miR-30c and miR-301a, respectively, in PlGF-treated cells as compared to untreated cells. However, the expression of miR-99a remained unaltered after PlGF stimulation. The expression of miR-299-5p and miR-609 was not detected in HPMVEC (data not shown) using the TaqMan microRNA assay. In order to identify the role of PlGF in the regulation of miR-30c, miR-301a and miR-99a expression, we utilized a siRNA approach for silencing endogenous PlGF expression. As shown in Figure 1B, HPMVEC transfected with PlGF siRNA showed elevated expression of both miR-30c and miR-301a by approximately 3.5-fold and 2.6-fold, respectively when compared to PlGF scRNA. However, PlGF siRNA did not change the expression of miR-99a in HPMVEC. It is pertinent to mention that transfection with PlGF siRNA almost completely abolished PlGF mRNA expression by approximately 95%, compared with untransfected cells, while PlGF scRNA had no effect on PlGF mRNA expression (data not shown). These results showed that endogenously expressed PlGF regulated basal expression of miR-30c and miR-301a in endothelial cells.
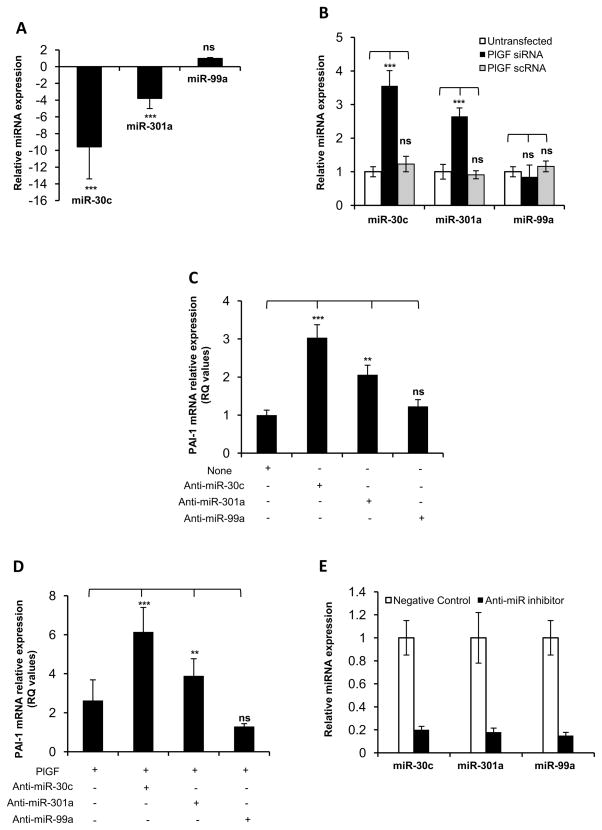
Figure 1. PlGF downregulates the levels of miRNA-30c and miR-301a, leading to increased PAI-1 mRNA expression.
A) HPMVEC were treated with PlGF for 2 hr, followed by isolation of total RNA. Levels of indicated miRNA candidates were analyzed using specific TaqMan MicroRNA assay for each miRNA as described in Materials and Methods. (B) HPMVEC were transfected with either PlGF siRNA or PlGF scRNA at 50 nM concentrations. Total RNA was isolated after 24 hr followed by qRT-PCR analysis for indicated miRNAs. (A and B) The relative expression levels of indicated miRNAs were normalized to 5S rRNA expression. The data are expressed as fold change values compared to control cells. (C) HPMVEC were transfected with indicated anti-miR oligonucleotides followed by total RNA isolation. The PAI-1 mRNA expression was estimated by qRT-PCR. (D) qRT-PCR analysis of PAI-1 mRNA expression in HPMVEC transfected with indicated anti-miR oligonucleotide, after 2 hr PlGF treatment. (C and D) PAI-1 mRNA levels were normalized to GAPDH mRNA expression. (E) Relative expression of indicated miRNA in HPMVEC transfected with corresponding anti-miR oligonucleotides. The level of miRNA expression observed in cells transfected with negative control oligonucleotide was considered “1”. The values shown are the mean ± S.E. of triplicate determinations at each time point from three independent experiments. *, p< 0.05, **, p< 0.01, ***, p< 0.001. ns, not significant.
Next, we investigated whether endogenous levels of miR-30c and miR-301a regulated expression of PAI-1 mRNA in HPMVEC, and were thus physiologically relevant. To address this, HPMVEC were transfected with specific antisense RNA oligonucleotides (i.e. anti-miRs). As shown in Figure 1C, transfection of HPMVEC with anti-miR-30c increased the basal expression of PAI-1 mRNA by 3-fold, while transfection with anti-miR-301a showed a 2-fold increase in PAI-1 mRNA expression. However, anti-miR-99a transfection did not affect PAI-1 mRNA expression. These results indicated that miR-30c and miR-301a, but not miR-99a, likely regulated endogenous levels of PAI-1 mRNA in HPMVEC. Subsequently, we examined whether reduced levels of miR-30c and miR-301a observed in HPMVEC, following PlGF treatment, contributed to increased expression of PAI-1 mRNA, which may be pathophysiologically relevant. As shown in Figure 1D, transfection of HPMVEC with anti-miR-30c augmented PlGF-mediated PAI-1 mRNA expression by 6-fold compared to a 2.5-fold increase in PAI-1 mRNA expression in response to PlGF alone. Similarly, transfection of anti-miR301a, followed by treatment with PlGF, resulted in a 4-fold increase in PAI-1 mRNA levels compared to a 2.5-fold increase in PAI-1 mRNA expression seen with PlGF alone. However, the overexpression of anti-miR99a did not change PlGF-mediated levels of PAI-1 mRNA (Figure 1D). It is pertinent to note that transfection of indicated anti-miRs showed significant reduction (~80%) in levels of individual miRNAs, as compared to negative control (Figure 1E). Taken together, these data showed that miR-30c and miR-301a, but not miR-99a, were involved in regulation of basal and PlGF-induced PAI-1 mRNA expression. Furthermore, these data indicated that PlGF-mediated reduction in the levels of miR-30c and miR-301a resulted in the accumulation of PAI-1 mRNA through a reduction in its degradation in HPMVEC.
miR-30C and miR-301a interact with the target binding sites in the 3′UTR of PAI-1 mRNA
In order to investigate whether miR-30c and miR-301a directly regulated PAI-1 mRNA expression, an experiment was performed in which the 3′UTR of the PAI-1 mRNA was inserted downstream of the luciferase open reading frame in pMIR-REPORT. As shown in schematics, Figure 2A, a region of the PAI-1 3′UTR (bases +292 to +746) relative to the translation stop codon, containing complementary binding sites for miR-30C, miR-301a and miR-99a, and adjacent surrounding sequences was used for reporter construction. Use of this segment also avoided potential interference from other putative miRNA binding sites. The resulting reporter construct (pMIR-PAI-1-3′UTR) was cotransfected with the pMIR-REPORT β-galactosidase control plasmid to monitor transfection efficiency in HPMVEC. Our results showed that basal reporter activity was drastically (>90%) reduced in cells transfected with pMIR-PAI-1-3′UTR when compared to cells transfected with control pMIR-REPORT construct; thus demonstrating the destabilizing effect of PAI-1-3′UTR on luciferase mRNA expression (Figure 2B). As shown in Figure 2B, transfection with pMIR-PAI-1-3′UTR reporter followed by treatment with PlGF reversed this effect and resulted in a time-dependent (0–2 hr) increase in luciferase activity. There was approximately a 4-fold increase in reporter activity by 2 hr. It is important to mention that PlGF did not change the reporter activity of a control plasmid, pMIR-REPORT (Figure 2B).
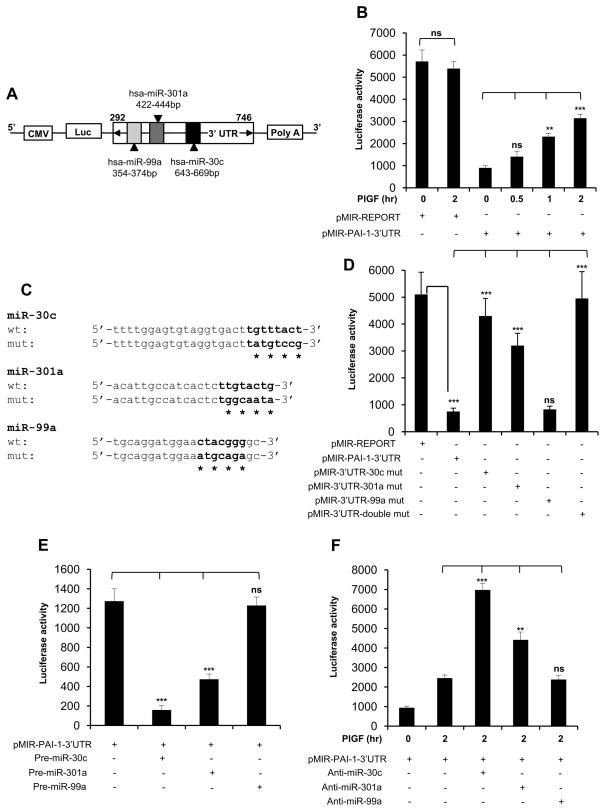
Figure 2. PlGF induced PAI-1-3′UTR reporter activity is abrogated by miR-30c and miR-301a, but not by miR-99a.
(A) Schematic representation of PAI-1-3′UTR luciferase reporter plasmid. The region between nucleotides 292 to 746 of PAI-1-3′UTR containing predicted target binding sites for miR-30c, miR-301a and miR-99a was cloned at the 3′end of luciferase reporter gene in pMIR-REPORT plasmid. (B) HPMVEC were transfected with either pMIR-REPORT control plasmid or pMIR-PAI-1-3′UTR construct. Following transfection, the cells were treated with PlGF for indicated time periods. (C) The sequences of predicted miR-30c, miR-301a and miR-99a binding sites within PAI-1-3′UTR. The bases in bold highlight “seed sequences” and the point mutations in seed sequence are indicated by asterisks. (D) HPMVEC were transfected with either pMIR-REPORT control plasmid, wild-type pMIR-PAI-1-3′UTR or indicated PAI-1-3′UTR mutant constructs for the binding of either miR-30c or miR-301a or miR-99a or both (miR-30c and miR-301a). 24 hr post-transfection, cells were lysed in reporter lysis buffer as per manufacturer’s instructions for luciferase assay. (E) HPMVEC were transfected with pMIR-PAI-1-3′UTR plasmid and indicated pre-miRNA precursor oligonucleotide. (F) HPMVEC were transfected with pMIR-PAI-1-3′UTR plasmid and indicated anti-miR followed by PlGF treatment for 2 hr. (B, D, E and F) Each construct was used at 0.5 μg. All luciferase reporters were co-transfected with β-galactosidase plasmid for normalization of transfection efficiency. The luciferase and_β-galactosidase activities were estimated as described in Materials and Methods. Data are representative of three independent experiments. *, p< 0.05, **, p< 0.01, ***, p< 0.001. ns, not significant.
Next, we determined which target sites in PAI-1-3′UTR were responsible for the dramatic reduction in reporter activity. For this, we introduced four point mutations in the corresponding “seed sequences” in pMIR-PAI-1-3′UTR for each miRNA candidate to eliminate any possible binding interactions (Figure 2C). In addition, double mutations were created in the PAI-1-3′UTR to jointly eliminate miR-30c and miR-301a binding. As shown in Figure 2D, the suppression of the reporter activity was significantly relieved in cells transfected with PAI-1-3′UTR-miR-30c mutant compared to the wild-type pMIR-PAI-1-3′UTR plasmid. The PAI-1-3′UTR-miR-301a-mutant showed partial recovery of reporter activity in comparison to the wild-type reporter. By contrast, no suppression in activity of the reporter construct, pMIR-PAI-1-3′UTR-miR-99a mutant was observed (Figure 2D), as expected. More interestingly, the double, miRNA site mutant completely rescued the repression of reporter activity (Figure 2D). Therefore, miR-30c and miR-301a, but not miR-99a, are jointly responsible for post-transcriptional inhibition of PAI-1 expression.
The above results were further corroborated in cells co-expressing specific miRNA precursor molecules and pMIR-PAI-1-3′UTR reporter plasmid. As shown in Figure 2E, overexpression of miR-30c precursor with pMIR-PAI-1-3′UTR reporter construct resulted in significant reduction (~90%) in luciferase activity. Similarly, miR-301a precursor transfection attenuated luciferase activity by approximately 60%. In contrast, transfection with miR-99a precursor showed no reduction in luciferase activity. To determine the specificity of these active miRs for PAI-1 mRNA, we used a complementary approach utilizing anti-miR oligonucleotides. As shown in Figure 2F, cotransfection of anti-miR-30c with PAI-1-3′UTR reporter augmented luciferase activity by 3-fold (lane 3 vs lane 2), while anti-miR-301 showed a 2-fold increase in reporter activity (lane 4 vs lane 2), following PlGF treatment for 2 hr. As expected, transfection with anti-miR99a had no effect on luciferase reporter activity in the presence of PlGF. Taken together; these data indicated that miR-30c and miR-301a directly target the 3′ UTR of PAI-1 mRNA for turnover, subsequently resulting in downregulation of PAI-1 expression.
Overexpression of precursor miRs downregulates PlGF-induced endogenous PAI-1 mRNA and protein expression
Since HPMVEC transfected with either anti-miR-30c or anti-miR-301a showed increased PlGF-mediated PAI-1 mRNA expression, we performed the converse experiment wherein cells were transfected with precursor molecules for miR-30c or miR-301a or miR-99a. Transfection with pre-miR-30c expression plasmid reduced PlGF-mediated PAI-1 mRNA expression to basal level when compared to cells treated with PlGF alone (Figure 3A, lane 3 vs lane 2). Similarly, overexpression of pre-miR-301a resulted in approximately an 80% reduction in PlGF-mediated PAI-1 mRNA expression as compared to PlGF-treated control cells (Figure 3A, lane 4 vs lane 2). However, transfection with pre-miR99a did not affect PlGF-mediated expression of PAI-1 mRNA (Figure 3A, lane 5 vs lane 2). Subsequently, we examined whether these miRNAs affected the expression of PAI-1 protein. As shown in Figure 3B, PlGF treatment of HPMVEC for 3 hr led to significant increase in PAI-1 protein levels, which was completely abrogated when the cells were transfected with pre-miR-30c expression plasmid (lane 6 vs lane 2). Similarly, overexpression of miR-301a in HPMVEC reduced PlGF-induced PAI-1 protein expression by ~80% (Figure 3B, lane 5 vs lane 2). However, overexpression of miR-99a did not alter PlGF-induced PAI-1 protein levels (Figure 3B, lane 4 vs lane 2).
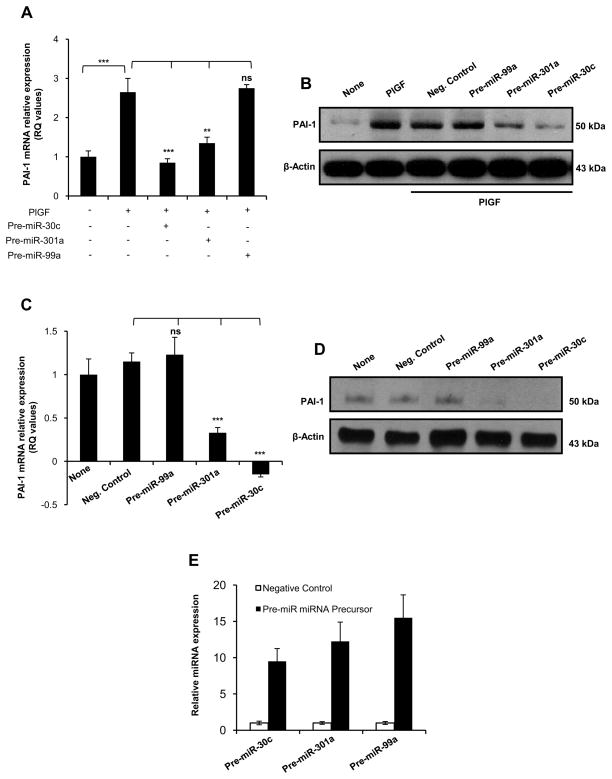
Figure 3. miR-30c and miR-301a affect PlGF-induced PAI-1 mRNA and protein levels.
(A and C) HPMVEC were transfected with indicated precursor miRNA; 24 hr post-transfection cells were incubated in the presence (A) or in the absence (C) of human recombinant PlGF for 2 hr. Total RNA was isolated using TriZOL reagent and subjected to qRT-PCR analysis of PAI-1 and GAPDH mRNA levels. (B and D) Immunoblot analysis of PAI-1 protein in HPMVEC transfected with indicated precursor miRNA oligonucleotide and incubated 3 hr in the presence (B) or in the absence (D) of PlGF. The same membrane was stripped and reprobed for β-actin. (E) HPMVEC were transfected with indicated precursor miRNA oligonucleotide and the relative expression of indicated miRNA was estimated using TaqMan MicroRNA assay. The results are indicated as the mean ± S.E. of triplicate determinations at each time point from three independent experiments. Indicated miRNA levels were normalized to 5S rRNA expression.*, p< 0.05, **, p< 0.01, ***, p< 0.001. ns, not significant. The data are representative of three independent experiments.
In addition, we investigated the effect of pre-miR-30c or miR-301a or miR-99a overexpression on endogenous PAI-1 mRNA and protein expression, in the absence of PlGF stimulation of HPMVEC. As shown in Figure 3C, transfection of pre-miR-30c and pre-miR-301 -singly, resulted in significant reduction of PAI-1 mRNA expression by approximately 120% and 70%, respectively compared to untransfected cells. However, overexpression of pre-miR-99a or control precursor molecules had no effect on PAI-1 mRNA level. The effect of precursor molecules for the above miRNA candidates on PAI-1 expression at protein levels was confirmed by western blotting. PAI-1 was significantly reduced in cells transfected with either pre-miR-30c or pre-miR-301a but not with pre-miR-99a when compared to cells transfected with negative control precursor molecules (Figure 3D). These results established that miR-30c and miR-301a regulate endogenous levels of PAI-1 expression in HPMVEC. We confirmed the overexpression of each of these pre-miRs (i.e. miR-30a, miR-301a, and miR-99a) in HPMVEC. As shown in Figure 3E, transfection of HPMVEC with pre- miR-30c, pre-miR-301a and pre-miR-99a expression plasmids resulted in 10-fold, 12-fold and 15-fold, respective increases in the corresponding miRNA levels. Taken together, the data showed that miR-30c and miR-301a, but not miR-99a, reduced endogenous basal and PlGF-induced PAI-1 mRNA and protein expression.
Downregulation of miR-30c and miR-301a levels in plasma from SCA subjects
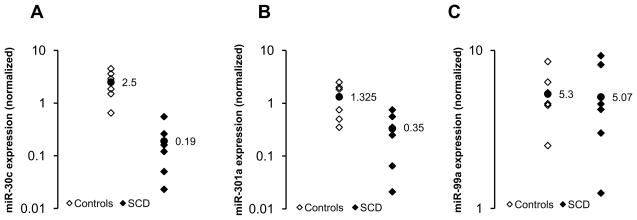
We have previously shown that levels of PlGF are elevated in patients with SCA [9, 40] and increased PAI-1 expression in SCA is transcriptionally upregulated by PlGF via HIF-1α and AP-1 [9]. Our present study shows that PlGF additionally down-regulates miR-30c and miR-301 expression. Hence, we wanted to determine whether levels of miR-30c and miR-301a were reduced in plasma obtained from SCA subjects. In order to identify any correlation between the elevated levels of PAI-1 and miRNA concentrations in plasma of SCA subjects, we used the same set of plasma samples for both SCA and healthy controls, which were previously used for PAI-1 analysis [9]. According to the earlier report [9], plasma levels of PAI-1 were significantly higher in SCA subjects compared to controls (17±3.2 ng/mL vs. 3.7±1.5 ng/mL). In the present study we found that circulating levels of miR-30c, miR-301a and miR-99a were indeed detectable in both samples from SCA patients (n=6) and healthy controls (n=6). The concentrations of both miR-30c and miR-301a were significantly lower, by approximately 13-fold and 3.8-fold, respectively, in plasma from SCA patients when compared to controls (Figure 4). However, there was no significant difference in the concentration of miR-99a between SCA patients and controls (Figure 4). Thus, our results showed inverse correlation between the levels of miR-30c and miR-301a, and PAI-1 in SCA.
Figure 4. Estimation of miR-30c, miR-301a and miR-99a levels in plasma samples from normal and SCA subjects.
The plasma concentrations of (A) miR-30c, (B) miR-301a, and (C) miR-99a were quantified from healthy controls (n=6) and SCA patients (n=6) using specific TaqMan MicroRNA assay for each miRNA candidate. The relative expression levels of individual miRNAs (log10 scale at y-axis) were normalized to RNU6B expression. Each data point represents values from a single subject. The mean value data point for healthy controls and SCA patients is displayed and represented as a filled circle (●).
DISCUSSION
PlGF is a member of the VEGF family of angiogenic growth factors and was originally shown to be released from placental trophoblast cells and umbilical vein endothelial cells [36]. However, recent studies show that PlGF is also produced by bone marrow erythroid cells, but not by other mature hematopoietic cells [37]. The levels of PlGF are significantly elevated in patients with chronic hemolytic anemia, such as in SCA and -thalassemia, due to increased compensatory erythropoiesis in response to anemia [38, 39]. Our previous studies identified the role of PlGF in the increased expression of many other genes, such as cytochemokines [38], 5-lipoxygenase activating protein for leukotriene synthesis [29], and endothelin-1 and its receptor [39]. These genes are involved in different pathologies manifested by SCA, including pulmonary hypertension [40] and lung injury.
Recently, we demonstrated that PlGF-induced PAI-1 mRNA in HPMVEC via transcriptional mechanisms, which involved activation of HIF-1α and AP-1 complex binding to cis- elements proximal to the promoter of PAI-1 [9]. In addition, we made the observation that PlGF increased PAI-1 mRNA levels as early as 1 hr post-stimulation with PlGF, which likely involved post-transcriptional mechanisms such as regulation of mRNA stability. In the present study, we ruled out the possibility of transcriptional regulation of PlGF-mediated PAI-1 mRNA accumulation at early time intervals (1–2 hr) in HPMVEC. PlGF failed to drive PAI-1 promoter-luciferase-reporter activity at 1 and 2 hr, post-induction; by contrast, a 5-6-fold increase in luciferase reporter activity was observed at 4 and 6 hr of treatment. Moreover, PlGF-induced PAI-1 mRNA expression at 2 hr was not reduced in cells transfected with HIF-1α siRNA or c-Jun siRNA, whereas significant reduction in PAI-1 mRNA was observed at 4 and 6 hr. The results of both experiments indicate involvement of post-transcriptional mechanisms in PlGF-induced PAI-1 mRNA expression during early induction. Our results from actinomycin-D inhibition during PlGF induction confirmed an increase in PAI-1 mRNA stability by 2-fold at 2 hr post-stimulation. The kinetic data of PAI-1 mRNA stability showed that in untreated cells there was a biphasic decline in PAI-1 mRNA, indicative of a second order degradation process, consistent with a miRNA-dependent mechanism. By contrast, the first order nature of PAI-1 mRNA decay in PlGF-treated cells suggested that under these conditions the turnover of PAI-1 mRNA was dependent only on mRNA concentration, subject to normal turnover of bulk cytoplasmic mRNA.
The regulation of PAI-1 expression is mainly achieved at the transcriptional level based on the presence of several binding sites for transcription factors within its promoter [8–10]. By contrast, the mechanisms involved in the post-transcriptional regulation of PAI-1 are poorly understood. Physiological stimuli such as transforming growth factor-β [12] and insulin [11] have been shown to regulate PAI-1 levels by involvement of post-transcriptional processes. PAI-1 mRNA harbors several 3′-AREs, which are involved in rapid mRNA destabilization and degradation [41]. cAMP has been shown to regulate PAI-1 mRNA stability through its AREs, however the identities of putative mRNA binding proteins remain unknown [42]. A recent study identified the role for ubiquitous RNA binding protein human-antigen R in angiotensin II mediated PAI-1 stabilization by binding to the ARE in a rat model of kidney fibrosis [43]. Nonetheless, the role of miRNAs in PAI-1 mRNA stability remains to be established.
MicroRNAs play an important role in stabilization and degradation of mRNAs [14–16, 44]. Herein, we successfully identified candidate miRNAs, which showed complementarity to binding sites within the 3′UTR of PAI-1 mRNA. The miRNA database revealed approximately 20 candidate miRNA binding sites in the PAI-1 mRNA. We selected five miRNA candidates (miR-30c, miR-301a, miR-99a, miR-299-5p and miR-609), which had the highest degree of species conservation with respect to potential target sites in PAI-1 mRNA 3′UTR, for experimental validation. In order to determine which of these candidate miRNAs actually regulated the expression of PAI-1 mRNA, we first determined the level of expression of these miRNAs in HPMVEC following PlGF treatment after 2 hr. The levels of miR-30c and miR-301a were significantly reduced in response to PlGF treatment, compared to untreated cells. The presence of miR299-5p and miR-609 was not detected by assay in HPMVEC under these conditions, while miR-99a showed no change. We further addressed the role of endogenous PlGF in the physiological regulation of these miRNAs by studying the consequences of reducing PlGF expression in the cells using PlGF siRNA. Our results showed increased levels of miR-30c and miR-301a in cells transfected with PlGF siRNA, supporting the concept that endogenous PlGF regulates basal expression of miR-30c and miR-301a. Thus, three miRNA candidates (miR-30c, miR-301a and miR-99a) were chosen for further study to determine whether they had a biological function in mediating post-transcriptional repression of PAI-1 expression. The effect of miRNA downregulation on PAI-1 mRNA was examined by transfection of HPMVEC with anti-miR-30c and anti-miR-301a. This resulted in augmented basal PAI-1 mRNA expression, while anti-miR-99a had no such effect. Consistent with these results, overexpression of these miRNAs in HPMVEC was performed by transfection with pre-miR-30c, pre-miR-301a or pre-miR-99a, singly. In the absence of PlGF, overexpression of miR-30c and miR-301, but not of miR-99a, resulted in reduced PAI-1 expression. In PlGF-induced cells, PAI-1 mRNA and protein expression induction was antagonized by overexpression of miR-30c and miR-301a.
Luciferase reporter assays were performed to confirm the predicted target sites of miR-30c, miR-301 on PAI-1 mRNA. The effect of individual miRNAs on luciferase reporter activity was antagonized by corresponding mutations within the PAI-1- 3′UTR of the luciferase reporters. A double mutation of the PAI-1-3′UTR construct for both miR-30c and miR-301a binding sites completely abrogated their destabilizing effect on reporter activity, indicating the involvement of both miRNAs in regulating PAI-1mRNA stability. However, as expected, the mutation of the putative miR-99a target site had no effect on reporter activity. Thus, our present study shows that miR-30c and miR-301a target PAI-1-3′UTR in a direct and sequence specific manner. Our results were further supported by miRNA overexpression; miR-30c and miR-301a, but not miR-99a, efficiently reduced reporter expression while corresponding anti-miR oligonucleotides antagonized endogenous miRNA activity and allowed upregulation of luciferase reporter activity. Our findings are in accordance with those of Natarajan and coworkers [44], who reported S100b mediated COX-2 mRNA stabilization at early time points occurred by reduction of miR-16 expression in THP-1 monocytic cells. Our results also parallel miR-409-3p mediated regulation of fibrinogen β mRNA expression, another important molecule in the blood coagulation cascade [45]. Moreover, studies are now emerging regarding the cellular signaling mechanisms and transcription factors involved in expression of miRNAs [46], which would provide another avenue for fine-tuning gene expression in cellular processes at the post-transcriptional level.
To understand the regulation of miR-30c and miR-301a, we examined their gene loci in the human genome (Genome Reference Consortium, version 37). miR-30c is located within a 3′-proximal intron of the NFYC gene (a transcription factor) on chromosome 1 in p34.2, while miR-301a is located in the 1st intron of SKA2 (spindle and kinetochore-associated protein 2) gene located on chromosome 17 at q22. The locations and orientation of miR-30c and miR-301a, are depicted in gene schematics shown in Supplementary Figure S3A and S3B, respectively. These show that both miRNAs can be co-synthesized from the respective NFYC and SKA2 transcription units, Thus, in the absence of any post-transcriptional regulation, synthesis of these miRNAs would be affected by transcriptional regulators of NFYC and SKA2. Indeed miR-301 has been reported to be expressed from a SKA2 transcript, and is observed to be a transcriptional regulator of SKA2 itself [47]. SKA2 protein is involved in mitotic spindle checkpoint silencing [47]. Thus, it will be of interest to determine how miR-301a and SKA2 are co-regulated by cis-elements in the SKA2 promoter.
NF-Y, a multimeric transcription factor, is regulated by cellular redox state, and plays an important role in the regulation of MHC class II gene expression [48]. NF-YC, one of the subunits of NF-Y, recognizes a CCAAT box motif found in promoter and enhancer elements and functions as a corepressor of agonist-bound mineralocorticoid receptor [49]. An in silico analysis of NF-YC intron where miR-30c and miR-30eare collocated, indicates the presence of several cryptic promoters and possible transcription factor binding sites (Supplementary Figure S3A). Thus, it is possible that both miR-30c and miR-30e originate from a single transcription unit, and are regulated independently of NFYC. Further studies are warranted to determine the synthetic origin of miR-30c, i.e. whether it originates from the NFYC transcription unit or from a smaller, intronic transcription unit encoding miR-30e and miR-30c.
In conclusion, PAI-1 expression can be regulated by a number of distinct transcription and post-transcriptional mechanisms. Our studies show that PlGF significantly reduces the levels of miR-30c and miR-301a in HPMVEC. The data obtained with inhibitor or mimics for miR-30c and miR-301a suggest that these miRs specifically promote degradation of PAI-1 mRNA in normal cells, while PlGF stabilizes PAI-1 mRNA expression by inhibiting expression of miR-30c and miR-301a. At present, we do not know whether PlGF increases degradation of these miRNAs (miR-30c and miR-301) or decreases their transcription to stabilize PAI-1 mRNA. Since a single miRNA potentially has many targets, it is possible that these identified miRNAs affect other distinct signaling molecules in signaling pathways, thus potentially regulating the expression of PAI-1. Because increased levels of PAI-1 in SCA can be one of the factors that contribute to a prothrombotic state, potentially predisposing patients to stroke and pulmonary hypertension, our studies uncovered a novel mechanism for the increased expression of PAI-1.
One possible scenario that may lead to abnormally high levels of PAI-1 in SCA would be that PlGF elaborated from erythroid cells keeps levels of miR-30c and miR-301a low. Indeed, our data in plasma of SCA subjects demonstrated significant down-regulation of miR-30c and miR-301a but not of miR-99a compared to healthy controls. In the same set of plasma, we observed a reciprocal relationship between PAI-1 and miR-30c and miR-301a levels in SCA. It will be interesting to validate these results in a larger sample population in order to develop these miRNAs as biomarkers for hypercoagulability in SCA patients. A precedent for this has been observed for specific miRNAs that are secreted during tumorigenesis [27]. Future studies in this direction are warranted. As a final thought, RNAi-based therapeutic approaches may be useful in ameliorating stroke and pulmonary hypertension in patients with SCA. Clinical trials using this approach for the purpose of attenuating viral infections and cancer in humans have ensued [50], suggesting the feasibility of a similar approach in SCA.
Supplementary Material
Acknowledgments
We thank Dr. Trine Fink (University of Aalborg, Aalborg, Denmark) for kindly providing PAI-1 promoter plasmid. We thank Dr. Gangning Liang (Keck School of Medicine, USC) for providing TaqMan MicroRNA assay for RNU6B. We thank the Hematology Repository at Cincinnati Children’s Hospital Medical Center for help with collection of patient blood samples and appropriate controls. We also thank the Institutional Core of USC Research Center for Liver Disease for the use of a sequence detection instrument (supported by National Institutes of Health Grant P30-DK 048522).
FUNDING
This work was partially supported by National Institute of Health Grant [CSCC-HL-070595] and [HL-079916] in addition to institutional support.
Abbreviations
- 3′UTR
3′ untranslated region
- AREs
adenosine- and uridine-rich elements
- HPMVEC
Human pulmonary microvascular endothelial cells
- miRNA
microRNAs
- PAI-1
Plasminogen activator inhibitor-1
- PlGF
Placenta Growth factor
- qRT-PCR
Quantitative real-time reverse transcription-PCR
- SCA
Sickle cell anemia
Footnotes
DISCLOSURES
The authors declare no competing financial interests.
AUTHOR CONTRIBUTION
Nitin Patel and Vijay Kalra devised the study and wrote the manuscript. Nitin Patel performed all of the experimental work. Stanley Tahara analyzed genomic locations of microRNA-30c and -301a. Punam Malik collected samples from SCA patients and normal controls. All authors edited the manuscript prior to submission.
References
- 1.Eddy AA. Plasminogen activator inhibitor-1 and the kidney. Am J Physiol Renal Physiol. 2002;283:F209–F220. doi: 10.1152/ajprenal.00032.2002. [DOI] [PubMed] [Google Scholar]
- 2.Fay WP. Plasminogen activator inhibitor 1, fibrin, and the vascular response to injury. Trends Cardiovasc Med. 2004;14:196–202. doi: 10.1016/j.tcm.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Ataga KI, Key NS. Hematology. American Society Hematology Educational Program; 2007. Hypercoagulability in sickle cell disease: new approaches to an old problem; pp. 91–96. [DOI] [PubMed] [Google Scholar]
- 4.Ataga KI, Moore CG, Hillery CA, Jones S, Whinna HC, Stayhorn D, Sohier C, Hinderliter A, Praise LV, Orringer EP. Coagulation activation and inflammation in sickle cell disease-associated pulmonary hypertension. Haematologica. 2008;93:20–26. doi: 10.3324/haematol.11763. [DOI] [PubMed] [Google Scholar]
- 5.Nsiri B, Gritli N, Mazigh C, Ghazouani E, Fattoum S, Machghoul S. Fibrinolytic response to venous occlusion in patients with homozygous sickle cell disease. Hematol Cell Ther. 1997;39:229–232. doi: 10.1007/s00282-997-0229-7. [DOI] [PubMed] [Google Scholar]
- 6.Hillery CA, Panepinto JA. Pathophysiology of stroke in sickle cell disease. Microcirculation. 2004;11:195–208. doi: 10.1080/10739680490278600. [DOI] [PubMed] [Google Scholar]
- 7.Dupont DM, Madsen JB, Kristensen T, Bodker JS, Blouse GE, Wind T, Andreasen PA. Biochemical properties of plasminogen activator inhibitor-1. Frontiers in Bioscience. 2009;14:1337–1361. doi: 10.2741/3312. [DOI] [PubMed] [Google Scholar]
- 8.Fink T, Kazlauskas A, Poellinger L, Ebbesen P, Zachar V. Identification of a tightly regulated hypoxia-response element in the promoter of human plasminogen activator inhibitor-1. Blood. 2002;99:2077–2083. doi: 10.1182/blood.v99.6.2077. [DOI] [PubMed] [Google Scholar]
- 9.Patel N, Sundaram N, Yang M, Madigan C, Kalra VK, Malik P. Placenta growth factor (PlGF), a novel inducer of plasminogen activator inhibitor-1 in sickle cell disease (SCD) J Biol Chem. 2010;285:16713–16722. doi: 10.1074/jbc.M110.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao H, Hyman MC, Lawrence DA, Pinsky DJ. Molecular regulation of the PAI-1 gene by hypoxia: contributions of Egr-1, HIF-1{alpha}, and C/EBP{alpha} FASEB J. 2007;21:935–949. doi: 10.1096/fj.06-6285com. [DOI] [PubMed] [Google Scholar]
- 11.Kietzmann T, Samoylenko A, Roth U, Jungermann K. Hypoxia-inducible factor-1 and hypoxia response elements mediate the induction of plasminogen activator inhibitor-1 gene expression by insulin in primary rat hepatocytes. Blood. 2003;101:907–914. doi: 10.1182/blood-2002-06-1693. [DOI] [PubMed] [Google Scholar]
- 12.Keeton MR, Curriden SA, van Zonneveld AJ, Loskutoff DJ. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 13.Vulin AI, Stanley FM. Oxidative Stress Activates the Plasminogen Activator Inhibitor Type 1 (PAI-1) Promoter through an AP-1 Response Element and Cooperates with Insulin for Additive Effects on PAI-1 Transcription. J Biol Chem. 2004;279:25172–25178. doi: 10.1074/jbc.M403184200. [DOI] [PubMed] [Google Scholar]
- 14.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 Regulates Human Angiotensin II Type 1 Receptor Expression in Fibroblasts. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 18.Ambros V. The function of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-{kappa}B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi Y, Liu G, Yang R. MicroRNAs: Novel regulators during the immune response. J Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 21.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 22.Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Ann N Y Acad Sci. 2010;1183:183–194. doi: 10.1111/j.1749-6632.2009.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A MicroRNA Signature of Hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the innate immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 27.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonsalves CS, Kalra VK. Hypoxia-Mediated Expression of 5-Lipoxygenase-Activating Protein Involves HIF-1{alpha} and NF-{kappa}B and MicroRNAs 135a and 199a-5p. J Immunol. 2010;184:3878–3888. doi: 10.4049/jimmunol.0902594. [DOI] [PubMed] [Google Scholar]
- 29.Patel N, Gonsalves CS, Yang M, Malik P, Kalra VK. Placenta growth factor induces 5-lipoxygenase-activating protein to increase leukotriene formation in sickle cell disease. Blood. 2009;113:1129–1138. doi: 10.1182/blood-2008-07-169821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang J, Ramu S, Lee S, Aguilar B, Ganesan SK, Yoo J, Kalra VK, Koh CJ, Hong YK. Phosphate-buffered saline-based nucleofection of primary endothelial cells. Anal Biochem. 2009;386:251–255. doi: 10.1016/j.ab.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel N, Kalra VK. Placenta growth factor (PlGF)-induced early growth response 1(Egr-1) regulateshypoxia-inducible factor-1a in endothelial cells. J Biol Chem. 2010;285:20570–20579. doi: 10.1074/jbc.M110.119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 34.Long J, Wang Y, Wang W, Chang BHJ, Danesh FR. Identification of MicroRNA-93 as a Novel Regulator of Vascular Endothelial Growth Factor in Hyperglycemic Conditions. J Biol Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Gazzar M, McCall CE. MicroRNAs Distinguish Translational from Transcriptional Silencing during Endotoxin Tolerance. J Biol Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauser S, Weich HA. A heparin-binding form of placenta growth factor (PlGF-2) is expressed in human umbilical vein endothelial cells and in placenta. Growth Factors. 1993;9:259–268. doi: 10.3109/08977199308991586. [DOI] [PubMed] [Google Scholar]
- 37.Tordjman R, Delaire S, Plouet J, Ting S, Gaulard P, Fichelson S, Romeo PH, Lemarchandel V. Erythroblasts are a source of angiogenic factors. Blood. 2001;97:1968–1974. doi: 10.1182/blood.v97.7.1968. [DOI] [PubMed] [Google Scholar]
- 38.Selvaraj SK, Giri RK, Perelman N, Johnson C, Malik P, Kalra VK. Mechanism of monocyte activation and expression of proinflammatory cytochemokines by placenta growth factor. Blood. 2003;102:1515–1524. doi: 10.1182/blood-2002-11-3423. [DOI] [PubMed] [Google Scholar]
- 39.Patel N, Gonsalves CS, Malik P, Kalra VK. Placenta growth factor augments endothelin-1 and endothelin-B receptor expression via hypoxia-inducible factor-1{alpha} Blood. 2008;112:856–865. doi: 10.1182/blood-2007-12-130567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundaram N, Tailor A, Mendelsohn L, Wansapura J, Wang X, Higashimoto T, Pauciulo MW, Gottliebson W, Kalra VK, Nichols WC, Kato GJ, Malik P. High levels of placenta growth factor in sickle cell disease promote pulmonary hypertension. Blood. 2010;116:109–112. doi: 10.1182/blood-2009-09-244830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montouri N, Rossi G, Ragno P. Post-transcriptional regulation of gene expression in the plasminogen activation system. Biol Chem. 2002;383:47–53. doi: 10.1515/BC.2002.005. [DOI] [PubMed] [Google Scholar]
- 42.Tillmann-Bogush M, Heaton J, Gelehrter T. Cyclic nucleotide regulation of PAI-1 mRNA stability. Identification of cytosolic proteins that interact with an a-rich sequence. J Biol Chem. 1999;274:1172–1179. doi: 10.1074/jbc.274.2.1172. [DOI] [PubMed] [Google Scholar]
- 43.Doller A, Gauer S, Sobkowiak E, Geiger H, Pfeilschifter J, Eberhardt W. Angiotensin II induces renal plasminogen activator inhibitor-1 and cyclooxygenase-2 expression post-transcriptionally via activation of the mRNA-stabilizing factor human-antifen-R. Am J Pathol. 2009;174:1252–1263. doi: 10.2353/ajpath.2009.080652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shanmugam N, Reddy MA, Natarajan R. Distinct Roles of Heterogeneous Nuclear Ribonuclear Protein K and microRNA-16 in Cyclooxygenase-2 RNA Stability Induced by S100b, a Ligand of the Receptor for Advanced Glycation End Products. J Biol Chem. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fort A, Borcel C, Migliavacca E, Antonarakis SE, Fish RJ, Neerman-Arbez M. Regulation of fibrinogen production by microRNAs. Blood. 2010;116:2608–2615. doi: 10.1182/blood-2010-02-268011. [DOI] [PubMed] [Google Scholar]
- 46.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 Modulates Wnt Signaling in Human Osteoblasts through a Positive Feedback Loop. J Biol Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396:978–982. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 48.Nakshatri H, Bhat-Nakshatri P, Currie RA. Subunit Association and DNA Binding Activity of the Heterotrimeric Transcription Factor NF-Y Is Regulated by Cellular Redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 49.Murai-Takeda A, Shibata H, Kurihara I, Kobayashi S, Yokota K, Suda N, Mitsuishi Y, Jo R, Kitagawa H, Kato S, Saruta T, Itoh H. NF-YC Functions as a Corepressor of Agonist-bound Mineralocorticoid Receptor. J Biol Chem. 2010;285:8084–8093. doi: 10.1074/jbc.M109.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.