Abstract
Recent genome-wide analyses have indicated that almost all primary transcripts from multi-exon human genes undergo alternative pre-mRNA splicing (AS). Given the prevalence of AS and its importance in expanding proteomic complexity, a major challenge that lies ahead is to determine the functional specificity of isoforms in a cellular context. A significant fraction of alternatively spliced transcripts are regulated in a tissue- or cell-type-specific manner, suggesting that these mRNA variants likely function in the generation of cellular diversity. Complementary to these observations, several tissue-specific splicing factors have been identified, and a number of methodological advances have enabled the identification of large repertoires of target transcripts regulated by these proteins. An emerging theme is that tissue-specific splicing factors regulate coherent sets of splice variants in genes known to function in related biological pathways. This review focuses on the recent progress in our understanding of neural-specific splicing factors and their regulatory networks and outlines existing and emerging strategies for uncovering important biological roles for the isoforms that comprise these networks.
Keywords: alternative splicing, RNA processing, gene regulation, genome-wide analysis
INTRODUCTION
The first application of cDNA microarrays to quantify mRNA levels represented a transformative milestone in gene expression research and marked the beginning of transcriptome analysis at an unprecedented scale (Schena et al. 1995). Initial experiments using microarrays in model organisms such as yeast revealed that the abundance of many transcripts can be regulated in a dynamic and concerted manner in various physiological contexts (DeRisi et al. 1997; Chu et al. 1998). The development of clustering approaches further revealed that genes exhibiting similar expression dynamics tend to be involved in similar biological processes, suggesting that potential functional correlations between genes could be inferred by analyzing coexpression patterns (Eisen et al. 1998). These concepts were subsequently extended to transcriptional networks in other organisms including mammals, highlighting the power of such approaches in understanding relationships between genes and their respective cellular pathways in diverse tissues and developmental states (Su et al. 2004; Zhang et al. 2004).
However, transcriptional control is only one of many modes in which the flow of genetic information can be regulated. Additional co- and post-transcriptional mechanisms can dictate how a pre-mRNA is processed, where it localizes as a mature transcript, and ultimately under what context it is translated (Moore and Proudfoot 2009). Genome-wide analyses of RNA-binding proteins (RBPs) and their target transcripts have revealed similar underlying principles to those found in the original microarray experiments. These and other observations have led to the development of the concept of RNA “regulons,” where transcripts from functionally related genes have evolved to be coordinately regulated by RBPs either co- or post-transcriptionally (Keene 2007). A better understanding of how these additional regulatory networks contribute to development will therefore be important.
Alternative splicing (AS) represents one such co- or post-transcriptional process that can increase proteomic complexity from a limited repertoire of genes (Chen and Manley 2009; Nilsen and Graveley 2010). Recent studies have suggested that AS is nearly ubiquitous in human transcripts and is frequently controlled in a tissue-specific manner (Pan et al. 2008; Wang et al. 2008). In particular, extensive tissue-specific AS has been observed in the mammalian nervous system and is thought to contribute to both its molecular and cellular complexity (Lipscombe 2005). It is thus not surprising that many studies of tissue-specific AS regulation have been directed at understanding splicing in the nervous system (Li et al. 2007). In conjunction with biochemical and molecular genetic methodologies, several genome-wide approaches have proven valuable in our understanding of how neural-specific splicing factors regulate AS in the nervous system (Blencowe 2006; Ule and Darnell 2007; Licatalosi and Darnell 2010). These approaches have also helped define some of the first AS regulatory networks, providing insights into their organizational principles and new insights regarding how specific groups of gene products function together in the developing and mature nervous system. This review discusses the progress in our understanding of neural splicing factors and their regulatory networks, and upcoming promises and challenges in this exciting area of gene expression research.
GENOME-WIDE APPROACHES FOR THE STUDY OF ALTERNATIVE SPLICING
Several microarray platforms developed in the past decade have enabled genome-scale analyses of pre-mRNA splicing. These platforms typically make use of combinations of short oligonucleotide probes that recognize exon and/or exon junction sequences (Calarco et al. 2007; Ben-Dov et al. 2008; Moore and Silver 2008). These microarrays have been instrumental in the large-scale profiling of splicing efficiency in wild-type and mutant strains of budding yeast (Clark et al. 2002; Pleiss et al. 2007), and in providing quantitative measurements of AS across tissues, during development, or upon disruption of splicing factor expression in metazoans (e.g., Johnson et al. 2003; Pan et al. 2004; Stolc et al. 2004; Blanchette et al. 2005; Ule et al. 2005; Sugnet et al. 2006; Boutz et al. 2007b; Fagnani et al. 2007; Barberan-Soler and Zahler 2008; Calarco et al. 2009; Hartmann et al. 2009). A number of general principles of AS regulation have been deduced from these studies, including evolutionary features associated with classes of commonly regulated alternative exons and comparisons of the dynamics and organization of AS and transcription networks (Blencowe 2006; Calarco et al. 2007).
More recently, next-generation sequencing technologies have been used to analyze polyadenylated transcripts at a remarkable degree of precision and sensitivity. In these approaches, cDNA fragments derived from mRNA populations are sequenced in a massively parallel manner (RNA-Seq) and aligned to reference transcriptomes (Pepke et al. 2009; Wang et al. 2009). RNA-Seq analyses profiling human cell lines and tissues reveal that >95% of multi-exon genes have more than one splice variant, and these isoforms are frequently differentially regulated across tissues (Pan et al. 2008; Wang et al. 2008). Additional recent studies have used this approach to identify novel and developmentally regulated splice variants in Drosophila and Caenorhabditis elegans (Gerstein et al. 2010; Graveley et al. 2010; Ramani et al. 2011). Advantages of RNA-Seq over microarray-based approaches are that novel exons and splice junctions can be detected without a priori knowledge, and cross-hybridization artifacts are not an issue. However, the alignment of short oligonucleotide reads to the transcriptome is not trivial, and RNA-Seq analyses are currently less cost-effective than microarray experiments. Despite these current limitations, read length and depth of coverage are continuing to improve, and new software for analyzing splicing from RNA-Seq data is constantly being developed. Currently, both microarray and RNA-Seq approaches are actively used for genome-wide analysis of AS (Fig. 1A), but RNA-Seq is quickly becoming the method of choice (Blencowe et al. 2009; Wang et al. 2009).
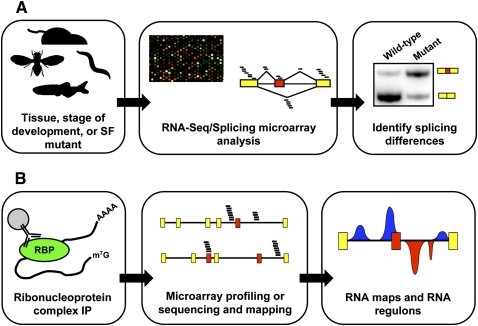
FIGURE 1.
Experimental approaches for genome-wide analysis of alternative splicing. (A) Quantitative splicing microarray profiling and RNA-Seq (middle panel) can be used to measure splicing efficiency and relative isoform usage across different tissue types, stages of development, in response to environmental stimuli, or in wild-type and mutant organisms (left panel). Predictions stemming from either approach can then be confirmed with experimental techniques such as semi-quantitative reverse transcription and PCR (RT-PCR; right panel). In microarray profiling experiments, short oligonucleotide probes annealing to exon body and exon junction sequences are used to monitor alternative splice site or exon usage. In RNA-Seq, short oligonucleotide reads are aligned to exon body and junction sequences, and the number of mapped reads can then be quantified to assess alternative splicing patterns. SF stands for splicing factor. (B) Immuno-affinity-based approaches such as ribonucleoprotein immunoprecipitation (RIP) and cross-linking and immunoprecipitation (CLIP) can be used to purify RNA-binding protein:pre-mRNA complexes (left panel). The purified RNA can then be sequenced, followed by alignment of the resulting short nucleotide reads to a reference genome of interest (middle panel). These genome-wide binding data provide a snapshot of the repertoire, or “regulon,” of a particular RNA-binding protein and can be used to infer functional relationships among genes encoding target transcripts. Alternatively, these data can be combined with RNA-Seq and microarray profiling data to obtain RNA maps (right panel) that correlate binding site positions with splicing regulatory differences observed upon loss or depletion of a given splicing factor.
Complementary to the approaches described above, AS regulatory networks can also be uncovered by identifying transcripts physically associated with specific splicing factors and ribonucleoprotein (RNP) complexes (Fig. 1B). In these approaches, an RNA-binding protein and its associated RNP complex is immunoprecipitated from a cell lysate, followed by purification and detection of bound RNA transcripts. RNP complexes can be purified either under native conditions (Tenenbaum et al. 2000) or under more stringent conditions if cross-linking methods are used (Niranjanakumari et al. 2002; Ule et al. 2003). The former approach has the advantage that all transcripts in an RNP complex can be identified. However, this approach runs the risks of losing low-affinity yet specific in vivo interactions and also transcript “reassociation,” in which transcripts that do not normally associate with an RNP complex bind during or following cell extraction (Mili and Steitz 2004). Methods involving cross-linking circumvent this problem because stringent purification conditions can be used but, depending on the cross-linking reagent employed, may only afford detection of transcripts that are directly bound by the factor. In the most recent incarnation of these latter methods, known as cross-linking and immunoprecipitation (CLIP), the associated RNA is partially digested during the protocol, leaving short associated fragments that represent in vivo binding sites recognized by the RNA-binding protein of interest (Jensen and Darnell 2008). Additional modifications to this protocol have further increased its resolution (Hafner et al. 2010; Konig et al. 2010). Recently, these approaches have been coupled with microarray profiling or high-throughput sequencing to obtain genome-wide snapshots of the repertoire of transcripts and binding sites associated with specific splicing factors (e.g., Keene et al. 2006; Licatalosi and Darnell 2010).
Bioinformatics methods play an important role in the analysis of the large catalog of splicing events identified by the experimental approaches described above. Such analyses have facilitated the discovery of splicing enhancer and silencer elements associated with constitutive and alternative exons (Fairbrother et al. 2002; Zhang and Chasin 2004; Yeo et al. 2007). Computational approaches have also been successful in identifying cis-regulatory elements enriched in tissue-specific AS events (Sugnet et al. 2006; Fagnani et al. 2007). Additional studies have also begun to identify relationships between groups of cis-elements, other factors influencing regulated splicing, and insights into the evolutionary dynamics of AS (Yeo et al. 2004; Xing and Lee 2006; Xiao et al. 2007; Friedman et al. 2008; Barash et al. 2010; Ke and Chasin 2010; Keren et al. 2010; Zhang et al. 2010). More recently, the utility of computational approaches has been further exemplified in the analysis of genome-wide chromatin immunoprecipitation (ChIP) data sets, which have revealed interesting connections between histone modifications and splicing (Kolasinska-Zwierz et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009; Huff et al. 2010; see below).
Collectively, the above techniques have become an indispensable set of tools in the analysis of splicing regulators. In the following section, we discuss how these approaches have been used to advance our understanding of tissue-specific splicing factors and the sets of AS events they regulate in the nervous system.
NOVA-1 AND NOVA-2
The first neuro-oncological ventral antigen (Nova-1) protein was originally identified as a factor recognized by antibodies produced in patients with paraneoplastic opsoclonus myoclonus ataxia (POMA), a motor control disorder resulting from an autoimmune response (Buckanovich et al. 1993). The Nova-1 antigen displays restricted expression in the nervous system, specifically in the hypothalamus, ventral midbrain, hindbrain, and spinal cord. Nova-2, a second antigen highly related to Nova-1, was subsequently identified and found to be expressed specifically in brain subregions, but with a reciprocal pattern of expression to Nova-1 (Yang et al. 1998). Both Nova proteins share sequence homology with the RNA-binding protein hnRNP K (Buckanovich et al. 1993), alluding to their role in RNA processing.
Initial biochemical, crystallographic, and molecular genetic-based approaches indicated that the Nova proteins bind YCAY repeat sequences, and a small number of target transcripts were identified based on the presence of these elements (Buckanovich and Darnell 1997; Lewis et al. 2000; Dredge and Darnell 2003). A significant breakthrough was brought forth by the development of a Nova-1 knockout mouse. These knockout mice die shortly after birth from motor deficiencies caused by apoptosis of brainstem and spinal cord neurons and display a number of phenotypes similar to those found in POMA (Jensen et al. 2000). Intriguingly, the loss of Nova-1 altered the neuron-specific alternative splicing patterns of target transcripts containing YCAY repeat elements. These results established Nova-1 as the first tissue-specific splicing regulator contributing to a developmental program and further implicated this factor as a causative agent in neurological disorders.
The functional analysis of the Nova proteins was greatly facilitated by the development of CLIP (Ule et al. 2003). The initial application of CLIP identified 34 target mRNAs associated with the Nova proteins. These target transcripts were enriched in genes with known roles at the synapse, in particular, inhibitory synaptic transmission. Ule and colleagues further analyzed mRNA samples from brain and immune system tissues of wild-type and Nova-2 knockout mice using AS microarrays (Ule et al. 2005). More than 50 AS events from the neocortex were identified that required Nova-2 for proper regulation, corresponding to ∼7% of all brain-specific AS events detected in their profiling experiments. This further suggested that Nova-2 plays a significant role in regulating brain-specific AS. In agreement with the CLIP analysis, a significant proportion of genes with Nova-2-regulated AS events were associated with functions at the synapse, in particular, synaptic development, synaptic transmission, cell–cell signaling, and cortical actin organization. Intriguingly, many of the target genes with regulated AS events interact with each other in protein complexes, suggesting that Nova-2 regulates a network of synaptic proteins and that it plays an important role in affecting physical interactions between these factors.
Using this set of AS events, a bioinformatics approach was used to examine the effects on exon inclusion resulting from the positional binding of the Nova proteins to YCAY repeats in regions surrounding these alternative exons (Ule et al. 2006). The resulting “RNA map” predicted that Nova could act as either an enhancer or a repressor of exon inclusion depending on the locations of the YCAY elements in pre-mRNA transcripts. These predictions were subsequently validated experimentally and led to testable hypotheses regarding the mechanism by which Nova regulates AS. Specifically, when bound to exonic YCAY elements, Nova blocks U1 snRNP association with the 5′ splice site. However, when bound in the intronic region downstream from an alternative exon, Nova stimulates splicing complex formation. These results indicated how genome-wide approaches can provide key regulatory insights into splicing mechanisms. As new technologies become available, our ability to uncover novel insights continues. For example, Licatalosi et al. (2008) subsequently coupled the CLIP protocol with high-throughput sequencing (HITS-CLIP, also referred to as CLIP-Seq) and also used a more comprehensive AS microarray to identify an extensive Nova regulatory network. In addition to confirming and further refining the bioinformatically predicted RNA map, the investigators identified a new role for Nova in regulating alternative polyadenylation. This new regulatory role for Nova appears distinct from its role in AS, since transcripts regulated at the level of poly(A) site selection generally do not have Nova-regulated alternative exons.
PTB and PTBP2
The presence of a tissue-restricted counterpart of the ubiquitously expressed polypyrimidine tract binding protein (PTB, also known as PTBP1 or hnRNP I) was initially detected in rat brain extracts based on its different electrophoretic mobility but similar RNA-binding activity to PTB (Ashiya and Grabowski 1997). Two independent studies subsequently identified and cloned PTBP2 (also known as nPTB and brPTB), confirming that it is a tissue-restricted paralog of PTB (Markovtsov et al. 2000; Polydorides et al. 2000). Markovtsov et al. (2000) identified PTBP2 as a key factor playing a role in the neuronal-specific splicing of the N1 alternative exon from the c-src transcript. Polydorides et al. (2000) identified PTBP2 as a protein that interacts with Nova-1/2, and further demonstrated its ability to antagonize the stimulatory effect of Nova proteins in the splicing of exon E3A in GlyRα2 transcripts. Both studies revealed an enriched expression pattern of PTBP2 in brain and testis, suggesting that this factor might act as a key modulator of alternative splicing patterns in these tissues.
Although PTBP2 has high sequence similarity to PTB, by virtue of its restricted expression pattern and other properties, it has the ability to establish tissue-specific splicing programs. Data from in vitro splicing assays indicate that PTBP2 can act either neutrally or as a weaker repressor than PTB in preventing c-src N1 exon inclusion (Markovtsov et al. 2000). Additionally, PTBP2 binds to an intronic region of the c-src pre-mRNA critical for tissue-specific splicing of exon N1, known as the distal control sequence, with higher affinity than PTB, and this binding is stimulated by generally expressed cofactors such as hnRNP H (Markovtsov et al. 2000). These results and others have suggested that a combination of differential intrinsic RNA-binding affinity and interactions with coregulators likely play a role in the distinct properties of PTB and PTBP2 (Charlet et al. 2002; Gromak et al. 2003). Further mechanistic studies on the repressive role of PTB in a non-neuronal context have revealed that intron-bound PTB can block pre-spliceosomal E complex formation by preventing 5′-splice-site-dependent recruitment of U2AF, and also blocking the transition from exon definition to intron definition (Sharma et al. 2005, 2008). In light of this evidence, it is possible that PTBP2 somehow acts distinctly at these steps in spliceosome assembly and commitment. Since the above studies have been based on specific model pre-mRNA substrates, additional work is required to help determine the generality of these principles.
Additional focused and genome-wide analyses of PTB and PTBP2 have shed light on the mechanism by which these factors regulate AS. Boutz et al. (2007b) demonstrated that both proteins display mutually exclusive expression patterns in the brain, with PTB being expressed in glial and non-neuronal cells, and PTBP2 in neurons. When PTB is present at sufficient levels, it represses splicing of exon 10 in PTBP2 transcripts, producing an isoform containing a premature termination codon (PTC) that is degraded by the nonsense-mediated mRNA decay (NMD) pathway (Boutz et al. 2007b; Spellman et al. 2007). PTB also appears to further inhibit PTBP2 expression or turnover at the protein level independent of its effect on splicing of exon 10. Thus, down-regulation of PTB should lead to the expression of functional PTBP2. In neurons, the silencing of PTB expression is achieved by the neuron-specific micro-RNA miR-124, which effectively derepresses PTBP2 exon 10 splicing, although efficient inclusion of this exon in neurons also requires positive-acting splicing regulators (Makeyev et al. 2007; Calarco et al. 2009; see below). This mutually exclusive expression pattern appears to be established after cells have committed to a neuronal fate, because neural progenitor cells appear to express both PTB and PTBP2 (Boutz et al. 2007b).
To assess functional consequences resulting from modulating PTB and PTBP2 levels in a neuronal context, Boutz et al. (2007b) performed AS microarray profiling experiments comparing the transcriptomes of neuroblastoma cells depleted of each splicing factor alone or in combination. These experiments revealed AS events with altered splicing patterns that depended solely on PTB or PTBP2 expression, as well as events that required both factors for regulation. Notably, events showing altered AS patterns undergo either increased inclusion or increased skipping, suggesting that these factors can act both as repressors or activators of splicing. Additional genome-wide experiments such as CLIP-Seq in the same neuroblastoma cells or even relevant neural tissues will help confirm whether these regulated events are direct targets. Additionally, a subset of the genes encoding transcripts with AS events regulated by PTB and PTBP2 have previously known roles in neuronal differentiation and function, providing a first glimpse into the splicing network regulated by these factors.
Two recent genome-wide analyses have provided further insights into the repertoire of transcripts controlled by PTB and PTBP2 and the mechanisms by which they operate. Xue et al. (2009) performed CLIP-Seq in HeLa cells to identify sites in the transcriptome where PTB binds in vivo. These binding sites were often found in introns, and enriched in pyrimidine-rich sequence elements. Intriguingly, an examination of the effect of shRNA-mediated knockdown of PTB revealed that PTB can also act as an enhancer of exon inclusion when its intronic binding sites closely flank neighboring constitutive exons. In another study, Llorian et al. (2010) knocked down PTB and PTBP2 expression in HeLa cells and used Affymetrix splicing microarrays to identify hundreds of AS events regulated by these factors. Analysis of these PTB/PTBP2-dependent AS events using an AS microarray data set from a separate study profiling diverse human tissues (Castle et al. 2008) revealed that they were often differentially regulated in nervous system and muscle tissues. These results are consistent with observations that pyrimidine-rich motifs are often associated with brain-specific AS events in mouse (Fagnani et al. 2007). Notably, these differentially regulated AS events are associated with genes enriched in G-protein signaling and regulation of the cytoskeleton and membrane traffic. An investigation of cis-elements enriched in PTB-regulated transcripts identified pyrimidine-rich motifs that exhibited positional bias depending on the regulatory effect. For instance, PTB was found to act as a repressor when bound in an intron upstream of an alternative exon or within the alternative exon itself, but can function as an activator when bound to an intronic element downstream from the alternative exon (Llorian et al. 2010). Finally, additional motifs belonging to the Fox and MBNL family of splicing regulators were also significantly associated with PTB-regulated AS events, suggesting that these regulators more generally act in combination with PTB and PTBP2, in agreement with previous studies on model pre-mRNA transcripts (Jin et al. 2003; Ladd et al. 2005; Underwood et al. 2005). The proposed RNA maps from Xue et al. (2009) and Llorian et al. (2010) do not perfectly overlap, likely due to the different methods from which they were derived. However, they have each provided novel mechanistic insights into PTB/PTBP2 AS regulation that warrant further investigation. Collectively, these studies have also elaborated the repertoire of tissue-specific AS events controlled by these factors.
nSR100/SRRM4
Members of the SR family and SR-related splicing factor proteins were first identified through genetic analyses of the sex-determination pathway in Drosophila, and subsequently by their ability to influence splicing activity in vitro in HeLa extracts (Amrein et al. 1988; Goralski et al. 1989; Ge and Manley 1990; Krainer et al. 1990). Since their initial discovery, many additional members have been identified, and SR and SR-related proteins currently represent a major class of constitutive and alternative splicing regulators (Lin and Fu 2007; Long and Caceres 2009). A hallmark of these splicing factors is a protein domain consisting of stretches of alternating arginine/serine dipeptide repeats (known as the RS domain), implicated in facilitating important protein–protein and protein–RNA interactions during spliceosome recruitment and catalysis (Wu and Maniatis 1993; Shen and Green 2004). SR proteins also possess one or two RNA recognition motifs (RRMs), allowing them to interact with pre-mRNAs in a sequence-specific manner. SR-related proteins like SRm160 and SRm300 do not possess canonical RRMs and, instead, are believed to function by bridging interactions across splice sites by binding to components of the splicing machinery and with other SR proteins (Blencowe 2000; Sharma et al. 2008). Nearly all SR family and SR-related proteins are expressed ubiquitously but at varying levels across tissues, and it has been proposed that the variation in expression of these factors contributes to tissue-specific splicing decisions (Zahler et al. 1992; Hanamura et al. 1998; Chen and Manley 2009). Recent evidence suggests that a subset of RS domain proteins with highly restricted expression patterns exists and regulates networks of tissue-specific AS events.
Using a computational approach, we created a database of RS-domain protein genes and mapped the expression profiles of these factors using existing microarray data sets (Calarco et al. 2009). The neural-specific SR-related protein of 100 kDa (nSR100, also known as SRRM4) was identified from this analysis on the basis of its specific and robust expression in brain and sensory organ tissues. We subsequently demonstrated that depletion of nSR100 prevented both neurite outgrowth in differentiating neuroblastoma cells as well as neural stem cell formation and maintenance. Consistent with these neural differentiation defects observed in vitro, reduced expression of nSR100 in developing zebrafish embryos impaired neurite outgrowth and nervous system development in vivo. Alternative splicing microarray profiling revealed that nSR100 regulates a network of exons that are spliced in a nervous-system-specific manner, and that it acts primarily as an activator of exon inclusion. Analysis of this splicing network revealed that genes regulated by nSR100 are enriched in processes associated with cytoskeleton regulation and neural differentiation, suggesting that the aberrant regulation of these events may be responsible for the observed phenotypes.
Further investigation of this network of AS events identified several cis-regulatory elements enriched in the intronic regions flanking nSR100-regulated exons (Calarco et al. 2009). Most prominently enriched were pyrimidine-rich elements, known to be also recognized by PTB and PTBP2. Interestingly, nSR100 promotes the inclusion of exon 10 in PTBP2 transcripts, which, as described above, modulates the levels of PTBP2 through an NMD-dependent mechanism (Boutz et al. 2007b; Spellman et al. 2007), indicting an intricate interplay between these AS regulatory factors. Consistent with these observations, both PTB and PTBP2 were found to regulate nearly all nSR100-dependent AS events. However, the effects of nSR100 on AS could not be explained solely by its influence on PTBP2 levels, because additional biochemical evidence demonstrated that nSR100 can directly and specifically promote the inclusion of its target alternative exons. Future work will be directed at better understanding how nSR100 works together with PTB, PTBP2, and other splicing regulators to regulate neural-specific AS in vivo.
The discovery that nSR100 acts as an AS regulator indicates that a subset of SR and SR-related proteins represent a novel class of tissue-specific splicing regulators. Indeed, our database of RS-domain protein genes has identified candidate additional members of this family that display cell- or tissue-restricted expression patterns (Calarco et al. 2009). Additional work aimed at understanding the role of these other RS-domain-containing genes in cellular differentiation and development will be important. Additionally, nSR100 is only conserved in vertebrate species, in stark contrast with other SR and SR-related proteins that are conserved across metazoans (Barbosa-Morais et al. 2006). The relatively recent evolution of nSR100 suggests that it may have evolved to contribute to more complex aspects of vertebrate nervous system development and function. It will be interesting whether other tissue-restricted RS-domain genes also display similar evolutionary features.
Fox splicing regulators
Unlike most cis-elements recognized by RNA-binding proteins that are typically degenerate, several studies identified the conserved hexanucleotide sequence (U)GCAUG enriched in introns downstream from brain-specific alternative exons (Brudno et al. 2001; Minovitsky et al. 2005). It was subsequently discovered that a family of tissue-specific RNA-binding proteins, homologous with the C. elegans feminizing on X (FOX-1) protein involved in dosage compensation, regulated tissue-specific AS events through interaction with this cis-element (Jin et al. 2003; Nakahata and Kawamoto 2005; Underwood et al. 2005; Ponthier et al. 2006). Both mammalian Fox-1 (also known as A2BP1) and Fox-2 (also known as Fxh or RBM9) show enriched or highly restricted expression in brain, heart, and skeletal muscle tissues. A third member of the family with similar RNA-binding specificity, Fox-3, has recently been identified as the antigen recognized by the anti-NeuN (neuronal nuclei) antibody, which for nearly two decades has served as a specific marker of most classes of post-mitotic neurons (Kim et al. 2009). A number of molecular genetic and biochemical studies on a small number of pre-mRNA substrates have indicated that the Fox family of splicing regulators can act as position-dependent splicing activator or repressor proteins (Jin et al. 2003; Nakahata and Kawamoto 2005; Underwood et al. 2005; Ponthier et al. 2006). However, it remained unclear whether these observations could generally be applied to Fox-dependent splicing regulation. An understanding of the network of transcripts regulated by these splicing regulators would yield novel insights into mechanisms that govern the development of neurons and muscle cells. Recent genome-wide analyses have begun to shed light on some of these questions.
Zhang et al. (2008a) mined 28 sequenced vertebrate genomes for Fox-binding sites in exons and 200 nt of flanking intron sequence. Thousands of putative targets were identified, hundreds of which are found in the vicinity of known alternative exons. AS patterns of candidate Fox-associated transcripts across tissues were further assessed by microarray profiling experiments, associating the location of Fox-binding sites with potential tissue-specific regulation in brain and muscle tissues. The results suggest that Fox-binding sites are more often enriched in intronic regions downstream from alternative exons, and when present allow Fox proteins to activate inclusion of alternative exons. However, when binding sites are present in upstream intronic regions, Fox generally acts as a splicing repressor. Intriguingly, complex regulatory patterns were also observed in which candidate Fox-dependent AS events displayed increased inclusion or skipping in either brain or muscle tissues, but not in both, indicating that combinatorial interactions with other splicing regulators play a role in regulating these events. GO enrichment analysis of the Fox splicing network revealed that genes with Fox-regulated AS events have a role in neuromuscular function, consistent with the tissues where these factors are enriched. Additionally, genes with Fox-dependent AS events were frequently associated with diseases, including several neurological, heart, and muscular disorders. Mutations in the Fox-1 gene or alteration of its expression has been associated with several neurological disorders and heart disease (Kaynak et al. 2003; Bhalla et al. 2004; Szatmari et al. 2007), thus making the identified splicing regulatory network an excellent data set to identify potentially interesting relationships between RNA targets and the etiology of some of these diseases.
In another study, Yeo et al. (2009) observed that Fox-2 is robustly expressed in human embryonic stem cells (hESCs). The investigators performed CLIP-Seq experiments to identify Fox-2-binding sites in vivo. Fox-2-binding clusters were identified in nearly 2000 protein-coding genes, suggesting a large network of targets potentially regulated by this factor in hESCs. Interestingly, further analysis of the CLIP tag clusters found enrichment of not only the UGCAUG consensus Fox-binding motif, but also other consensus elements, suggesting potential alternative modes of RNA recognition through their interactions with additional factors. Investigating the effects on splicing patterns of interacting transcripts, the authors found that downstream intronic binding sites lead to exon inclusion, while upstream intronic sites lead to repression by Fox-2, consistent with the results of Zhang et al. (2008a). Genes encoding transcripts bound by Fox-2 were enriched in splicing factors and kinases, suggesting that this protein may act to coordinate a network of splicing events controlled by multiple factors, and also signaling cascades important for maintenance of the ESC state. Supporting this notion, the investigators observed that knockdown of Fox-2 leads to a specific and dose-dependent increase in cell death of hESCs. Thus, the network of targets regulated by Fox-2 will likely provide insights into stem cell biology.
Two recent AS profiling studies have also found a connection between the Fox family of splicing regulators and breast and ovarian cancer (Venables et al. 2009; Lapuk et al. 2010). These studies implicate the altered regulation of Fox-dependent splicing networks in various cancers, and analysis of these networks should uncover how these isoforms contribute to the development and progression of the disease.
Sam68
Sam68 (Src-associated in mitosis, 68 kDa) was originally identified as a phosphorylation substrate of the tyrosine kinase Src during mitosis and subsequently found to be related to a family of RNA-binding proteins now known as STAR (signal transduction and activation of RNA) or GSG (GRP33, Sam68, GLD-1) domain proteins (Fumagalli et al. 1994; Taylor and Shalloway 1994; Vernet and Artzt 1997). Sam68 is proposed to play roles in numerous aspects of mRNA metabolism, including AS, mRNA export and localization, and translation regulation (Lukong and Richard 2003). Several studies have also suggested links between this factor and various cancers (Rajan et al. 2008). Although expressed in multiple tissues, Sam68 expression is induced at sites of neurogenesis in mice (Lim et al. 2006). Mice lacking Sam68 display motor coordination deficits and male sterility, suggesting that this factor is involved in aspects of neuronal function and fertility (Lukong and Richard 2008; Paronetto et al. 2009). Given these intriguing phenotypes, it has been an important goal to identify the network of transcripts regulated by Sam68. Several studies have identified mRNAs bound to Sam68 both in neurons and in spermatocytes (Grange et al. 2009; Paronetto et al. 2009). Additionally, a small number of Sam68-dependent AS events from single-gene analyses have been discovered (Matter et al. 2002; Paronetto et al. 2007; Pedrotti et al. 2010).
To better define the network of AS events regulated by Sam68, AS microarray profiling was recently performed in mouse neuroblastoma cells treated with shRNAs targeting Sam68 expression (Chawla et al. 2009). From more than 1000 profiled AS events, about 30 Sam68-dependent AS events were identified. Consistent with other studies of splicing regulators, Sam68 appears to function both as an enhancer or repressor of exon inclusion. A search for previously identified AU-rich motifs known to bind Sam68 revealed an enrichment of these cis-elements in intronic regions flanking Sam68-regulated alternative exons. Mutagenesis experiments studying a Sam68-regulated exon in epsilon sarcoglycan (Sgce) transcripts demonstrated that multiple Sam68-binding sites are important in order to achieve full regulation by the RNA-binding protein.
Given that Sam68 is activated during neurogenesis, Chawla et al. (2009) used P19 cells, which can be differentiated into neuronal-like cells, to assess the role of Sam68 in neuronal differentiation. Sam68 expression was found to be induced during P19 cell differentiation, and AS events from the identified Sam68 regulatory network were found to undergo splicing changes during this process. Intriguingly, depletion of Sam68 inhibits P19 differentiation and alters the regulation of these differentiation-dependent AS events. Thus, this work has uncovered an important network of isoforms that contributes to aspects of neuronal differentiation. It will be interesting to determine whether any of these targets are causal in the motor coordination deficits in Sam68 null mice.
Additional neural-specific splicing regulators
Several additional neural-restricted RNA-binding proteins have been found to regulate AS, but to date no genome-wide analyses of these factors have been conducted in tissues of the nervous system. Mammalian Hu proteins, homologs of the Drosophila splicing regulator ELAV, are represented by four family members, three of which (HuB, HuC, and HuD) are highly expressed in nervous system tissues (Hinman and Lou 2008). Similar to the Nova proteins, the Hu proteins were also identified as neuronal antigens recognized by antibodies produced in patients with paraneoplastic disorders (Szabo et al. 1991). Furthermore, HuD knockout mice display a number of phenotypes both during development and in adulthood, including impaired motor and reflex skills, and aberrant control of neural progenitor cell proliferation and differentiation (Akamatsu et al. 2005). Work on these factors has indicated that they recognize AU-rich sequence elements and are involved in many aspects of mRNA metabolism, including AS (Hinman and Lou 2008).
Two additional classes of RNA-binding proteins with family members expressed specifically in nervous system and/or muscle tissues are the muscleblind-like (MBNL) and CUGBP/ETR-like (CELF; also known as Bruno-like) factors (Barreau et al. 2006; Pascual et al. 2006). These factors have been implicated in neurological disorders and in CUG trinucleotide expansion diseases such as myotonic dystrophy (Cooper et al. 2009; Gallo and Spickett 2010). These factors are also involved in several aspects of mRNA metabolism, including AS (Ladd et al. 2001; Ho et al. 2004). Several studies using model pre-mRNA substrates have demonstrated that MBNL and CELF bind CUG-repeat and UG-rich elements, respectively, and typically antagonize each other and other factors such as PTB in AS regulation (Charlet et al. 2002; Ho et al. 2004). Recent genome-wide analyses of regulated AS during heart development and myogenic differentiation have revealed that MBNL and CELF proteins can be dynamically and reciprocally regulated (Kalsotra et al. 2008; Bland et al. 2010). The altered expression of these factors helps coordinate a network of splicing transitions that take place between embryonic and adult heart tissue or between undifferentiated and differentiated myoblasts. Work from Dembowski and Grabowski has further demonstrated that CUGBP2 (also known as NAPOR, ETR-3, CELF2, and BRUNOL3) can regulate neural-specific skipping of the NI exon in NMDA R1 receptor transcripts by binding to UG-rich cis-elements flanking the branchpoint sequence upstream of the alternative exon (Dembowski and Grabowski 2009). Subsequent computational analyses identified additional exons regulated according to this branchpoint “perimeter” binding model, suggesting that this mode of regulation by CELF factors may be a general mechanism by which they regulate exon skipping in the brain.
More than a decade ago, the Quaking (qk) gene was identified as the locus mutated in Quaking mice, which exhibit rapid tremors due to a deficiency in myelination of the nervous system (Ebersole et al. 1996). The qk locus encodes several alternative isoforms of QKI, which are related to the STAR family of RNA-binding proteins. Several lines of evidence have connected QKI proteins with regulating AS of transcripts associated with myelination. For instance, the nuclear isoform QKI-5 represses the inclusion of exon 12 in myelin-associated glycoprotein (MAG) transcripts, which encode a transmembrane protein known to play a role in the initiation and maintenance of the myelin sheath (Wu et al. 2002). Additionally, QKI proteins can indirectly regulate AS through the modulation of other splicing regulators. Specifically, QKI-6, a cytoplasmic variant encoded by the qk locus, represses the translation of hnRNPA1, which, in turn, regulates MAG exon 12 AS (Zhao et al. 2010). Several other transcripts encoded by genes relevant to myelin formation and maintenance display altered splicing patterns in Quaking mice (Wu et al. 2002). It will be interesting to determine whether similar regulatory mechanisms also apply to other QKI targets. As aberrant regulation of QKI in humans has been linked to schizophrenia (Aberg et al. 2006a,b), an in-depth analysis of the repertoire of transcripts regulated by these factors may provide helpful insights into the disease.
Taken together, an understanding of the network of splicing events regulated by these proteins and a better characterization of the mechanisms by which they regulate AS will help to elucidate the physiological roles of these RNA-binding proteins in nervous system development and function.
FUTURE PROMISES AND CHALLENGES AHEAD
Identification of new neural splicing regulators
Although several neural-restricted splicing regulators have been identified, it is unlikely that these factors alone account for the prominent complexity generated by AS in the nervous system. Additional factors that regulate AS in the nervous system likely exist, and their detailed characterization will be important. Several strategies have emerged over recent years to facilitate the identification of new AS regulators. These general strategies are applicable to studies of neural AS regulation. For instance, large-scale gene expression and in situ hybridization studies have been conducted in a number of mammalian and vertebrate organisms (Su et al. 2004; Thisse et al. 2004; Zhang et al. 2004; McKee et al. 2005; Lein et al. 2007). Intriguingly, in one of these studies, it was found that many RNA-binding proteins are variably expressed in different brain regions, suggesting specialized roles for these factors (McKee et al. 2005). These studies have provided valuable data sets that can be mined for RNA-binding proteins and other proteins involved in pre-mRNA processing with restricted expression patterns in the nervous system, and will help focus efforts on characterizing additional AS regulators of interest.
Several luciferase- and fluorescent protein-based reporters that provide a readout of AS patterns for specific transcripts have recently been developed (Wang et al. 2004; Levinson et al. 2006; Orengo et al. 2006; Stoilov et al. 2008; Warzecha et al. 2009; Moore et al. 2010; Younis et al. 2010). These reporters have facilitated large-scale RNAi, cDNA overexpression, or small-molecule screens to uncover proteins and compounds that can modulate AS under a variety of conditions and cellular states. These reporters have also been used in model organisms such as mouse and C. elegans with the goal of identifying factors regulating tissue- and developmental-stage-specific AS events (Bonano et al. 2007; Kuroyanagi et al. 2010). We have recently applied these fluorescent reporters to analyze AS in the C. elegans nervous system and have observed a remarkable degree of regulatory complexity, with differential splicing between individual neuronal subtypes (JA Calarco, M Zhen, and BJ Blencowe, unpubl.). Large-scale forward or reverse genetic screens making use of such reporters in model organisms, as well as in cellular models of neural differentiation, will serve as excellent platforms for future discovery of neural splicing regulators. Moreover, the unbiased nature of these screens will help identify additional factors that may directly or indirectly contribute to splicing regulation, such as chromatin-modifying factors, signaling proteins, and non-coding RNAs, which are all known to influence AS (Lynch 2007; Allemand et al. 2008; Khanna and Stamm 2010; Luco et al. 2010; Tripathi et al. 2010).
Finally, biochemical analyses of RNP complexes have been greatly facilitated by advances in mass spectrometry over the past few years. These techniques have allowed the identification of components of RNP complexes assembled on in vitro–synthesized transcripts, including the spliceosome itself (Hartmuth et al. 2002; Jurica et al. 2002; Zhou et al. 2002; Sharma et al. 2005). Using related approaches, model RNA substrates containing cis-elements associated with neural-specific AS events could be incubated with extracts derived from neural cells, and any bound proteins could be purified and theoretically identified (Butter et al. 2009). The above approaches will be instrumental in uncovering novel neural-specific splicing factors.
Determining the role of tissue-specific isoforms
Networks of isoforms regulated by specific splicing factors can provide informative functional relationships among gene products (Fig. 2A). For instance, several studies have suggested that proteins encoded by transcripts with AS events regulated by a given splicing factor physically associate with each other through protein–protein interactions (PPIs) (Ule et al. 2005; Calarco et al. 2009; Warzecha et al. 2010). These observations suggest that tissue-specific AS networks may have evolved in part to fine-tune PPIs for a given cell or tissue type and are consistent with data indicating that a significant proportion of alternative exons map to the surfaces of proteins, potentially influencing PPIs (Wang et al. 2005). Approaches that can be performed on a large scale, such as yeast two-hybrid assays (Bruckner et al. 2009) or high-throughput mammalian coimmunoprecipitation assays (Barrios-Rodiles et al. 2005), can be used to systematically test how PPIs are affected by individual splice variants in these networks. We are applying such approaches to study the influence of nSR100 on PPI networks in the nervous system (J Ellis, M Barrios-Rodiles, JA Calarco, J Wrana, and BJ Blencowe, unpubl.).
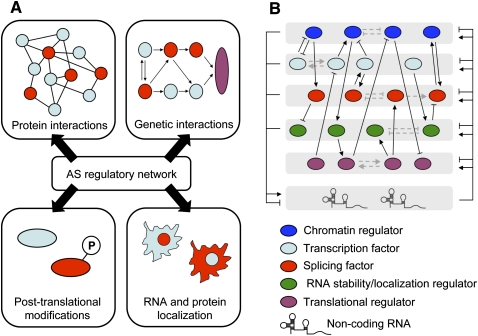
FIGURE 2.
Regulatory outcomes in alternative splicing networks and cross-talk between factors acting at multiple layers of gene regulation. (A) Coordinated alternative splicing events from an alternative splicing network (isoforms regulated by a specific splicing factor are displayed in red in the top and bottom panels) could result in the modification of protein–protein interactions (top left panel), modulation of genetic interactions within biological pathways (top right panel), altered post-translational modification sites in targeted proteins (bottom left panel), and altered RNA or protein localization in the cell (bottom right panel). Both single candidate gene studies and high-throughput approaches can be used to understand the functional consequences of these regulatory events. (B) Cross-regulation (black lines and arrows) has been observed between various classes of trans-acting regulators, such as chromatin-modifying proteins (blue), splicing factors (red), transcription factors (light blue), RNA stability or localization factors (green), translational regulators (purple), and non-coding RNAs. Cross-regulatory interactions have also been observed between members acting in the same process (gray dashed lines and arrows). The diagram displays both known regulatory events as well as theoretical events. A detailed understanding of how each of these factors interact and regulate each other will be essential for a complete understanding of the underlying regulatory networks they control.
Neural-specific AS events could also influence the localization of RNA or protein isoforms by inserting or removing critical localization signals. Theoretically, each isoform in a given regulatory network could be tagged with a fluorescent or other detectable reporter such that its localization pattern can be analyzed. Recent advances in high-throughput microscopy (Walter et al. 2010) should allow systematic analysis of isoform-specific localization. Additionally, regulated exons could overlap with post-translational modification sites in protein isoforms. For instance, a recent study revealed that Nova-regulated alternative exons encode putative phosphorylation sites more often than constitutive or other alternative exons (Zhang et al. 2010). It will be interesting to determine the biological significance of the altered phosphorylation of targeted proteins and whether the selective insertion or removal of phosphorylation sites by AS is a general hallmark of all splicing regulatory networks.
Although the techniques described above will be informative, the most definitive way to determine the functional consequences of altered splicing of a regulated exon will ultimately involve molecular genetic approaches. The selective elimination or introduction of specific isoforms in transgenic animals has proven a definitive and effective means of assessing the functional contribution of these variants (Beffert et al. 2005; Demir and Dickson 2005). Similar approaches can be used to interrogate isoforms in AS regulatory networks, and recent examples emerging from genetic analysis of Nova-regulated AS events illustrate their power. For example, phenotypic analyses of Nova knockout mice have revealed defects in neuromuscular junction (NMJ) morphology and impaired neuronal migration during development (Ruggiu et al. 2009; Yano et al. 2010). The aforementioned approaches used to identify Nova-regulated isoforms identified target exons in agrin and disabled-1 transcripts, which are genes with known roles in NMJ formation and neuronal migration, respectively. Transgenes expressing the specific isoforms affected by loss of Nova significantly rescued the NMJ and neuronal migration defects, suggesting that these splice variants are indeed required for nervous system development.
Such genetic analyses in mammalian or even vertebrate model organisms demand significant time investments. It is therefore not feasible to systematically assess the function of all isoforms identified in a regulatory network in this manner. The complementary use of RNAi-based approaches in cellular models of neuronal physiology, development, and differentiation may help prioritize which isoforms should be further analyzed in transgenic animal models. Alternatively, in other model organisms used in neurobiology such as C. elegans and Drosophila (Sattelle and Buckingham 2006; Sengupta and Samuel 2009), transgenic strains can be made relatively faster and on a larger scale, making such genetic analyses more tractable. Ultimately, to obtain a complete landscape of isoform functions in a splicing regulatory network, a combination of these approaches should be used to maximize both the scale and level of detail required to uncover novel biological insights.
Examining relationships among splicing factors
Numerous biochemical studies have revealed that tissue-specific splicing factors act in concert with other tissue-specific or ubiquitously expressed splicing regulators to regulate AS (Chen and Manley 2009). It is therefore becoming apparent that integrating regulatory networks of individual splicing factors is necessary in order to achieve an understanding of combinatorial control of tissue-specific AS patterns. Another emerging theme is that altered expression or activity of factors defined as “core” components of the spliceosome can lead to specific changes in AS patterns (Park et al. 2004; Yu et al. 2008; Zhang et al. 2008b; Saltzman et al. 2011). A better understanding of how these core splicing regulators can interact with the landscape of other tissue-specific and generally expressed auxiliary factors will be important.
As mentioned above, splicing factors can be dynamically regulated during development or in response to certain stimuli. Temporal regulation of the expression of CELF and MBNL regulators during heart development has been shown to play an integral role in establishing embryonic or adult splicing patterns (Kalsotra et al. 2008). Depolarization-induced changes in the nuclear accumulation of hnRNPA1 and Fox-1 in neurons lead to modulation of exon inclusion in target transcripts encoding synaptic proteins (An and Grabowski 2007; Lee et al. 2009). A recent study has also demonstrated that SR proteins can dynamically alter the repertoire of target transcripts they interact with during neuronal differentiation (Anko et al. 2010). These examples indicate that most genome-wide analyses of splicing regulatory networks have only provided a snapshot of the landscape of transcripts bound to and influenced by a given splicing factor. Large-scale analyses over developmental time courses and under different physiological perturbations will be informative to address some of these limitations.
Splicing factors frequently regulate AS events within their own transcripts and in those encoding other RNA-binding proteins. These auto- and cross-regulatory events often insert or remove PTC-containing exons that lead to the selective degradation of spliced transcripts by the NMD pathway, ultimately influencing the overall abundances of these factors (Lareau et al. 2007b; Ni et al. 2007; Spellman et al. 2007; Saltzman et al. 2008). Alternatively, the activity of splicing regulators was also found to be influenced by AS events in their own transcripts creating dominant negative isoforms or variants with altered post-translational modification sites (Dredge et al. 2005; Damianov and Black 2010). Collectively, these results suggest that altering the function of any single splicing factor may result in the perturbation of a network of factors. Understanding the functional relationships in these regulatory cascades will greatly improve our ability to identify programs of exons controlled during specific developmental processes.
Exploring connections between splicing factors and other regulators of gene expression
In addition to the complex functional relationships between splicing regulators, extensive connections between AS and other gene regulatory processes have been identified. For instance, splicing and other RNA processing events occur while RNA polymerase II (Pol II) actively transcribes pre-mRNA (Maniatis and Reed 2002). The emergence of a cotranscriptional splicing mechanism has led to the evolution of numerous interactions between the splicing and transcriptional machineries, mediated largely through the carboxy-terminal domain (CTD) of the largest subunit of Pol II (de Almeida and Carmo-Fonseca 2008; Pandit et al. 2008). The CTD of Pol II consists of repeats of the heptapeptide sequence YSPTSPS, which can be post-translationally modified in a dynamic manner during transcription (Egloff and Murphy 2008). Recent genome-wide analyses in yeast have demonstrated that certain CTD modifications can be regulated in a gene-specific manner during transcription (Kim et al. 2010; Tietjen et al. 2010). It will be interesting to determine if similar principles also extend to tissue-specific modifications of the CTD during transcription in metazoans and whether such modifications could lead to differential recruitment of AS regulators at specific splice sites.
The CTD and other factors can also influence the elongation rate of Pol II, which, in turn, can impact the time nascent splice sites are exposed to the splicing machinery, with the potential to influence AS (Munoz et al. 2010). Alternatively, an altered rate of Pol II elongation could also affect the association of splicing regulators with Pol II during cotranscriptional splicing (Munoz et al. 2010). Recent global analyses have demonstrated that changing the elongation rate of Pol II can influence AS of transcripts encoding genes involved in apoptosis and RNA metabolism (Munoz et al. 2009; Ip et al. 2011). However, it is currently not known whether Pol II elongation rates can be influenced differentially across tissues and whether any such differences can lead to tissue-specific AS regulation. Future studies will hopefully shed light on these interesting possibilities.
Numerous links between chromatin regulation and splicing have also emerged over recent years. For instance, a number of chromatin-remodeling factors have been found to interact with components of the spliceosome and other regulators of splicing (Allemand et al. 2008). In one interesting study, it was demonstrated that the SWI/SNF catalytic subunit Brm facilitates the accumulation of Pol II with modified CTD phosphorylation patterns over a regulated alternative exon in CD44 transcripts (Batsche et al. 2006). Brm also associates with components of the spliceosome and Sam68, supporting its role in integrating signals from chromatin, the transcriptional machinery, and the spliceosome. A series of recent studies has demonstrated that specific histone modifications in nucleosomes are preferentially enriched or depleted in exon and intron sequences (Kolasinska-Zwierz et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009; Huff et al. 2010). These results suggest that particular histone marks could be instructive or permissive in directing splicing decisions, and raise interesting mechanistic questions. For instance, can different histone modifications in the vicinity of exons recruit splicing factors, or are they generated as a result of splicing? Are histone marks associated with cell- and tissue-specific alternative exons different between tissues? A recent paper has suggested that, for at least some regulated AS events, histone modifications can display biased patterns of enrichment over exons in different cell types, and the factors interacting with these marks can potentially serve to recruit splicing regulators (Luco et al. 2010). A better understanding of how these signals are interpreted by chromatin regulators and the spliceosome in order to generate tissue-specific AS patterns will be an important goal.
In addition to the direct links between AS and other gene regulatory processes described above, there is also significant indirect cross-regulation. The coupling of alternative splicing with the NMD pathway serves both as a mechanism for quality control and in the regulation of transcript levels as described above (Lareau et al. 2007a). Many transcription factors are alternatively spliced (Taneri et al. 2004), and knockdown of splicing regulators leads to altered AS of transcription factor isoforms (Boutz et al. 2007b; Calarco et al. 2009). Additionally, miRNAs target splicing factors and, by altering their expression levels, indirectly modulate developmental AS programs (Boutz et al. 2007a; Makeyev et al. 2007; Kalsotra et al. 2010). Most studies of gene regulatory networks have thus far focused only on individual processes. The extent of cross-talk between trans-acting proteins at various stages of gene regulation suggests that analyses of the effects of splicing regulators should be extended to other modes of regulation. Furthermore, a detailed understanding of the interplay between transcription factors, splicing factors, miRNAs, and other pleiotropic regulators in a given developmental paradigm will help identify functional relationships among groups of genes targeted by these factors (Fig. 2B).
The generation of predictive models governing tissue-specific splicing
From a theoretical perspective, as all of the above-mentioned analyses are being performed, their cumulative integration provides resources to generate predictive models of tissue-specific splicing regulation. Recent work from the Frey and Blencowe groups demonstrated that machine learning approaches can be used to infer combinations of features, such as cis-regulatory sequences, exon/intron lengths, splice site strengths, and others, that are maximally predictive of tissue-specific AS patterns (Barash et al. 2010). The utility of this inferred tissue-dependent “splicing code” was demonstrated through its ability to identify associations between sets of features, provide testable hypotheses on critical sequence elements involved in regulating AS patterns, and uncover novel tissue-specific AS events from an analysis of pre-mRNA sequence alone (Barash et al. 2010). Recently, Zhang et al. (2010) combined several genome-wide data sets studying Nova-regulated alternative exons to establish parameters that could predict Nova-dependent AS events through a machine learning approach. This approach was able to identify many more transcripts regulated by the Nova proteins than could be identified by each experimental data set in isolation, predict combinatorial regulation with the Fox splicing factors, and infer novel principles by which these regulators function.
These studies would not be possible without the generation of empirically derived genome-wide data sets, from which the algorithms are usually trained, and decades of intensive research from the splicing community, from which parameters have been derived. As more detailed AS profiling data become available, new trans-acting factors and cis-regulatory elements are uncovered, and other features such as chromatin states are included in new algorithms, the predictive power of computational models will continue to improve.
CONCLUDING REMARKS
The inception of large-scale approaches to analyze gene expression has ushered in a new era in which biological phenomena can be studied both on a gene-by-gene basis and from a global perspective. A principle that has emerged from genome-wide transcriptome analyses is that master regulators have evolved to control networks of transcripts involved in common developmental and functional pathways. Neural splicing regulatory networks also appear to have evolved under similar organizing principles. Investigations of these networks have served two key purposes. First, they have enabled new insights into the mechanisms by which splicing factors regulate AS. Second, they have identified novel functional associations between gene products that would otherwise not have been illuminated by studies of transcription factor or miRNA regulatory networks. As we move forward, work in the future will be directed at understanding exactly how individual isoforms in splicing regulatory networks work together to achieve biological outputs.
Finally, although this review has focused on splicing factors in the nervous system, similar attributes have been identified in other tissues and cellular pathways (e.g., Warzecha et al. 2009, 2010; Moore et al. 2010). Analyses of splicing factor regulatory networks in many distinct tissue types will undoubtedly reveal novel insights in diverse molecular pathways. Collectively, these studies will lead to a comprehensive picture into how cellular complexity is achieved through the interplay of multiple modes of gene regulation, and how the perturbation of these processes lead to various diseases.
ACKNOWLEDGMENTS
We thank Nuno Barbosa-Morais and Arneet Saltzman for critical reading of the manuscript. Our research is supported by grants from the CIHR (to M.Z. and B.J.B.), the NCIC/CCS (to B.J.B.), and from Genome Canada through the Ontario Genomics Institute (to B.J.B. and others). J.A.C. has been supported by an Alexander Graham Bell Canada Graduate Scholarship from NSERC.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2603911.
REFERENCES
- Aberg K, Saetre P, Jareborg N, Jazin E 2006a. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci 103: 7482–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, Jazin E 2006b. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am J Med Genet B Neuropsychiatr Genet 141B: 84–90 [DOI] [PubMed] [Google Scholar]
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, et al. 2005. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci 102: 4625–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Batsche E, Muchardt C 2008. Splicing, transcription, and chromatin: a menage a trois. Curr Opin Genet Dev 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Amrein H, Gorman M, Nothiger R 1988. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell 55: 1025–1035 [DOI] [PubMed] [Google Scholar]
- An P, Grabowski PJ 2007. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol 5: e36 doi: 10.1371/journal.pbio.0050036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Morales L, Henry I, Beyer A, Neugebauer KM 2010. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat Struct Mol Biol 17: 962–970 [DOI] [PubMed] [Google Scholar]
- Ashiya M, Grabowski PJ 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3: 996–1015 [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Barberan-Soler S, Zahler AM 2008. Alternative splicing regulation during C. elegans development: Splicing factors as regulated targets. 4: e1000001 doi: 10.1371/journal.pgen.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S 2006. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB 2006. Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie 88: 515–525 [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. 2005. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307: 1621–1625 [DOI] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13: 22–29 [DOI] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, et al. 2005. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron 47: 567–579 [DOI] [PubMed] [Google Scholar]
- Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J 2008. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem 283: 1229–1233 [DOI] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF 2004. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet 49: 308–311 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev 19: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA 2010. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res 38: 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 25: 106–110 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ 2006. Alternative splicing: New insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Ahmad S, Lee LJ 2009. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev 23: 1379–1386 [DOI] [PubMed] [Google Scholar]
- Bonano VI, Oltean S, Garcia-Blanco MA 2007. A protocol for imaging alternative splicing regulation in vivo using fluorescence reporters in transgenic mice. Nat Protoc 2: 2166–2181 [DOI] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL 2007a. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev 21: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL 2007b. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U 2009. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci 10: 2763–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG 2001. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res 29: 2338–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Darnell RB 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol 17: 3194–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB 1993. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11: 657–672 [DOI] [PubMed] [Google Scholar]
- Butter F, Scheibe M, Morl M, Mann M 2009. Unbiased RNA–protein interaction screen by quantitative proteomics. Proc Natl Acad Sci 106: 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Saltzman AL, Ip JY, Blencowe BJ 2007. Technologies for the global discovery and analysis of alternative splicing. Adv Exp Med Biol 623: 64–84 [DOI] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. 2009. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138: 898–910 [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet BN, Logan P, Singh G, Cooper TA 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Chawla G, Lin CH, Han A, Shiue L, Ares M Jr, Black DL 2009. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol 29: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Black DL 2010. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 16: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Carmo-Fonseca M 2008. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett 582: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Dembowski JA, Grabowski PJ 2009. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet 5: e1000595 doi: 10.1371/journal.pgen.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, Dickson BJ 2005. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121: 785–794 [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Dredge BK, Darnell RB 2003. Nova regulates GABAA receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol 23: 4687–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB 2005. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J 24: 1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K 1996. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet 12: 260–265 [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S 2008. Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, et al. 2007. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol 8: R108 doi: 10.1186/gb-2007-8-6-r108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Friedman BA, Stadler MB, Shomron N, Ding Y, Burge CB 2008. Ab initio identification of functionally interacting pairs of cis-regulatory elements. Genome Res 18: 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA 1994. A target for Src in mitosis. Nature 368: 871–874 [DOI] [PubMed] [Google Scholar]
- Gallo JM, Spickett C 2010. The role of CELF proteins in neurological disorders. RNA Biol 7: 474–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL 1990. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62: 25–34 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralski TJ, Edstrom JE, Baker BS 1989. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell 56: 1011–1018 [DOI] [PubMed] [Google Scholar]
- Grange J, Belly A, Dupas S, Trembleau A, Sadoul R, Goldberg Y 2009. Specific interaction between Sam68 and neuronal mRNAs: Implication for the activity-dependent biosynthesis of elongation factor eEF1A. J Neurosci Res 87: 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2010. The developmental transcriptome of Drosophila melanogaster. Nature doi: 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, Rideau A, Southby J, Scadden AD, Gooding C, Huttelmaier S, Singer RH, Smith CW 2003. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J 22: 6356–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR Jr, Krainer AR 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4: 430–444 [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Castelo R, Blanchette M, Boue S, Rio DC, Valcarcel J 2009. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol 10: R11 doi: 10.1186/gb-2009-10-1-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci 99: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA 2004. Muscleblind proteins regulate alternative splicing. EMBO J 23: 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Plocik AM, Guthrie C, Yamamoto KR 2010. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol 17: 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res (in press) doi: 10.1101/gr.111070.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB 2008. CLIP: Crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol 488: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25: 359–371 [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 22: 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA 2008. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci 105: 20333–20338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Wang K, Li PF, Cooper TA 2010. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev 24: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi-Meskishvili V, Hetzer R, et al. 2003. Genome-wide array analysis of normal and malformed human hearts. Circulation 107: 2467–2474 [DOI] [PubMed] [Google Scholar]
- Ke S, Chasin LA 2010. Intronic motif pairs cooperate across exons to promote pre-mRNA splicing. Genome Biol 11: R84 doi: 10.1186/gb-2010-11-8-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307 [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Khanna A, Stamm S 2010. Regulation of alternative splicing by short non-coding nuclear RNAs. RNA Biol 7: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S 2009. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem 284: 31052–31061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D 1990. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62: 35–42 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M 2010. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc 5: 1495–1517 [DOI] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Stenberg MG, Swanson MS, Cooper TA 2005. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn 233: 783–793 [DOI] [PubMed] [Google Scholar]
- Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, et al. 2010. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res 8: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE 2007a. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 623: 190–211 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE 2007b. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL 2009. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 23: 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176 [DOI] [PubMed] [Google Scholar]
- Levinson N, Hinman R, Patil A, Stephenson CR, Werner S, Woo GH, Xiao J, Wipf P, Lynch KW 2006. Use of transcriptional synergy to augment sensitivity of a splicing reporter assay. RNA 12: 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 8: 819–831 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, Alvarez-Buylla A 2006. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci 31: 131–148 [DOI] [PubMed] [Google Scholar]
- Lin S, Fu XD 2007. SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623: 107–122 [DOI] [PubMed] [Google Scholar]
- Lipscombe D 2005. Neuronal proteins custom designed by alternative splicing. Curr Opin Neurobiol 15: 358–363 [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Richard S 2003. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta 1653: 73–86 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S 2008. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav Brain Res 189: 357–363 [DOI] [PubMed] [Google Scholar]
- Lynch KW 2007. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol 623: 161–174 [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T 2007. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20: 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695 [DOI] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA 2005. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol 5: 14 doi: 10.1186/1471-213X-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10: 1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res 33: 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Silver PA 2008. Global analysis of mRNA splicing. RNA 14: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Munoz MJ, de la Mata M, Kornblihtt AR 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci 35: 497–504 [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res 33: 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Orengo JP, Bundman D, Cooper TA 2006. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res 34: e148 doi: 10.1093/nar/gkl967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD 2008. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 20: 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci 101: 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, et al. 2009. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol 185: 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R 2006. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74: 65–80 [DOI] [PubMed] [Google Scholar]
- Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C 2010. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J 29: 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A 2009. Computation for ChIP-seq and RNA-seq studies. Nat Methods (Suppl)6: S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5: e90 doi: 10.1371/journal.pbio.0050090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci 97: 6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. 2006. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem 281: 12468–12474 [DOI] [PubMed] [Google Scholar]
- Rajan P, Gaughan L, Dalgliesh C, El-Sherif A, Robson CN, Leung HY, Elliott DJ 2008. Regulation of gene expression by the RNA-binding protein Sam68 in cancer. Biochem Soc Trans 36: 505–507 [DOI] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Herbst R, Kim N, Jevsek M, Fak JJ, Mann MA, Fischbach G, Burden SJ, Darnell RB 2009. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci 106: 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 25: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelle DB, Buckingham SD 2006. Invertebrate studies and their ongoing contributions to neuroscience. Invert Neurosci 6: 1–3 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Sengupta P, Samuel AD 2009. Caenorhabditis elegans: a model system for systems neuroscience. Curr Opin Neurobiol 19: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL 2005. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell 19: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell 16: 363–373 [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27: 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci 105: 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655–660 [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol 2: e4 doi: 10.1371/journal.pcbi.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM 1991. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell 67: 325–333 [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. 2007. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39: 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneri B, Snyder B, Novoradovsky A, Gaasterland T 2004. Alternative splicing of mouse transcription factors affects their DNA-binding domain architecture and is tissue specific. Genome Biol 5: R75 doi: 10.1186/gb-2004-5-10-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D 1994. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368: 867–871 [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci 97: 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C 2004. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77: 505–519 [DOI] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. 2010. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB 2007. Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain. Adv Exp Med Biol 623: 148–160 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 25: 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. 2009. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 16: 670–676 [DOI] [PubMed] [Google Scholar]
- Vernet C, Artzt K 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet 13: 479–484 [DOI] [PubMed] [Google Scholar]
- Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, et al. 2010. Visualization of image data from cells to organisms. Nat Methods Suppl7: S26–S41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845 [DOI] [PubMed] [Google Scholar]
- Wang P, Yan B, Guo JT, Hicks C, Xu Y 2005. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc Natl Acad Sci 102: 18920–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP 2010. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 29: 3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Maniatis T 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Wu JI, Reed RB, Grabowski PJ, Artzt K 2002. Function of quaking in myelination: Regulation of alternative splicing. Proc Natl Acad Sci 99: 4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Burge CB 2007. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci 104: 18583–18588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Lee C 2006. Alternative splicing and RNA selection pressure—evolutionary consequences for eukaryotic genomes. Nat Rev Genet 7: 499–509 [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB 1998. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci 95: 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB 2010. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66: 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Hoon S, Venkatesh B, Burge CB 2004. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci 101: 15700–15705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand EL, Liang TY 2007. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet 3: e85 doi: 10.1371/journal.pgen.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G 2010. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 30: 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847 [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. 2004. The functional landscape of mouse gene expression. J Biol 3: 21 doi: 10.1186/jbiol16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ 2008a. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008b. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Mandler MD, Yi H, Feng Y 2010. Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc Natl Acad Sci 107: 19061–19066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]