Abstract
Post-transcriptional control of mRNA stability and translation is central to multiple developmental pathways. This control can be linked to cytoplasmic polyadenylation in certain settings. In maturing Xenopus oocytes, specific mRNAs are targeted for polyadenylation via recruitment of the Cytoplasmic Polyadenylation Element (CPE) binding protein (CPEB) to CPE(s) within the 3′ UTR. Cytoplasmic polyadenylation is also critical to early embryonic events, although corresponding determinants are less defined. Here, we demonstrate that the Xenopus ortholog of the poly(rC) binding protein αCP2 can recruit cytoplasmic poly(A) polymerase activity to mRNAs in Xenopus post-fertilization embryos, and that this recruitment relies on cis sequences recognized by αCP2. We find that the hα-globin 3′ UTR, a validated mammalian αCP2 target, constitutes an effective target for cytoplasmic polyadenylation in Xenopus embryos, but not during Xenopus oocyte maturation. We further demonstrate that the cytoplasmic polyadenylation activity is dependent on the action of the C-rich αCP-binding site in conjunction with the adjacent AAUAAA. Consistent with its ability to target mRNA for poly(A) addition, we find that XαCP2 associates with core components of the Xenopus cytoplasmic polyadenylation complex, including the cytoplasmic poly(A) polymerase XGLD2. Furthermore, we observe that the C-rich αCP-binding site can robustly enhance the activity of a weak canonical oocyte maturation CPE in early embryos, possibly via a direct interaction between XαCP2 and CPEB1. These studies establish XαCP2 as a novel cytoplasmic polyadenylation trans factor, indicate that C-rich sequences can function as noncanonical cytoplasmic polyadenylation elements, and expand our understanding of the complexities underlying cytoplasmic polyadenylation in specific developmental settings.
Keywords: cytoplasmic polyadenylation, mRNA stability, αCP2, CPEB, poly(A) polymerase, α-globin
Introduction
Cytoplasmic polyadenylation constitutes a well-described mechanism of post-transcriptional control (Richter 1999). This pathway is of maximal importance in settings where transcription is inadequate to dictate rapid developmental and/or physiologic responses. While the components and mechanisms of cytoplasmic polyadenylation have been examined in detail in the context of Xenopus laevis oocyte maturation, the prevalence and mechanisms of this pathway at other developmental stages and in other species remain less well understood.
The process of meiotic maturation of Xenopus oocytes is dependent on the controlled activation of deadenylated and translationally-inert maternal mRNAs. Translational activation is mediated by the re-lengthening of the poly(A) tails of these mRNAs. Two well-described cytoplasmic polyadenylation determinants are the Musashi Binding Element (MBE; [G/A]U1-3AGU), which appears to act on early (mos-independent) mRNAs, and the maturation-type Cytoplasmic Polyadenylation Element (CPE; U4-6A1-2U1-2), which generally acts on both early and late (mos-dependent) mRNAs (Radford et al. 2008; MacNicol and MacNicol 2010). These 3′ UTR motifs serve as binding sites for corresponding trans factors: Musashi (Charlesworth et al. 2006) and cytoplasmic polyadenylation element binding protein (CPEB) (Hake and Richter 1994), respectively. CPEB is postulated to nucleate the assembly of a poly(A) addition core complex, the “G complex,” that contains the scaffold protein symplekin, cleavage and polyadenylation specificity factor (CPSF), and the cytoplasmic poly(A) polymerase (PAP), XGLD2 (Barnard et al. 2004). Recent models propose that this complex is preassembled on a target mRNA prior to meiotic activation but is unable to effectively extend the poly(A) tail due to the presence in the complex of the 3′-5′ poly(A) ribonuclease (PARN) (Kim and Richter 2006). Translational repression of these target mRNAs may be further reinforced by CPEB recruitment of maskin, an eIF4E binding protein that blocks formation of the eIF4F translation initiation complex (Radford et al. 2008). Upon induction of oocyte meiotic maturation, CPEB is phosphorylated, resulting in the release of PARN and remodeling of maskin protein–protein interactions. This alteration in complex composition is thought to relieve cap occlusion and allow the resident GLD2, now unopposed by PARN, to elongate the poly(A) tail (Kim and Richter 2006). This CPEB phosphorylation event also promotes binding of embryonic poly(A) binding protein (ePAB) to the newly elongated poly(A) tail. These various modifications of mRNP constituents are thought to act in concert to activate mRNA translation by stabilizing a translation-enhancing “closed loop” configuration of the mRNA via a 3′-5′ interaction between ePAB and the cap-associated eIF4G (Kim and Richter 2007).
Although the pathway and components of cytoplasmic polyadenylation are most clearly defined during progesterone-stimulated Xenopus oocyte maturation, it is important to note that a variety of additional cis elements and corresponding trans factors appear to be active in other settings. For example, analyses of post-transcriptional controls during gametogenesis in Caenorhabditis elegans have identified three factors, GLD-3, RNP-8, and GLS-1, that recruit cytoplasmic polyadenylation to subsets of mRNAs (Wang et al. 2002; Kim et al. 2009; Schmid et al. 2009). Each factor is likely to recognize distinct sets of cis elements, and each is nonorthologous with CPEB. Recent studies of the Drosophila Toll mRNA have identified a noncanonical 3′ UTR “polyadenylation region” that governs embryonic cytoplasmic polyadenylation independently of the AAUAAA cleavage and polyadenylation site (Coll et al. 2010). Thus, cytoplasmic polyadenylation mechanisms are complex and are regulated by an assortment of cis elements and trans factors.
Subsequent to fertilization, the initial development of the newly formed Xenopus embryo is programmed to a large extent by the translational activation of dormant maternal mRNAs (Richter 1999). A cohort of mRNAs activated during this early embryonic period was found to contain one or more embryonic-specific poly(U) tract CPEs within their 3′ UTRs (Simon et al. 1992; Simon and Richter 1994; Simon et al. 1996). ElrA, the Xenopus ortholog of HuR, has been implicated as the corresponding trans factor (Wu et al. 1997; Slevin et al. 2007). However, this assignment has yet to be confirmed by functional studies, and necessary cofactors have not yet been identified. Furthermore, XGLD2, although expressed in embryos (Rouhana et al. 2005), has not been linked to cytoplasmic poly(A) addition in this setting. In sum, the targeting mechanisms of cytoplasmic polyadenylation in early Xenopus embryos appear to be distinct in critical attributes from those in the maturing oocyte and remain to be more completely defined.
The potential complexity of sequence determinants and activities is highlighted by a study of the pp2Ac mRNA. Cytoplasmic polyadenylation of pp2Ac in the early Xenopus embryo relies on the combined actions of a C14 tract in cis with a classical U/A-rich oocyte maturation CPE. RNA affinity purification identified the Xenopus ortholog of the KH-domain RNA binding protein αCP2 (XαCP2) as the putative C14 binding protein (Paillard et al. 2000). While of potential interest, these studies failed to demonstrate whether XαCP2 did, in fact, bind in vivo to the pp2Ac mRNA target and/or interact with the cytoplasmic polyadenylation machinery. The possibility that αCP2 plays a direct role in targeting cytoplasmic polyadenylation is of particular interest because this same protein, also known as heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2)/poly(rC) binding protein 2 (PCBP2), has been implicated in multiple post-transcriptional control pathways in mammalian cells, including splicing (Ji et al. 2007), 3′ end formation (X Ji and SA Liebhaber, in prep.), translational control (Makeyev and Liebhaber 2002), and stabilization of long-lived mRNAs via poly (A) tail maintenance (Kong and Liebhaber 2007).
In the present article, we explore the linkage between Xenopus αCP2 and cytoplasmic polyadenylation by assessing in vivo occupancy of αCP2 on an endogenous target and by asking whether a well-known αCP2 binding target, the human α-globin 3′ UTR, can be effectively targeted for cytoplasmic polyadenylation in the X. laevis embryo.
Results
αCP2 associates in vivo with the pp2Ac mRNA, a previously established target of cytoplasmic polyadenylation
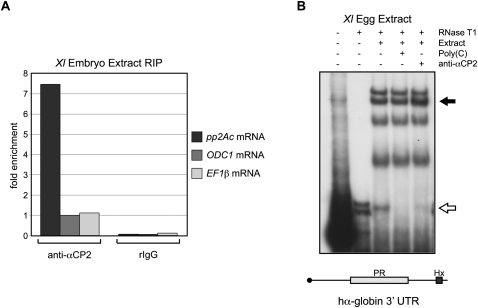
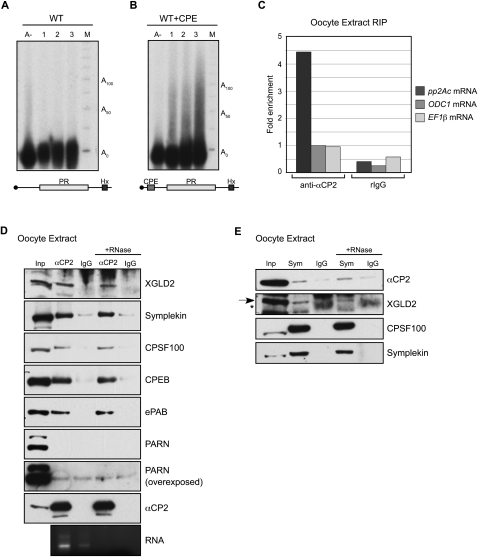
Previously, a C14 cytoplasmic polyadenylation element identified in the pp2Ac mRNA enriched for the Xenopus paralog of αCP2 (XαCP2) in an in vitro affinity purification assay (Paillard et al. 2000). To explore a potential role for αCP2 as a recruiter of cytoplasmic polyadenylation machinery, we first asked whether XαCP2 associated with the pp2Ac mRNA in vivo. We performed RNA coimmunoprecipitation experiments (RIPs) using cytoplasmic extract from post-fertilization embryos. These experiments revealed that the endogenous pp2Ac mRNA was enriched in anti-αCP2 RIPs compared with two control mRNAs: ornithine decarboxylase 1 (ODC1) mRNA, which contains a polyuridine eCPE (Osborne et al. 1991), and elongation factor 1β (EF1β) mRNA, which lacks any recognizable CPEs (Fig. 1A). Thus, we concluded that XαCP2 associates in vivo with the pp2Ac mRNA and that it may constitute the potential trans factor that recruits cytoplasmic polyadenylation machinery to pp2Ac.
FIGURE 1.
Xenopus αCP2 associates in vivo with the pp2Ac mRNA and can also recognize the human α-globin 3′ UTR, a previously established αCP2 binding target. (A) In vivo association of XαCP2 with pp2Ac mRNA. RNP complexes were immunoprecipitated from cytoplasmic embryo extract with anti-αCP2 or a preimmune rabbit control sera (rIgG). RNAs present in the αCP2 IP and in the control IP were assayed by real-time RT-PCR for pp2Ac, ODC1 (ornithine decarboxylase 1), and EF1β (elongation factor 1β) mRNAs. ODC1 and EF1β are predicted nonbinding controls (see text). Results are all normalized to the level of ODC1 mRNA in the anti-αCP2 IP pellet (defined as arbitrary unit = 1). One representative RIP experiment is depicted here. Two independent repeat experiments which confirm significant enrichment of pp2Ac mRNA in the anti-αCP2 RIP are presented in Supplemental Figure S3. (B) RNA electrophoretic mobility shift assay (RNA-EMSA) of the 32P-labeled hα-globin 3′ UTR incubated with Xenopus egg cytoplasmic extract. The open arrow denotes the position of the poly(C)-sensitive 3′ UTR RNP complex (α-complex). The closed arrow denotes the position of the labeled complex supershifted by the αCP2 antisera.
The 3′ UTR of the hα-globin mRNA is targeted for cytoplasmic polyadenylation in X. laevis embryos
To further explore a role for αCP2 in recruiting cytoplasmic polyadenylation, we assayed cytoplasmic polyadenylation of a mammalian mRNA that is a well-defined αCP2 target. The human α-globin 3′ UTR contains a sequence of three C-rich repeats that are required for efficient αCP binding in mammalian cells (Fig. 2A; Kiledjian et al. 1995; Weiss and Liebhaber 1995). This sequence, referred to as the αCP protected region (PR), is located 18 nucleotides (nt) upstream of the AAUAAA cleavage and polyadenylation site (Waggoner and Liebhaber 2003b). It is of note that this distance is consistent with an effective spacing between a CPE-type sequence and the AAUAAA (Richter 1999; Pique et al. 2008). Since the hα-globin 3′ UTR lacks any of the previously reported cytoplasmic polyadenylation cis elements, it appears to be an ideal construct to explore αCP2-mediated cytoplasmic polyadenylation.
FIGURE 2.
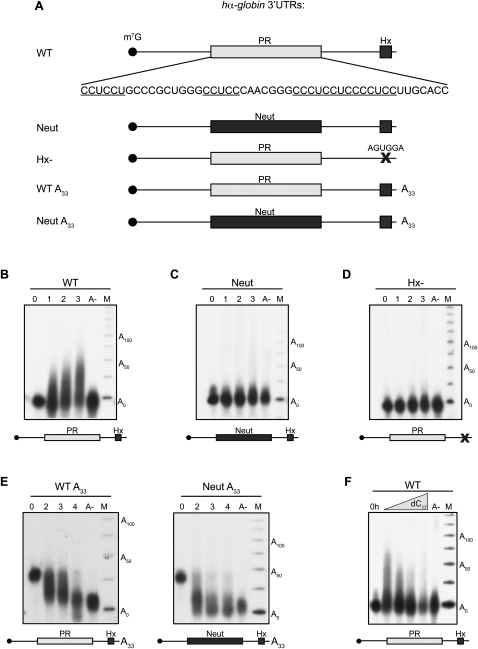
The hα-globin 3′ UTR is a substrate for αCP2-mediated cytoplasmic polyadenylation when injected into Xenopus embryos. (A) α-globin 3′ UTR RNAs used in this figure. PR denotes the 42-base C-rich stability determinant in the hα-globin 3′ UTR that is bound by αCP (Waggoner and Liebhaber 2003b). Neutral denotes a 42-base segment originating from the coding region of the α-globin mRNA that is substituted in place of the PR. Hx denotes the AAUAAA cleavage and polyadenylation hexanucleotide. X denotes the mutation of the cleavage and polyadenylation hexanucleotide from AAUAAA to AGUGGA. A33 denotes a 33-base (A) tail that is present on the synthetic RNA at the time of injection. All RNAs are synthesized in vitro in the presence of m7G cap and are [32P] internally labeled. (B–E) Polyadenylation assays. The specified 3′ UTR construct was injected into stage II embryos. RNA was extracted from aliquots of embryos after a 0-, 1-, 2-, or 3-h incubation, and the size of the labeled hα-globin 3′ UTR was determined on a denaturing acrylamide gel. An aliquot of RNA from the 3-h sample was also treated with RNase H in the presence of oligo dT to remove the poly(A) tail prior to analysis (lane A-). The marker lane (M) contains a [32P] end-labeled 25-nt single-stranded DNA ladder. (B) WT. The WT α-globin 3′ UTR is a substrate for cytoplasmic polyadenylation. (C) Neut. Replacement of the PR by a neutral sequence abolished cytoplasmic polyadenylation. (D) Hx-. Mutation of the AAUAAA cleavage and polyadenylation site to AGUGGA abolished cytoplasmic polyadenylation. (E) Deadenylation assays show that the presence of the PR sequence results in a net slowing of poly(A) tail shortening. WT A33 (left) and Neut A33 (right) α-globin 3′ UTRs containing A33 tails were injected into two-cell embryos in the same manner as carried out for the polyadenylation assays. RNA was isolated at hourly intervals over 4 h and sized on a denaturing gel. The input A33 RNA and the deadenylated input (A-) were loaded in the lanes 1 and 5, respectively, for size comparison. (F) Polyadenylation of WT hα-globin 3′ UTR is blocked by co-injection of unlabeled oligo dC. The WT 3′ UTR was assayed for polyadenylation in Xenopus embryos in the presence of increasing amounts of co-injected oligo dC17. RNA was isolated from each sample 3 h after injection. Experiment was carried out as in B.
To explore whether the α-globin 3′ UTR was indeed a suitable target for the Xenopus αCP2 ortholog, we first assessed the sequence conservation between the mammalian and X. laevis orthologs. Xenopus αCP2 (XαCP2) is remarkably conserved with its mammalian ortholog; the primary structure of the three RNA-binding KH domains (each, 69 amino acids) diverge by only 1, 0, and 2 conservative substitutions, respectively, from the corresponding human structure (Gravina et al. 2002). Thus, we predicted that RNA binding specificities should be similarly conserved. Mammalian αCP2 has a strong preferential binding to single-stranded cytosine-rich nucleic acid sequences (Makeyev et al. 2002) and binds in a 1:1 molar ratio with the hα-globin PR element forming the “α-complex” (Chkheidze et al. 1999). RNA EMSA using Xenopus egg cytoplasmic extract confirmed that XαCP2 can assemble the α-complex in vitro on the hα-globin 3′ UTR (Fig. 1B). This RNP complex is sensitive to competition with unlabeled excess poly(C), and incubation with an anti-αCP2 antibody produces a supershifted band and concurrent loss in intensity of the original α-complex band. Thus, XαCP2 is predicted to recognize the hα-globin 3′ UTR construct in Xenopus embryos and to serve as a reasonable target with which to investigate αCP2-mediated cytoplasmic polyadenylation.
The 3′ UTR segment of the hα-globin 3′ UTR (109 nt) (Fig. 2A, WT) lacking a poly(A) tail was synthesized in vitro in the presence of m7G cap and [α32P] CTP and injected into the animal poles of stage II X. laevis embryos. The injected embryos were incubated at room temperature, and total RNA was isolated from subsets of embryos at hourly intervals (0, 1, 2, 3) post-injection. The size of the labeled RNA was directly assessed on a denaturing gel (Fig. 2B). These data revealed that the hα-globin 3′ UTR was elongated over the 3-h period by 50–75 nt. The extension was quantitatively removed by RNase H treatment in the presence of oligo dT18 but not in the presence of other homodeoxyribooligomers (Fig. 2B; Supplemental Fig. S1). These results are consistent with this extension representing a poly(A) tail. The extension was similar to lengths reported for Xenopus embryonic Cl1 and Cl2 mRNAs that contain canonical poly(U) eCPEs (Simon et al. 1992; Simon and Richter 1994). These data lead us to conclude that the 3′ UTR sequence of the hα-globin mRNA can effectively recruit PAP activity in early Xenopus embryos.
Polyadenylation of the hα-globin 3′ UTR in Xenopus embryos is dependent on the C-rich αCP PR
We next asked whether the αCP cognate binding site, the PR, was involved in targeting the hα-globin 3′ UTR for cytoplasmic polyadenylation in Xenopus embryos. The PR was replaced with an equal sized “neutral” sequence derived from the coding region of the hα-globin mRNA (Kong et al. 2003). This substitution is known to abolish αCP binding in mammalian erythroid cells, resulting in defects in stability (Kong et al. 2003), splicing (Ji et al. 2007), and 3′end formation (X Ji and SA Liebhaber, in prep.). The hαNeut 3′ UTR (Fig. 2A, Neut) failed to undergo discernible extension during the 3-h incubation (Fig. 2C). These data support a model where the PR can act in a similar fashion as a CPE in Xenopus embryos. This C-rich element, henceforth referred to as the C-CPE, is structurally distinct from the U/A-rich maturation CPE, the MBE, and the U-rich eCPE (see Introduction).
To further define the cis-acting determinants required for C-CPE function, we introduced an inactivating mutation at the AAUAAA that converted it to AGUGGA (Fig. 2A, Hx-). This mutation, which has been shown to effectively block AAUAAA-mediated 3′ end processing (Wilusz et al. 1989; Sheets et al. 1990), completely abolished cytoplasmic polyadenylation of the otherwise intact hα-globin 3′ UTR (Fig. 2D). The observed dependence of the C-CPE activity on the AAUAAA is reminiscent of the dependence of the canonical maturation CPE on this element and the corresponding interaction of CPEB with cytoplasmic CPSF (Dickson et al. 1999; Barnard et al. 2004). These data lead us to conclude that the C-CPE and the AAUAAA act in concert to recruit cytoplasmic PAP to the hα-globin 3′ UTR.
To further test the actions of the PR C-CPE, we asked whether it could effectively slow down the rate of poly(A) shortening of a pre-adenylated RNA substrate. When the wild-type (WT) hα-globin 3′ UTR containing an A33 tail (Fig. 2A, WT A33) was injected into embryos, the poly(A) tail was gradually shortened over time (Fig. 2E, left panel). A parallel control study (Fig. 2E, right panel) of the same 3′ UTR lacking the C-rich stability determinant (Fig. 2A, Neut A33) revealed a marked acceleration of poly(A) tail shortening. These data are consistent with a model in which the C-rich stability element recruits PAP to counter the actions of endogenous poly(A) nucleases, resulting in a net slowing in the rate of poly(A) shortening on a target mRNA.
Recruitment of PAP activity to the hα-globin 3′ UTR is dependent on the action of the αCP2 RNP complex (“α-complex”)
We next tested the functional linkage between XαCP2 binding to the C-CPE and recruitment of PAP activity. Based on the sensitivity of the α-complex to poly(C) competition (Fig. 1B), we attempted to block XαCP2 binding to the α-globin 3′ UTR by co-injecting an unlabeled oligo dC17 competitor along with the labeled hαWT 3′ UTR (WT). This co-injection specifically diminished cytoplasmic polyadenylation of WT hα-globin 3′ UTR in a concentration-dependent manner (Fig. 2F). In contrast, the parallel injection of a nonspecific homodeoxyribooligomer had no appreciable effect (Supplemental Fig. S2). These data support an essential role for αCP2 in PAP recruitment.
XαCP2 interacts with the cytoplasmic polyadenylation core complex
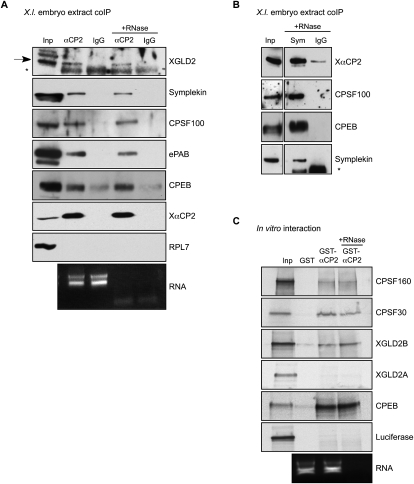
The preceding studies support the conclusion that the hα-globin 3′ UTR is targeted for cytoplasmic polyadenylation in X. laevis embryos and that this activity is dependent on the concerted actions of the C-rich CPE and the adjacent AAUAAA. To explore the biochemical basis for C-CPE action, we sought to identify relevant proteins in the Xenopus embryo that interact with XαCP2 in vivo. The endogenous XαCP2 was immunoprecipitated from cytoplasmic extracts prepared from pre-midblastula transition embryos, and the presence of associated proteins was assessed by Western analysis. These studies revealed enrichment for the Xenopus proteins symplekin, CPSF100, and the cytoplasmic PAP XGLD2 (Fig. 3A). These three proteins are integral components of the core cytoplasmic polyadenylation “G complex” (Barnard et al. 2004). These interactions were resistant to RNase treatment, suggesting that the noted associations are not dependent on RNA tethering. A reciprocal co-immunoprecipitation (co-IP) using an antibody to symplekin confirmed a subset of the noted interactions (Fig. 3B). These data are consistent with the conclusion that XαCP2 mediates cytoplasmic polyadenylation in Xenopus embryos by recruitment of the core cytoplasmic polyadenylation G complex to target mRNAs.
FIGURE 3.
XαCP2 interacts with proteins involved in the cytoplasmic polyadenylation pathway. (A) Co-IP analysis of XαCP2-associated proteins in Xenopus embryo extract. XαCP2 and associated proteins were immunoprecipitated with anti-αCP2 antibody in the presence or absence of RNase. Protein enrichment was assessed relative to the input (2.5% of total) and to their content in a parallel immunoprecipitation carried out with preimmune sera (IgG). The enrichment for XαCP2 confirmed the effectiveness of the immunoprecipitation. The data reveal selective enrichment of the G-complex proteins XGLD2, symplekin, CPSF100, and CPEB, as well as the embryonic PABP isoform (ePAB). In each case, the association with XαCP2 was RNA independent. There was no appreciable enrichment of the ribosomal protein, RPL7, used as a non-interacting control. The black arrow in the XGLD2 Western denotes the XGLD2 band, while the asterisk (*) denotes the position of the immunoglobulin heavy-chain in the immunoprecipitates. Aliquots of total RNA extracted from the nonbinding supernatants of each co-IP were run on an Agarose gel to confirm effective RNase treatment as monitored by loss of 18S and 28S ribosomal RNA bands. (B) A reciprocal co-IP using anti-symplekin antibody confirms the interactions with XαCP2, CPSF100, and CPEB. The asterisk in the symplekin Western denotes unreduced immunoprecipitation antibody. (C) GST pull-down studies. Recombinant GST-XαCP2 and GST were used as the two affinity reagents. RRL in vitro translation reactions containing [35S]-met were charged with templates encoding CPSF160, CPSF30, XGLD2A, XGLD2B, CPEB, or Luciferase. Each in vitro translation reaction was incubated with glutathione-Sepharose beads conjugated with recombinant GST or recombinant GST-XαCP2. One GST-XαCP2 pulldown was treated with RNase to distinguish RNA-dependent interactions. Each input lane represents 5% of total input. The bottom panel is a sample of total RNA from the supernatant of a representative GST pulldown; RNase treatment was monitored as described in A.
The αCP2 co-IP was subsequently probed for several other proteins of interest. A prominent finding was the association between αCP2 and CPEB1. This is of note because it has been recently shown that during late oocyte maturation, the proteasomal degradation of CPEB1 (Mendez et al. 2002) is paralleled by translational activation of CPEB4 (Igea and Mendez 2010). CPEB1 levels remain low and CPEB4 levels remain high post-fertilization (Igea and Mendez 2010) and serve as the primary CPE-binding factor. Thus, one potential function of the αCP2-CPEB1 interaction might be to increase local CPEB1 activity at target mRNAs in this environment. Due to a lack of specific antisera, we currently do not know whether αCP2 interacts with CPEB4.
Analyses of the XαCP2 coIP from embryo extract also revealed an enrichment for ePAB (Fig. 3A). This association was partially resistant to RNase treatment. This finding is of note as it diverges from an extant mechanistic model of canonical CPEB actions in which ePAB is only directly associated with the G complex prior to the activation of maturation-type CPE-mediated cytoplasmic polyadenylation (Kim and Richter 2007). While it is possible that there exists a subset of “inactive” XαCP2-associated G complex (XαCP2/G complex) containing directly associated ePAB post-fertilization, it must also be considered that the mechanisms governing XαCP2–ePAB interaction could be distinct from those governing the CPEB–ePAB interaction.
Protein–protein interactions within the novel XαCP2/G complex were confirmed and extended by GST pull-down studies (Fig. 3C). Interactions were observed between XαCP2 and two additional components of the CPSF complex, CPSF 160 and 30, as well as between XαCP2 and XGLD2B, one of the isoforms that encode XGLD2 (Rouhana et al. 2005). These data are consistent with the results of the coIP studies and support the formation of an XαCP2-associated G complex in the Xenopus embryo. It is of note that we did not observe an interaction between XαCP2 and the XGLD2A isoform, suggesting that the two XGLD2 isoforms may constitute distinct cytoplasmic polyadenylation complexes.
The C-CPE acts synergistically with a canonical oocyte maturation CPE in the early embryo
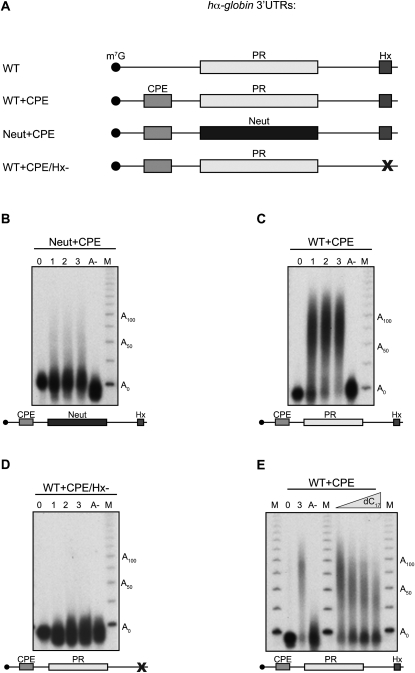
The activities of the previously described canonical CPEs in X. laevis appear to be largely developmentally specific. The maturation CPE is particularly robust during meiotic maturation, with its specific timing reflecting the number and positioning of CPEs relative to the AAUAAA and the presence or absence of associated Pumilio binding elements (PBEs) (Pique et al. 2008). In contrast, the U12-27 eCPE appears to selectively recruit PAP activity at the early embryonic stage of development. There is, however, evidence that overlap and interactions among the CPE elements at the meiotic maturation and embryonic stages may occur. For example, maximal embryonic recruitment of PAP to the pp2Ac mRNA 3′ UTR requires both the C14-CPE element and a maturation-type (U/A rich) CPE. To explore the capacity of the C-CPE to serve as an auxiliary element, we asked whether it could functionally interact with an oocyte maturation-type CPE in the early embryo. Removal of the C-CPE (PR) from the hα-globin 3′ UTR completely ablates PAP recruitment (Fig. 2C, Neut). Insertion of a maturation-type CPE (UUUUUAU) 67 nt upstream of the AAUAAA into this αNeut 3′ UTR (Fig. 4A, Neut+CPE) results in low levels of poly(A) addition (Fig. 4B). Remarkably, insertion of this maturation CPE in cis to the PR C-CPE (Fig. 4A, WT+CPE) results in a dramatic enhancement of PAP recruitment over that of the CPE alone (Fig. 4C). Compared with the activity with the C-CPE alone (i.e., the native hα-globin 3′ UTR), it is apparent that the C-CPE and this distal U/A CPE act in a synergistic fashion in the embryonic context (cf. Figs. 2B, 4B with Fig. 4C). As is the case for the action of the C-CPE and maturation-type (U/A) CPE when assessed individually, the robust activity in the presence of both elements is completely dependent on concerted action with the adjacent AAUAAA (Fig. 4D). The direct contribution of the C-CPE to this robust synergy was confirmed by the partial inhibition of this activity by oligo dC competition; both the size of the resulting poly(A) tail and the amount of polyadenylated substrate decrease substantially in the presence of a specific competitor (Fig. 4E; Supplemental Fig. S2).
FIGURE 4.
The C-CPE can synergize with a classical maturation-type CPE to promote robust cytoplasmic polyadenylation in X. laevis embryos. (A) RNAs assayed for cytoplasmic polyadenylation in Xenopus embryos. Each study (B–D) was carried out as in Figure 2, B–E. CPE denotes the classical maturation CPE UUUUUAU. For other notations, see legend for Figure 2A. (B) hα-globin 3′ UTR RNA containing an inserted maturation CPE but lacking the endogenous C-rich PR element (Neut+CPE). This 3′ UTR undergoes modest polyadenylation. (C) WT α-globin 3′ UTR containing an inserted 5′ maturation CPE (WT+CPE). This 3′ UTR undergoes robust polyadenylation. (D) WT α-globin 3′ UTR + maturation CPE lacking a functional AAUAAA (mutated to AGUGGA; WT+CPE/Hx-). This mutation abolishes all cytoplasmic polyadenylation. (E) WT α-globin 3′ UTR containing an inserted 5′ maturation CPE (WT+CPE) in the increasing presence or absence of unlabeled oligo dC17 competitor. The competition highlights the contribution of XαCP2 to the synergistic cytoplasmic polyadenylation seen with the WT+CPE 3′ UTR. 0 indicates RNA from the zero hour time point; 3 indicates a 3-h time point sample with no oligo dC competition. RNAs were harvested 3 h after injection for all competed samples.
The molecular basis of the observed synergy between the C-CPE and U/A CPE may reflect the association of XαCP2 with CPEB1 (Fig. 3A). Since CPEB1 levels undergo a dramatic decrease following meiotic maturation, the noted synergy between the C-CPE and the maturation CPE in Xenopus embryos might reflect a productive interaction between their two cognate binding proteins whereby the interaction with XαCP2 increases the effective local concentration of CPEB on a target RNA. An alternative model that remains untested is that this synergy may involve the trans factor, CPEB4, and not CPEB1.
αCP2-mediated cytoplasmic polyadenylation is inactive in maturing Xenopus oocytes despite recognition of a target mRNA and assembly of the core αCP2/G complex
To further explore the characteristics of the hα-globin PR C-CPE, its activity was assessed in maturing Xenopus oocytes. In contrast to its activity in the Xenopus embryo, the C-CPE is unable to recruit cytoplasmic polyadenylation when injected into stage VI Xenopus oocytes subsequently activated by progesterone (Fig. 5A). This lack of activity is consistent with the reported lack of activity of the pp2Ac C14 CPE in the maturing oocyte (Paillard et al. 2000). As expected, insertion of a maturation CPE into the hα-globin 3′ UTR triggers robust polyadenylation in the same oocyte environment (Fig. 5B). In an effort to explore the basis for the embryonic specificity of the C-CPE, we asked if XαCP2 is present in the oocytes and, if so, whether it is capable of binding to mRNA in this environment. Immunoprecipitation of endogenous XαCP2-RNP complexes from oocyte cytoplasmic extract revealed a marked enrichment for the pp2Ac mRNA (Fig. 5C). Thus XαCP2 is present in maturing oocytes (see also Fig. 5D) and is bound to a C-CPE containing mRNA even though it is not able to mediate polyadenylation on this bound mRNA.
FIGURE 5.
The C-rich CPE is inactive during Xenopus oocyte maturation despite the binding of XαCP2 to the target mRNA and the assembly of an XαCP2/G cytoplasmic polyadenylation complex. (A,B) Xenopus oocyte polyadenylation assays. The 32P-labeled WT α-globin 3′ UTR (A) and labeled WT+CPE α-globin 3′ UTR (B) were injected into stage VI oocytes. Hourly time points were taken relative to a 1-h time point that was taken 1 h after a 16-h progesterone treatment. (C) Immunoprecipitation of XαCP2 mRNP complexes from oocytes as in Figure 1B. RNAs bound to XαCP2 were isolated and assayed by qRT-PCR. One representative RIP experiment is depicted here. Two independent repeat experiments that show similar enrichment are presented in Supplemental Figure S3. (D) Immunoprecipitation of XαCP2-associated proteins from X. laevis stage V and VI oocyte cytoplasmic extracts. The studies reveal RNase-resistant associations of XαCP2 with XGLD2, symplekin, CPSF100, CPEB, and ePAB. PARN was not specifically enriched in the XαCP2 co-immunoprecipitates. (E) Immunoprecipitation of symplekin-associated proteins from Xenopus oocytes. Oocyte extract was immunoprecipitated with antibody to symplekin or normal mouse IgG, and Western analysis was carried out with antibodies to each of the indicated proteins. The arrow in the GLD2 Western indicates the GLD2 band; the asterisk indicates a nonspecific band present in mouse IgG IPs.
One possible explanation for the above findings would be that XαCP2, although bound to an mRNA, is unable to assemble the XαCP2/G complex in the oocyte environment. This was tested by analysis of an XαCP2 co-IP for components of the core G-complex components. Remarkably, the Western analyses revealed enrichment for XGLD2, symplekin, CPSF100, and ePAB, as well as for CPEB (Fig. 5D). This enrichment was, in each case, resistant to RNase. A reciprocal immunoprecipitation study using an antibody for symplekin confirmed interactions with XαCP2, XGLD2, and CPSF (Fig. 5E). These immunoprecipitation studies reveal that XαCP2 is present in the oocyte and that it associates with the G complex in as robust a fashion as observed in embryos as judged by the co-IP studies (cf. Fig. 5D,E and Fig. 3A,B). Despite this evidence for XαCP2-associated G-complex assembly and association of XαCP2 with a target mRNA, αCP2-mediated cytoplasmic polyadenylation appears to be inactive in the oocyte.
It has been reported that CPEB assembles the G complex on target mRNAs prior to oocyte maturation and that this complex only becomes active after progesterone stimulation. This switch from a prepositioned repressive complex to an active complex appears to be related to the presence of the poly(A) nuclease PARN on the former and its release at the time of activation, allowing unfettered action of the retained XGLD2 PAP (Kim and Richter 2006). Remarkably, the Western analysis of the immuno-enriched αCP2 complexes failed to reveal enrichment for PARN within the oocyte despite the abundant levels of PARN in the input extract and the abundant levels of the other members of the XαCP2/G complex in the coIP (Fig. 5D). Thus the developmental specificity of the C-CPE activity does not appear to reflect association with PARN in the oocytes and may instead reflect a novel controlling mechanism that remains to be further explored.
Discussion
On the basis of the foregoing studies, we conclude that Xenopus αCP2 nucleates a cytoplasmic polyadenylation “G complex” that is active in early X. laevis embryos. This complex is recruited by a noncanonical class of embryonic cis elements: the C-CPEs (C-rich cytoplasmic polyadenylation elements). The C-rich hα-globin PR acts as a C-CPE in the Xenopus embryo and demonstrates the same dependence on the AAUAAA determinant as that seen with the maturation CPE. This cooperative action of the C-CPE and AAUAAA is consistent with the observed interaction between XαCP2 and CPSF. In addition, we demonstrate that the C-CPE can act in a synergistic fashion with an oocyte U/A-rich maturation-type CPE to target PAP activity in the early embryo. This synergy between the C-CPE and the U/A CPE may reflect the observed direct interaction between corresponding binding proteins XαCP2 and CPEB1. While the relationship of these findings to post-transcriptional controls in mammalian cells remains to be explored, the current data are consistent with a model whereby XαCP2 recruits the cytoplasmic polyadenylation machinery to target mRNAs as a method of controlling gene expression in early Xenopus embryos.
C-rich sequences can act as novel CPEs
The current study was initiated with the question of whether the previously established C14 motif in the Xenopus pp2Ac mRNA does in fact target embryonic cytoplasmic polyadenylation activity through recruitment of Xenopus αCP2 (Paillard et al. 2000). We first demonstrated that αCP2 binds to the pp2Ac mRNA in vivo, and then asked whether αCP2-mediated cytoplasmic polyadenylation could be recruited by other known targeting sequences. We focused on the C-rich PR element in the 3′ UTR of the hα-globin mRNA because of its established roles in poly(A) metabolism in mammalian cells (Kong and Liebhaber 2007). We have previously demonstrated in erythroid cells that the PR functions as a stability element that governs the rate of cytoplasmic poly(A) shortening, and have recently shown that the PR functions as an upstream sequence element during nuclear 3′ end formation (X Ji and SA Liebhaber, in prep.). The current studies confirm that the WT hα-globin 3′ UTR RNA can be targeted for cytoplasmic polyadenylation activity in early Xenopus embryos (Fig. 2B). Of importance, this activity was ablated when the PR element was replaced by an unrelated neutral sequence (Fig. 2C). Furthermore, recruitment of the cytoplasmic PAP by the PR element was specifically sensitive to co-injection with a oligo dC17 competitor (Fig. 2F). Taken together, these data suggest that the C-rich PR can govern cytoplasmic polyadenylation.
It should be noted that the primary structure of the C-CPE in the pp2Ac 3′ UTR differs significantly from the PR sequence in the hα-globin 3′ UTR. Although both are C-rich, the pp2Ac C-CPE consists of a homopolymeric stretch (C14), while the PR is more complex, containing a series of single-stranded C-rich patches in a distinct secondary structure (see Fig. 2A; Weiss and Liebhaber 1995; Thisted et al. 2001). These differences, which appear to represent a higher level of structural divergence than seen with the canonical maturation and embryonic CPEs, suggest the C-CPE class may encompass a structurally and functionally divergent set of cytoplasmic control determinants. Further studies of endogenous C-CPEs should reveal whether the exact sequence and positioning of the C-CPE(s) within the 3′ UTR affects timing, efficiency, or other characteristics of polyadenylation, such as independence versus co-dependence on other types of CPEs.
XαCP2 is a novel mediator of the cytoplasmic polyadenylation machinery
In vitro and in vivo approaches in the present report revealed that XαCP2 exists in a complex(es) with proteins that play a central role in cytoplasmic polyadenylation. Prominent among these proteins are CPSF, symplekin, and XGLD2. The in vivo association of XαCP2 with various subunits of the CPSF complex is consistent with the functional studies demonstrating a mutual dependency between the action of the C-CPE and the adjacent AAUAAA in targeting cytoplasmic polyadenylation to a particular mRNA. The presence of symplekin in the complex is consistent with its function as a scaffold for a number of 3′ end formation reactions, both nuclear and cytoplasmic (Mandel et al. 2008). The evidence that the major cytoplasmic PAP GLD2 also interacts with XαCP2 identifies the likely mediator of the polymerase action.
An observation that merits further note is the apparent association of XαCP2 with CPEB. This association is consistent with the observed synergistic actions of the C-CPE with the maturation CPE in both the pp2Ac (Paillard et al. 2000) and the hα-globin 3′ UTRs (Fig. 4). XαCP2 may facilitate recruitment of the CPEB1 complex and/or stabilize the associated polyadenylation machinery on the target mRNA, which might serve to overcome the low levels of CPEB1 in the early embryo. Additionally, it is an open question whether CPEB1 is present in all αCP2/G complexes and is thus recruited to C-CPE–containing 3′ UTRs regardless of the presence of a maturation-type CPE(s).
An unexpected finding in the present study was the observation that the XαCP2 cytoplasmic polyadenylation complex exists in both oocytes and embryos. This was surprising since our studies and studies from Paillard et al. (2000) both revealed that C-CPE mediates polyadenylation activity in fertilized Xenopus embryos and not in meiotically activated oocytes. A survey of multiple proteins in the XαCP2/G complex(es) failed to reveal any critical differences that might account for the selective activity in the embryonic stage of development (Fig. 5D,E). Therefore, it is possible that a novel mechanism governs developmental activation of this complex.
What are the mechanisms that govern regulation of αCP2-mediated cytoplasmic polyadenylation?
We have shown that in mature oocytes, XαCP2 effectively binds its endogenous target pp2Ac and is complexed with cytoplasmic polyadenylation machinery. However, XαCP2-mediated cytoplasmic polyadenylation is inactive at this time. This raises the question of how this type of polyadenylation is activated after fertilization. Although phosphorylation plays a regulatory role in activation of the CPEB/G complex (Radford et al. 2008), it is unclear whether αCP2-associated cytoplasmic polyadenylation is regulated in the same way. While prior studies of αCP2 have identified phosphorylation events that regulate its stability and its affinity for RNA (Leffers et al. 1995; Chang et al. 2007), any phosphorylation events that control protein–protein interactions remain elusive. Additionally, the mobility of XαCP2 by SDS-PAGE appears to be unaltered after fertilization (Gravina et al. 2002; data not shown), implying that XαCP2 is not phosphorylated after fertilization. These observations suggest that distinct mechanism(s) should be considered to explain the developmental specificity of the XαCP2/G complex.
Does αCP2-mediated cytoplasmic polyadenylation exist in mammalian somatic cells?
We have shown that human α2-globin mRNA, a tissue-specific mammalian mRNA, is targeted for cytoplasmic polyadenylation in Xenopus embryos. An obvious next question that we are currently pursuing is whether the C-rich PR stability element mediates a similar cytoplasmic polyadenylation activity in mammalian cells. It is of note in this regard that deletion of the PR decreases hα-globin mRNA stability in erythroid cells in parallel with an increase in the rate of net poly(A) tail shortening (Kong and Liebhaber 2007). The rate of poly(A) tail shortening and corresponding acceleration of mRNA decay is further accentuated by a naturally occurring translation anti-termination mutation (αCS) that allows the elongating ribosome to extend into the 3′ UTR before terminating within the poly(A) addition signal (AAUAAA) and presumably evicting any associated trans factors (Kong and Liebhaber 2007). Thus, in erythroid cell culture, both αCP2 and an AAUAAA binding factor, most likely cytoplasmic CPSF, may act in concert to maximally stabilize the poly(A) tail of the hα-globin mRNA via recruitment of PAP activity. This model is supported by the observations in this study, where αCP2-mediated cytoplasmic polyadenylation can slow the net rate of mRNA deadenylation (Fig. 2E). The question of whether the PR, or any other mRNA stability element(s), acts via recruitment of cytoplasmic polyadenylation activity in mammalian somatic cells remains to be explored. Current evidence suggests that cytoplasmic polyadenylation is an important means of gene regulation in mammalian somatic cells (Choi et al. 2004; Burns and Richter 2008; Novoa et al. 2010). However, studies of αCP2 involvement in mammalian cytoplasmic polyadenylation mechanisms are complicated by its roles in multiple nuclear and cytoplasmic pathways. While we have been able to demonstrate that the mammalian orthologs of αCP2 and GLD2 can interact in vitro (MR Vishnu and SA Liebhaber, unpubl.), the functional correlates of this interaction have yet to be formally tested. In this regard, it is worth noting that there is no phenotype associated with the homozygous deletion of GLD2 in the mouse (Nakanishi et al. 2007), implying that functional redundancy exists in mammalian cells. It will thus be necessary to develop new approaches and experimental models to overcome these difficulties in order to explore possible role(s) for αCP2 in mammalian somatic cytoplasmic polyadenylation and resulting cytoplasmic controls.
MATERIALS AND METHODS
Constructs/reagents
All substrates for in vitro transcription were generated by modification of parent plasmids pSP64T7-αWT and pSP64T7-αNeut. These originating plasmids were created by insertion of WT (αWT) or neutral type (αNeut) α-globin cDNA flanked with a T7 promoter and poly(A33) tail into the EcoRI and PstI sites of the pSP64 vector (Promega). The “neutral” version of α-globin replaces the C-rich PR motif with an equal-sized (42 bp) sequence taken from the hα-globin exon 3 coding region (Kong et al. 2003). All other mutations were introduced using the Quik Change II site-directed mutagenesis kit (Stratagene). The polyadenylation hexanucleotide AAUAAA was mutated to AGUGGA, generating the pSP64T7-αHx- plasmid. A canonical maturation CPE (UUUUUAU) was inserted into the 3′ UTRs by replacing the WT (7 bp) sequence 14 bp downstream from the stop codon and 67 bp upstream of the AAUAAA site of pSP64T7-αWT, pSP64T7-αNeut, and pSP64T7-αHx- to generate pSP64T7-αWT+CPE, pSP64T7-αNeut+CPE, and pSP64T7-αWT+CPE/Hx-. DNA templates for in vitro transcription of 3′ UTR constructs were PCR generated to include or exclude the poly(A) tail.
To create a GST-XαCP2 expressing clone, the ORF from IMAGE clone 5073503 was PCR amplified and inserted into the XhoI site of pET42b(+) (Novagen). To produce protein, pET42-XαCP2 was transformed into BL21-CodonPlus (DE3)-RIL competent cells (Stratagene) and induced with 1mM IPTG overnight at 30°C. Cells were lysed via sonication and lysozyme treatment. Recombinant protein was isolated on glutathione-Agarose beads (Sigma), washed, and eluted with buffer containing 10 mM glutathione. The purified protein was dialyzed against 20 mM Tris (pH 7.6), 100 mM NaCl, 1 mM EDTA, 20% glycerol, and 1 mM DTT prior to use.
Xenopus injection experiments
RNA transcripts were in vitro transcribed using the Maxiscript T7 kit (Ambion) in the presence of [32P]αCTP (Amersham) and 7mG(5′)ppp(5′)G cap structure analog (NEB) and then gel purified. Eggs were obtained from Xenopus females, in vitro fertilized, and stage II embryos were microinjected with labeled RNA substrate as described (Deardorff et al. 1998). After incubation at room temperature, batches of four embryos were disrupted in Trizol reagent (Invitrogen), and total RNA was extracted. A side aliquot of the RNA from the final time point was digested with RNase H/oligo dT. Equal Cherenkov counts of all samples were analyzed on a denaturing urea RNA gel and autoradiographed. Maturation and subsequent injection of oocytes was performed according to the method of Paillard et al. (2000). Briefly, stage VI oocytes were isolated, injected, and then treated for 16 h with progesterone. Three hourly time points were subsequently taken.
Electrophoretic mobility shift and supershift assays
RNA electrophoretic mobility shift and supershift assays (RNA-EMSA) were carried out as described previously (Chkheidze et al. 1999). Briefly, [32P] labeled αWT probe was incubated with 5 μL Xenopus cytoplasmic extract at room temperature for 30 min in 25 μL binding buffer. Unbound RNAs were degraded with RNase T1. Heparin was added to each reaction mixture prior to loading. Reactions were run on a 5% native polyacrylamide gel in 0.5% TBE buffer. Supershift assays were performed using anti-αCP2 antibody (Chkheidze et al. 1999).
Co-IPs
Cytoplasmic extract prepared from Xenopus embryos between stages 2 and 6 according to the method of Desai et al. (1999) or from stage V–VI Xenopus oocytes according to the method of Powers et al. (2001) were quick frozen and stored at −20°C. For each co-IP, ∼1 mg of protein was diluted into 1 mL with TMN buffer (10 mM Tris at pH 7.5, 2.5 mM MgCl2, 100 mM NaCl, 1× Protease Inhibitor Cocktail [Sigma], 1× Halt phosphatase inhibitor cocktail [Pierce], 1 mM PMSF) along with 4 μg of purified antibody. Antibodies used in these co-IP/Western studies included the following: anti-αCP2 (laboratory designation FF3; epitope is conserved in XαCP2) (Chkheidze et al. 1999), anti-symplekin (BD transduction), anti-XGLD2 (gift of M. Wickens, University of Wisconsin, Madison), anti-CPEB (gift of J. Richter, University of Massachusetts Medical School, Worcester), anti-ePAB (gift of J. Steitz, Yale University, New Haven, CT), anti-PARN (gift of M. Wormington, University of Virginia, Charlottesville), anti-CPSF100 (gift of J. Manley, Columbia University, New York), and anti-RPL7 (Ji et al. 2003). Normal rabbit or mouse IgG (Santa Cruz) was used in control co-IP reactions. After preincubation of the antibody with the extract, Protein G–Sepharose (GE Healthcare) was added. Aliquots of immunoprecipitates were treated with RNase cocktail (A and T1; Ambion) for the last hour of this binding step; then each sample was washed extensively with TMN wash buffer (TMN buffer + 0.01% Triton X-100). Immunoprecipitated proteins were eluted in reducing NuPAGE sample buffer (Invitrogen) and subjected to SDS-PAGE and Western analysis. HRP conjugated secondary antibodies used were anti-mouse (GE Healthcare) and anti-rabbit light-chain and anti-guinea pig (Jackson Immunoresearch).
For RNA co-IPs (RIPs), binding was carried out in modified TMN buffer (10 mM Tris at pH 7.5, 2.5 mM MgCl2, 150 mM NaCl, 1× Protease Inhibitor Cocktail [Sigma], 1× Halt phosphatase inhibitor cocktail [Pierce], 1 mM PMSF, 1 mM DTT, and 0.4U/μL RNasein [Promega]). Complexes were washed twice in binding buffer (20 mM HEPES at pH 7.9, 150 mM NaCl) + 0.05% Triton X-100 and then twice in binding buffer + 1% Triton X-100 (Waggoner and Liebhaber 2003a). RNAs were purified using Trizol (Invitrogen), DNase treated, and then reverse transcribed using Superscript III (Invitrogen) in the presence of a 1:1 mixture of oligo dT and random hexamers. MRNA levels were assayed by qRT-PCR using Sybr Green chemistry with the following primers: Pp2Ac forward primer, 5′CGGCAACTTCAGGGGAGCGT; Pp2Ac reverse primer, 5′CAGCATGCAAAGGACGGGGGT; ODC1 forward primer, 5′AAGCGCTCCCCCGTGTCACT; ODC1 reverse primer, 5′TCCGCTCGGGGGAAACTCCA; EF1β forward primer, 5′GGGTCCGTGACGAGCGCTTA; and EF1β reverse primer, 5′TCCCCACAACAAGCCGTCCA.
GST pulldowns
Three micrograms of recombinant GST protein was added to prewashed glutathione-Sepharose and allowed to bind in 1× PBS + 0.5% Triton X-100. Simultaneously, in vitro translation reactions were performed using the T7 or SP6 TNT Quick Coupled System (Promega). The following IMAGE clones were used as templates for in vitro translation: XGLD2A, IMAGE:5084876; XGLD2B, IMAGE:6643643; Xenopus CPEB, IMAGE:6861964; human CPSF160, IMAGE:4300196; and human CPSF30, IMAGE:6199123. The GST-protein conjugated beads were then washed and incubated with the 35S methionine–labeled in vitro translated protein in binding buffer (1× PBS, 2.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail). RNase cocktail (Ambion) was added to the indicated reactions at this point. Beads were then washed extensively with wash buffer (1× PBS, 2.5 mM MgCl2, and 0.25% Triton X-100) and once in binding buffer. Protein in each reaction was eluted in NuPAGE sample buffer, electrophoresed, and autoradiographed.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank M. Wickens, J. Richter, J. Steitz, J. Manley, and M. Wormington for generously providing antibodies. We thank members of the Liebhaber/Cooke laboratory and Klein laboratory for discussions and technical help. M.R.V. was supported by NIH T32-DK07780. P.S.K. was supported by NIH R01-GM076621. S.A.L. was supported by NIH R37-MERIT HL65449.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2587411.
REFERENCES
- Barnard DC, Ryan K, Manley JL, Richter JD 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651 [DOI] [PubMed] [Google Scholar]
- Burns DM, Richter JD 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev 22: 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Santhanam R, Trotta R, Neviani P, Eiring AM, Briercheck E, Ronchetti M, Roy DC, Calabretta B, Caligiuri MA, et al. 2007. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPα-driven myeloid differentiation. Blood 110: 994–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM 2006. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J 25: 2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA 1999. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol Cell Biol 19: 4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, Barash I, Rhoads RE 2004. Insulin and prolactin synergistically stimulate β-casein messenger ribonucleic acid translation by cytoplasmic polyadenylation. Mol Endocrinol 18: 1670–1686 [DOI] [PubMed] [Google Scholar]
- Coll O, Villalba A, Bussotti G, Notredame C, Gebauer F 2010. A novel, noncanonical mechanism of cytoplasmic polyadenylation operates in Drosophila embryogenesis. Genes Dev 24: 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Tan C, Conrad LJ, Klein PS 1998. Frizzled-8 is expressed in the Spemann organizer and plays a role in early morphogenesis. Development 125: 2687–2700 [DOI] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol 61: 385–412 [DOI] [PubMed] [Google Scholar]
- Dickson KS, Bilger A, Ballantyne S, Wickens MP 1999. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol Cell Biol 19: 5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina P, Campioni N, Loreni F, Pierandrei-Amaldi P, Cardinali B 2002. Complementary DNA analysis, expression and subcellular localization of hnRNP E2 gene in Xenopus laevis. Gene 290: 193–201 [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627 [DOI] [PubMed] [Google Scholar]
- Igea A, Mendez R 2010. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J 29: 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Kong J, Liebhaber SA 2003. In vivo association of the stability control protein αCP with actively translating mRNAs. Mol Cell Biol 23: 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Kong J, Carstens RP, Liebhaber SA 2007. The 3′ untranslated region complex involved in stabilization of human α-globin mRNA assembles in the nucleus and serves an independent role as a splice enhancer. Mol Cell Biol 27: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA 1995. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J 14: 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183 [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD 2007. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev 21: 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, Kimble J 2009. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell 16: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Liebhaber SA 2007. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3′ untranslated region. Nat Struct Mol Biol 14: 670–676 [DOI] [PubMed] [Google Scholar]
- Kong J, Ji X, Liebhaber SA 2003. The KH-domain protein αCP has a direct role in mRNA stabilization independent of its cognate binding site. Mol Cell Biol 23: 1125–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H, Dejgaard K, Celis JE 1995. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem 230: 447–453 [PubMed] [Google Scholar]
- MacNicol MC, MacNicol AM 2010. Developmental timing of mRNA translation–integration of distinct regulatory elements. Mol Reprod Dev 77: 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Eastmond DL, Liebhaber SA 2002. Targeting a KH-domain protein with RNA decoys. RNA 8: 1160–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD 2002. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 21: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Kumagai S, Kimura M, Watanabe H, Sakurai T, Kashiwabara S, Baba T 2007. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem Biophys Res Commun 364: 14–19 [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R 2010. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol 12: 447–456 [DOI] [PubMed] [Google Scholar]
- Osborne HB, Duval C, Ghoda L, Omilli F, Bassez T, Coffino P 1991. Expression and post-transcriptional regulation of ornithine decarboxylase during early Xenopus development. Eur J Biochem 202: 575–581 [DOI] [PubMed] [Google Scholar]
- Paillard L, Maniey D, Lachaume P, Legagneux V, Osborne HB 2000. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in Xenopus embryos. Mech Dev 93: 117–125 [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R 2008. A combinatorial code for CPE-mediated translational control. Cell 132: 434–448 [DOI] [PubMed] [Google Scholar]
- Powers M, Evans EK, Yang J, Kornbluth S 2001. Preparation and use of interphase Xenopus egg extracts. Curr Protoc Cell Biol 11: 11.10.1–11.10.24 [DOI] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH 2008. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 1779: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev 63: 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M 2005. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11: 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Kuchler B, Eckmann CR 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev 23: 824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18: 5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Richter JD 1994. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol Cell Biol 14: 7867–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Tassan JP, Richter JD 1992. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev 6: 2580–2591 [DOI] [PubMed] [Google Scholar]
- Simon R, Wu L, Richter JD 1996. Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol 179: 239–250 [DOI] [PubMed] [Google Scholar]
- Slevin MK, Gourronc F, Hartley RS 2007. ElrA binding to the 3′UTR of cyclin E1 mRNA requires polyadenylation elements. Nucleic Acids Res 35: 2167–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T, Lyakhov DL, Liebhaber SA 2001. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and αCP-2KL, suggest distinct modes of RNA recognition. J Biol Chem 276: 17484–17496 [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Liebhaber SA 2003a. Identification of mRNAs associated with αCP2-containing RNP complexes. Mol Cell Biol 23: 7055–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner SA, Liebhaber SA 2003b. Regulation of α-globin mRNA stability. Exp Biol Med 228: 387–395 [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316 [DOI] [PubMed] [Google Scholar]
- Weiss IM, Liebhaber SA 1995. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol Cell Biol 15: 2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J, Pettine SM, Shenk T 1989. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res 17: 3899–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Good PJ, Richter JD 1997. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol 17: 6402–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]