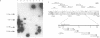Abstract
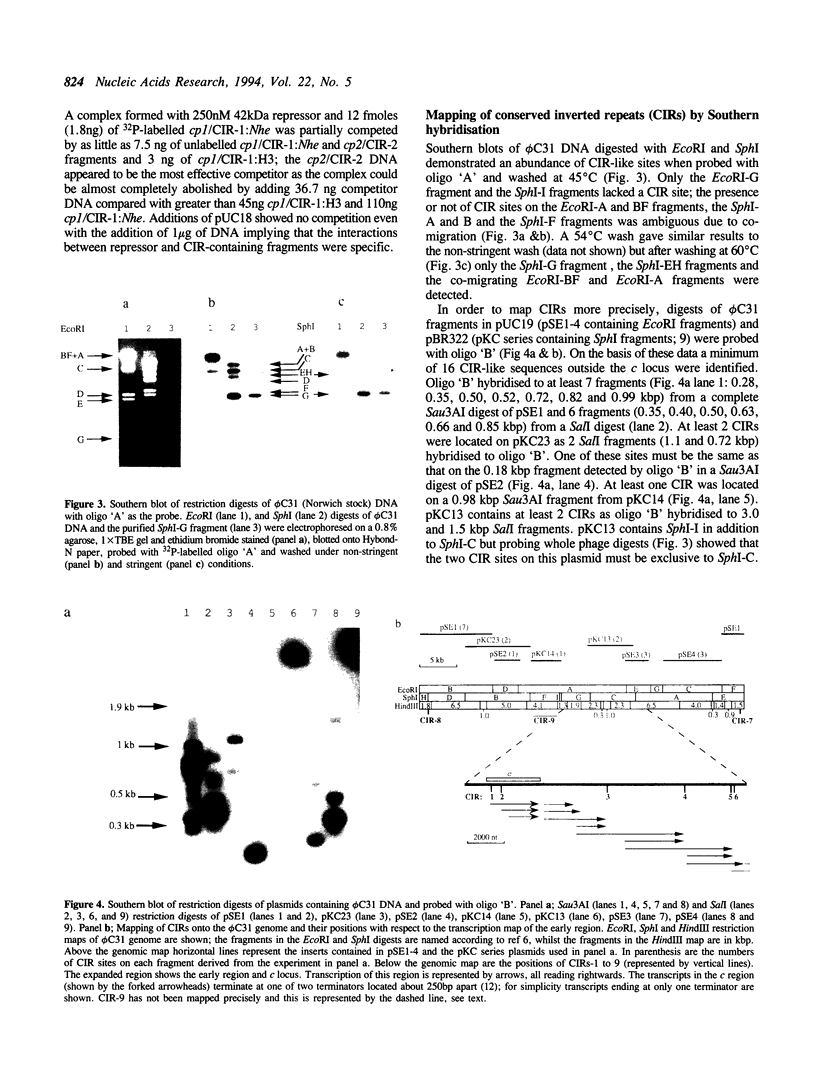
Previous reports have suggested that the repressor gene, c, of phiC31 is autoregulated and that likely operators are conserved inverted repeat sequences (CIRs1&2) located just upstream of the promoters, cp1 and cp2. Evidence is now presented that the CIRs 1&2 are indeed binding sites for one of the three inframe, N-terminally different protein isoforms of 42, 54 and 74 kDa produced by the c gene. A cp1-aphII fusion was repressed in a Streptomyces coelicolor A3(2) phiC31 lysogen and characterisation of an operator-constitutive (Oc) mutant showed a single mutation in CIR-1. CIR-1 containing fragments were retarded in electrophoresis gels by the 42 kDa repressor protein isoform and this retardation was inhibited by the addition of competing DNA fragments containing either CIR-1 or CIR-2. Using a combination of Southern blotting and analysis of available DNA sequence we also show that at least 18 copies of the CIRs are present throughout the phiC31 genome. Alignment of 9 CIR sequences showed that 8 contained a perfectly conserved 17 bp core whilst the exception had a single mismatch. The core includes a 16 bp inverted repeat (IR), and is usually part of a more extensive and less highly conserved palindrome. When superimposed on a previously derived transcription map of the early region, the CIRs lie in intergenic regions associated with transcription initiation and/or termination.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. Structural analysis of loci involved in pSAM2 site-specific integration in Streptomyces. Plasmid. 1989 Jan;21(1):59–70. doi: 10.1016/0147-619x(89)90087-5. [DOI] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989 Mar;8(3):973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton C. J., Guthrie E. P., Chater K. F. Phage vectors that allow monitoring of transcription of secondary metabolism genes in Streptomyces. Biotechnology (N Y) 1991 Jul;9(7):652–656. doi: 10.1038/nbt0791-652. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. E., Chater K. F., Bruton C. J., Piret J. M. The restriction mapping of c gene deletions in Streptomyces bacteriophage phi C31 and their use in cloning vector development. Gene. 1983 May-Jun;22(2-3):167–174. doi: 10.1016/0378-1119(83)90100-2. [DOI] [PubMed] [Google Scholar]
- Ingham C. J., Crombie H. J., Bruton C. J., Chater K. F., Hartley N. M., Murphy G. J., Smith M. C. Multiple novel promoters from the early region in the Streptomyces temperate phage phi C31 are activated during lytic development. Mol Microbiol. 1993 Sep;9(6):1267–1274. doi: 10.1111/j.1365-2958.1993.tb01256.x. [DOI] [PubMed] [Google Scholar]
- Ingham C. J., Smith M. C. Transcription map of the early region of the Streptomyces bacteriophage phi C31. Gene. 1992 Dec 1;122(1):77–84. doi: 10.1016/0378-1119(92)90034-m. [DOI] [PubMed] [Google Scholar]
- Kuhstoss S., Richardson M. A., Rao R. N. Plasmid cloning vectors that integrate site-specifically in Streptomyces spp. Gene. 1991 Jan 2;97(1):143–146. doi: 10.1016/0378-1119(91)90022-4. [DOI] [PubMed] [Google Scholar]
- Laity C., Chater K. F., Lewis C. G., Buttner M. J. Genetic analysis of the phi C31-specific phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2). Mol Microbiol. 1993 Jan;7(2):329–336. doi: 10.1111/j.1365-2958.1993.tb01124.x. [DOI] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Lomovskaya N. D., Chater K. F., Mkrtumian N. M. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol Rev. 1980 Jun;44(2):206–229. doi: 10.1128/mr.44.2.206-229.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Sinclair R. B., Bibb M. J. The repressor gene (c) of the Streptomyces temperate phage phi c31: nucleotide sequence, analysis and functional cloning. Mol Gen Genet. 1988 Aug;213(2-3):269–277. doi: 10.1007/BF00339591. [DOI] [PubMed] [Google Scholar]
- Sinclair R. B., Bibb M. J. Transcriptional analysis of the repressor gene of the temperate Streptomyces phage phi C31. Gene. 1989 Dec 28;85(2):275–282. doi: 10.1016/0378-1119(89)90419-8. [DOI] [PubMed] [Google Scholar]
- Sladkova I. A., Vasil'chenko L. G., Lomovskaia N. D., Mkrtumian N. M. Fizicheskoe kartirovanie DNK aktinofagov Streptomyces coelicolog A3(2). I. Lokalizatsiia c-oblasti aktinofaga phiC31. Mol Biol (Mosk) 1980 Jul-Aug;14(4):910–915. [PubMed] [Google Scholar]
- Smith M. C., Ingham C. J., Owen C. E., Wood N. T. Gene expression in the Streptomyces temperate phage phi C31. Gene. 1992 Jun 15;115(1-2):43–48. doi: 10.1016/0378-1119(92)90538-z. [DOI] [PubMed] [Google Scholar]
- Smith M. C., Owen C. E. Three in-frame N-terminally different proteins are produced from the repressor locus of the Streptomyces bacteriophage phi C31. Mol Microbiol. 1991 Nov;5(11):2833–2844. doi: 10.1111/j.1365-2958.1991.tb01992.x. [DOI] [PubMed] [Google Scholar]
- Suárez J. E., Clayton T. M., Rodríguez A., Bibb M. J., Chater K. F. Global transcription pattern of phi C31 after induction of a Streptomyces coelicolor lysogen at different growth stages. J Gen Microbiol. 1992 Oct;138(10):2145–2157. doi: 10.1099/00221287-138-10-2145. [DOI] [PubMed] [Google Scholar]
- Vasil'chenko L. G., Mkrtumian N. M., Lomovskaia N. D. Geneticheskoe kartirovanie i kharakteristika deletsionnykh mutantov aktinofaga fi C31 Streptomyces coelicolor A3(2), ne sposobnykh k lizogenizatsii. Genetika. 1981;17(11):1967–1974. [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]