Abstract
Genetic influences on the predisposition to complex behavioral or physiological traits can reflect genetic polymorphisms that lead to altered gene product function, and/or variations in gene expression levels. We have explored quantitative variations in an animal's alcohol consumption, using a genetical genomic/phenomic approach. In our studies, gene expression is correlated with amount of alcohol consumed, and genomic regions that regulate the alcohol consumption behavior and the quantitative levels of gene expression (behavioral and expression quantitative trait loci [QTL]) are determined and used as a filter to identify candidate genes predisposing the behavior. We determined QTLs for alcohol consumption using the LXS panel of recombinant inbred mice. We then identified genes that were: 1) differentially expressed between five high and five low alcohol-consuming lines or strains of mice; and 2) were physically located in, or had an expression QTL (eQTL) within the alcohol consumption QTLs. Comparison of mRNA and protein levels in brains of high and low alcohol consuming mice led us to a bioinformatic examination of potential regulation by microRNAs of an identified candidate transcript, Gnb1 (G protein beta subunit 1). We combined our current analysis with our earlier work identifying candidate genes for the alcohol consumption trait in mice, rats and humans. Our overall analysis leads us to postulate that the activity of the GABAergic system, and in particular GABA release and GABA receptor trafficking and signaling, which involves G protein function, contributes significantly to genetic variation in the predisposition to varying levels of alcohol consumption.
Keywords: alcohol consumption, candidate genes, genetical genomics, GABA release, GABA receptor, G protein
1. Introduction
Alcohol use disorders exact a large toll on society in terms of morbidity, mortality and economic losses (American Psychiatric Association, 1994; Hasin et al., 2007; McLellan et al., 2000; US Department of Health and Human Services, 2000). Alcohol use disorders can be characterized as pharmacogenetic disorders, since problems arise from an interaction of a xenobiotic with an organism, and the organism's response to the xenobiotic (ethanol) is determined almost equally by genetic and environmental factors. The CNS responses to ethanol (phenotypes) encompass acute and chronic toxicity, quantity of alcohol consumed, tolerance and physical dependence (Ehlers et al., 2010; Grant et al., 1988; Kril et al., 1997; Leeman et al., 2010; Tabakoff et al., 1986; Tabakoff and Hoffman, 2000). It is of value to consider these (and other) phenotypes separately when searching for either their biological or environmental determinants. Many studies blur the important distinctions between alcohol-related phenotypes, and such studies lead to confusion rather than clarification of the etiology of the various components of alcohol use disorders. At this point, it is also necessary to clarify the fact that the title of this manuscript does not reflect an oversight of alcohol use disorders in females. There is significant evidence in mice and humans for differences between males and females with regard to alcohol-related behavior and toxicity (Barker et al., 2010; Blednov et al., 2005; Devaud et al., 2006; Eagon, 2010; Schulte et al., 2009; Strong et al., 2010). Since we have not completed our studies with females, we should not attempt to extrapolate from the studies on males reported here to the determinants of alcohol use disorders in women.
In our current study, we focus on the genetic determinants of voluntary alcohol intake levels in non-dependent animals. Levels of alcohol intake do correlate positively with the incidence of alcohol dependence (Dawson et al., 2008), but, if nothing else, it immediately becomes obvious that drinking levels are a continuous variable, while alcohol dependence, as it is currently defined (American Psychiatric Association, 1994; Hasin et al., 2006), is a categorical diagnosis. Voluntary alcohol intake levels certainly do not reflect the severity of neuroadaptations which affect the magnitude of withdrawal signs in animals (Crabbe et al., 2010). Furthermore, we would posit that even if the negative reinforcing effects of ethanol can drive alcohol intake in a dependent animal (Koob and Le Moal, 1997), the study of voluntary alcohol intake in a non-dependent animal does not shed light directly on the mechanisms or consequences of alcohol dependence.
Lest we convince the reader that what we will describe is irrelevant to the societal problems that emanate from alcohol dependence, we only have to state that without the intake of alcohol there would be no alcohol dependence, that the propensity for alcohol dependence is related to the quantity of alcohol consumed, and that many societal problems with alcohol do not relate only to dependent individuals (e.g., violence and accidents, etc. while under the influence).
Thus, what are the biological, and particularly the genetically-influenced, predisposing factors to high or low levels of alcohol consumption by males? The experimental approaches many times used in studies of the neurochemical/molecular basis for proclivity to drink alcohol by non-human species, and particularly rodents, have been directed specifically at examining the function of one or another receptor, effector or metabolic enzyme. On the other hand, the assessments of the contributions of genetic loci to alcohol drinking behavior have proposed a minimum of 4 genetic loci in mice and rats (Belknap and Atkins, 2001; Crabbe et al., 2010), and certainly more “genes” which contribute to the genetic variance (approximately 50% of total variance) associated with an animal's level of voluntary ethanol consumption. To address this issue, it has been proposed that a “systems” approach is more appropriate to study polygenic (“complex”) traits such as alcohol preference. The introduction of “genetical genomics” (Jansen and Nap, 2001; Schadt et al., 2003), and the utilization of genetical genomics with the more classical behavioral QTL approaches (Bystrykh et al., 2005; Chesler et al., 2005; Hu et al., 2008; Hubner et al., 2005; Tabakoff et al., 2008; Tabakoff et al., 2009), has resulted in a process by which systems genomics can be applied to the examination of the etiologic factors contributing to complex behavioral (or other clinical) phenotypes.
We have used the genetical genomic/phenomic approach to examine the genetic determinants of alcohol preference in male mice. Specifically, we first identified QTLs for the alcohol drinking paradigm designated “drinking in the dark” (Rhodes et al., 2005; Rhodes et al., 2007), using the LXS panel of recombinant inbred (RI) mouse strains (Williams et al., 2004). We then used microarray analysis to determine transcripts that were physically localized in the QTL and/or had expression QTL (eQTL) (Jansen and Nap, 2001) that overlapped the alcohol drinking QTL, and that were differentially expressed in brains of mice that display high and low alcohol drinking behavior. The combination of the results from this approach, with results from previous genetical genomic/phenomic and genetic studies of rats and humans, respectively (Tabakoff et al., 2009), led to identification of candidate brain signaling systems that predispose to quantitative variation in alcohol consumption in mice, rats and men.
2. Material and Methods
2.1 Alcohol Consumption by LXS Recombinant Inbred Mice
2.1.1. Mice
The LXS recombinant inbred panel was generated from a cross between Inbred Long Sleep (ILS) and Inbred Short Sleep (ISS) mouse strains (Williams et al., 2004). Animal breeding and behavioral testing were conducted in the specific pathogen-free facility at the Institute for Behavioral Genetics, Boulder, CO. At 25 days of age, mice were weaned, and transferred to sex-specific cages. Prior to testing, mice were maintained on a normal 12-hour light/dark cycle, and were given food and water ad libitum.
2.1.2. Alcohol Consumption (“Drinking in the Dark”, DID)
“Drinking in the dark” is a paradigm that makes use of the rodent's normal patterns of eating and drinking, which occur during the nocturnal period. Mice are given limited access to an alcohol solution at the beginning of this dark period. We used male mice from 38 LXS RI strains (median n=9, <4 in LXS23 and LXS32 and 4-15 per strain in remaining strains), ILS and ISS mice (n=7 and 15, respectively), as well as C57BL/6J and DBA 2/J mice (n=25 and 10, respectively). One week prior to testing, animals (60-80 days old) were individually housed in a room set on a reverse light:dark cycle. After this week of acclimatization, mice were weighed one hour prior to lights out (0830), on each of three consecutive test days. Four hours after lights out (1230), water bottles were replaced with 10 mL drinking tubes containing 20% ethanol (v/v in tap water). Two hours after introduction of the alcohol-containing drinking tube (1430), the tubes were removed, the fluid levels were recorded, and the tubes were replaced with water bottles. Alcohol intake was calculated in g/kg for the two-hour interval on day 3, since alcohol intake has been shown to stabilize after the second day of DID testing (Rhodes et al., 2005).
All procedures followed the National Institutes of Health (NIH) guide for the care and use of laboratory animals, and were approved by the University of Colorado, Boulder, Institutional Animal Care and Use Committee (IACUC). All efforts were made to minimize animal suffering, and to utilize in vitro techniques, as required by IACUC protocols.
2.1.3. LXS Genotyping
SNP information on the LXS RI panel was collected by Dr. Gary Churchill and colleagues at The Jackson Laboratory using the Affymetrix Mouse Diversity Genotyping Array. Of the 314,865 SNPs retrieved, 40,158 SNPs indicated differing homozygous genotypes between parental strains (ILS and ISS), had a valid homozygous genotype call for more than 95% of the 38 LXS RI strains with DID information, and had a valid dbSNP identifier. These SNPs represent 3,663 unique strain distribution patterns (SDP) among the 38 RI strains.
2.1.4. QTL mapping
QTL mapping was executed using a weighted marker regression (Carlborg et al., 2005) on strain means for day 3 alcohol intake. Genome-wide p-values were calculated empirically through permutations limited to unique SDP (Churchill and Doerge, 1994). Confidence intervals for QTL location are represented by an 80% Bayesian credible interval (Sen and Churchill, 2001). All calculations were executed in the R/qtl package of R statistical software (Broman et al., 2003).
2.2. Gene Expression Measurements
2.2.1. Mice
High and low alcohol drinking mice were selected for brain gene expression comparison based on their differences in the DID phenotype and/or a 24-hour access/2-bottle choice paradigm (Crabbe et al., 2010). Based on our data, the two parental strains of the LXS panel (ILS and ISS) differ significantly in DID (p=0.003), as do the C57BL/6J and DBA/2J mice (p=1.0×10-6). Others have shown a similar result for C57BL/6J and DBA/2J mice (Rhodes et al., 2007). Both strain pairs, ILS vs ISS mice and C57BL/6J vs DBA/2J mice, have also shown significant differences in alcohol consumption in the 24-hour access/2-bottle choice paradigm (Crabbe et al., 2010) (Bennett & Carosone-Link, unpublished data). Six alcohol-naïve adult male C57BL/6J and DBA/2J mice were obtained from The Jackson Laboratory (Bar Harbor, ME), and five alcohol-naïve adult male ILS and ISS mice were obtained from the Institute for Behavioral Genetics in Boulder, CO for brain gene expression measurements.
Additionally, three replicate selected lines of high alcohol-preferring (HAP) and low alcohol-preferring (LAP) mice were obtained from Indiana University School of Medicine (Dr. Nicholas Grahame). Each independently selected line was generated by selective breeding for alcohol preference in a 24-hour/2-bottle choice paradigm from heterogeneous stock mice (Grahame et al., 1999). Four to six adult alcohol-naïve male mice from each replicate line (HAP1/LAP1, generation 24; HAP2/LAP2, generation 19; HAP3/LAP3, generation 13) were included in our gene expression experiments.
2.2.2. Microarray Processing for Measuring Gene Expression
Mice were sacrificed using CO2 exposure and whole brains were quickly removed, frozen on dry ice, and kept at -70°C until RNA extraction. Brains were divided sagittally and total RNA was extracted from the left half of the brain of each mouse using the RNeasy Midi kit (Qiagen, Valencia, CA) and the RNeasy Mini kit (Qiagen) for cleanup. cDNA from the brain of each individual mouse was hybridized to a separate Affymetrix GeneChip® Mouse Genome 430 2.0 array (Affymetrix, Santa Clara, CA) as previously described (Saba et al., 2006).
2.2.3. Statistical Analysis
Prior to normalization, probes of poor integrity were eliminated from further analysis. Poor integrity probes include probes that did not match the mouse genome exactly, matched the genome in more than one place, or contained a known SNP (see Hoffman et al., 2010 for more details). Overall, 111,297 (22%) probes were eliminated and 3,657 (8%) probesets were removed entirely from the analysis.
Data for each pair of strains/lines were normalized and summarized into probesets separately using RMA (Irizarry et al., 2003). All arrays were examined for quality, and arrays that did not meet our standards were eliminated from further analysis (see (Saba et al., 2006) for more detail on quality control standards).
Statistical analysis of differential expression was limited to probesets that were either located in one of the QTLs identified for DID in the LXS panel, or that had an eQTL whose maximum LOD score was within the QTLs identified for DID. Differential expression was calculated using the empirical Bayes method outlined in Smyth (2004). A false discovery rate was applied to account for multiple testing (Benjamini and Hocberg, 1995). eQTLs were calculated from the BXD recombinant inbred panel, using data obtained from the same Affymetrix microarray (Hoffman et al., 2010) and these data are available on http://Phenogen.ucdenver.edu.
2.3. Protein Expression Analysis
The only transcript that was differentially expressed (see Results) in all groups of high and low alcohol consuming animals was Gnb1 (guanine nucleotide binding protein (G protein), beta 1 subunit), but the differences in the expression of Gnb1 mRNA were not in the same direction across all high alcohol consuming and low alcohol consuming pairs. Therefore, we analyzed the protein levels of Gβ1 in brains of C57BL/6J, DBA/2J, HAP1, LAP1, ISS and ILS mice. Mice were sacrificed with CO2, brains were removed, and homogenates (1600×g supernatants) were prepared and subjected to immunoblotting as previously described (Snell et al., 1996). Protein samples (2.5-10 μg/lane) were separated on 4-12% Bis-Tris NuPage®Novex® gels or 14% Tris-glycine gels (NuPage) (HAP1, LAP1), followed by transfer to nitrocellulose membranes. Membranes were probed with antibodies for mouse Gβ1 (C57BL/6J, DBA/2J, ISS, ILS, ProteinTech Group Inc, Chicago, IL; HAP1, LAP1, Santa Cruz Biotechnology, Santa Cruz, CA; 1:1500), followed by goat anti-rabbit HRP conjugate (BioRad, Hercules, CA, 1:5000-1:20,000). Blots were developed with ECL reagent (Perkin Elmer, Waltham, MA) and exposed to X-ray film (Life Science Products, Frederick, CO). Membranes were stripped with Restore™ Western Blot Stripping Buffer (Thermo Scientific, Rockford, IL) and reprobed with an antibody against a protein loading control (β-actin, Sigma, St. Louis, MO, 1:1500 or β-tubulin, BD Pharmingen, San Diego, CA, 1 μg/ml), followed by secondary antibody and visualization with ECL reagent. Protein bands were quantitated using BioRad QuantityOne software, and values for the G protein subunit were normalized for protein loading by calculating a ratio of the Gβ1 to β-actin or β-tubulin. Statistical significance of differences between strains was determined with a one-sample t-test on the ratios of Gβ1/loading control levels between the high and low alcohol-consuming mice.
2.4. MicroRNA Target Prediction
The transcript and protein level results for Gnb1 and Gβ1 were consistent (i.e., same directionality of differences) across the C57BL/6J vs DBA/2J, and HAP1 vs LAP1comparisons, however, the results for mRNA and protein were in opposite directions in the ILS vs ISS mice (see Results). Therefore, we utilized available bioinformatics programs to assess Gnb1 transcript characteristics and to investigate potential microRNA involvement in the translation of Gnb1 mRNA to protein.
We used the University of California Santa Cruz Genome Browser (Rhead et al. 2010) and their compilation of genes based on RefSeq, UniProt, GenBank, CCDS, and comparative genomics to identify transcripts for Gnb1. To examine the possible presence of microRNA target sites in the Gnb1 mRNA, seven different computational prediction software packages were applied to the 3′-UTR of Gnb1: DIANA microT v3.0 (Maragkakis et al., 2009); http://diana.cslab.ece.ntua.gr/microT/), ElMMo release 3 (Gaidatzis et al., 2007); http://www.mirz.unibas.ch/ElMMo3/), microRNA.org (Betel et al., 2008); http://www.microrna.org/), PicTar (Krek et al., 2005); http://pictar.mdc-berlin.de/), MicroCosm (Griffiths-Jones et al., 2008); http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl), PITA (Kertesz et al., 2007); http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html), TargetScan (Friedman et al., 2009), http://www.targetscan.org/). Because of the high false positive rate of individual programs to predict miRNA targets, we looked for a consensus among the programs. All programs were run using default values, except for PITA where “Minimum seed conservation” was set to 0.1 to minimize the output. For EIMMo, results were filtered so that the “Expected score” was at least 0.3.
3. Results
3.1. Alcohol Consumption
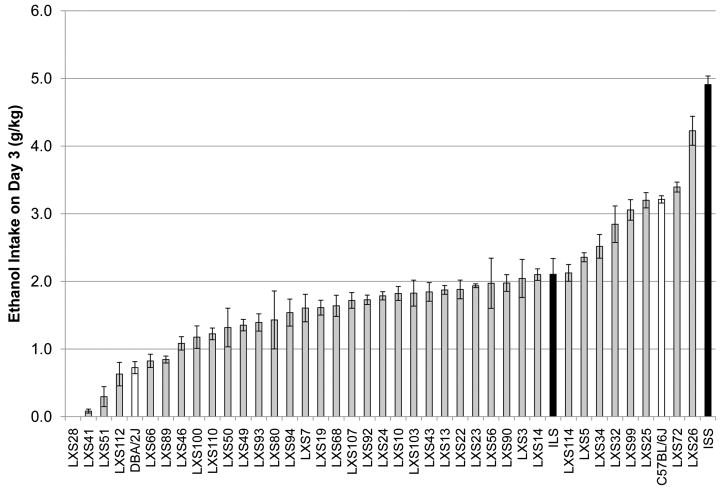
Analysis of variance showed a significant effect of strain (p < 0.0001; r2 = 0.36) on alcohol intake (Figure 1) by the LXS RI panel. ISS mice consumed more alcohol than any other strain, and as predicted by previous research (Rhodes et al 2007), C57BL/6J mice drank more than DBA/2J mice. In one RI strain (LXS28), none of the mice drank any ethanol at all on day 3, thus this strain is represented by a mean of 0 and a standard error of 0 in Figure 1.
Figure 1. Distribution of Drinking in the Dark Consumption across LXS Panel.
Strain means and standard errors displayed are for alcohol consumed in grams per kilogram of body weight during a 2-hour access period on day 3 of the testing paradigm. Only data from male mice have been included. The solid black bars represent the two parental strains of the LXS panel (ILS and ISS), the white bars represent C57BL/6J and DBA/2J inbred strains, and the gray bars represent the LXS recombinant inbred strains.
3.2. QTL mapping
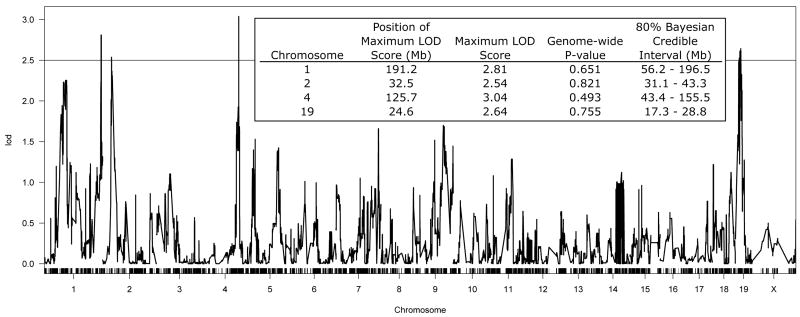
Four QTLs exceeded a LOD threshold of 2.5 (Figure 2). Of the four QTLs, 2 had suggestive genome-wide p-values (Lander and Kruglyak, 1995) (p<0.67)). Lander and Kruglyak proposed a LOD threshold of 1.9-2.8 for “suggestive” QTLs in a genome wide analysis, while Van Ooijon (1999) proposed a LOD threshold of 2.1 for suggestive QTLs in a recombinant inbred panel. As noted by Lander and Kruglyak (1995), a merely suggestive QTL may point to an important genomic region deserving investigation. Furthermore, although these QTLs did not meet strict standards for statistical significance (Lander & Kruglyak, 1995), all four QTL regions have been previously implicated in alcohol consumption in the two-bottle choice paradigm (Table 1; (Belknap and Atkins, 2001; Bice et al., 2009; Tabakoff et al., 2009)). Therefore, these QTLs can be considered as a confirmation of previous work, which reduces the probability that they represent false positive results.
Figure 2. LOD Profile of QTL for Ethanol Consumption (DID) across LXS Panel and Summary of Suggestive QTL.
Strain means from Figure 1 were used in a weighted marker regression to identify QTL for DID. The figure insert includes further information on all peaks with a LOD score greater than 2.5.
Table 1. Comparison of Drinking in the Dark QTLs to Previously Published QTLs for Alcohol Consumption in the Two-Bottle Choice Paradigm.
For the two-bottle choice QTL on chromosome 1, the Mb locations were determined from the physical location of the peak SNP (rs13476012) and extended 50 Mb on either side (50 Mb is approximately equal to 20 cM on chromosome 1 according to http://cgd.jax.org/mousemapconverter/ (Cox et al., 2009)). For the QTLs from Belknap and Atkins (2001), cM positions were converted to Mb locations using the same website.
| Chromosome | QTL for DID in LXS Panel | QTL for Alcohol Consumption in Two-Bottle Choice Paradigm | Reference for Two-Bottle Choice Paradigm | Mapping Population for Two-Bottle Choice Paradigm |
|---|---|---|---|---|
| 1 | 56.2 - 196.5 Mb | 51.8 - 151.8 Mb | (Bice et al., 2009) | populations mainly derived from HAP1/LAP1 |
| 2 | 31.1 - 43.3 Mb | 11.7 - 81.7 Mb | (Belknap and Atkins, 2001) | meta-analysis of populations derived from C57BL/6 and DBA/2 |
| 4 | 43.4 - 155.5 Mb | 125.2 - 141.5Mb | (Belknap and Atkins, 2001) | meta-analysis of populations derived from C57BL/6 and DBA/2 |
| 19 | 17.3 - 28.8 Mb | 11 - 35 Mb (syntenic to rat Chr 1: 213 - 228 Mb) | (Tabakoff et al., 2009) | HXB/BXH recombinant inbred rat panel |
3.2.1. Differentially Expressed Transcripts
Of the 41,380 probesets (16,425 known protein coding regions; 11,317 of these expressed in brain) available on the Affymetrix GeneChip®Mouse Genome 430 2.0 array after the masking procedure, 4,687 probesets (2,081 known protein coding regions) were either physically located in a DID QTL (n=4,279) and/or had a significant eQTL (p<0.10) within one of the DID QTL (n=836). Of the 836 probesets which represented transcripts with eQTLs within bQTLs, 428 transcripts were the products of genes located within the bQTLs (potentially cis eQTLs). When the 4,687 probesets were examined for differential expression among inbred strains and selected lines, the comparison of C57BL/6J to DBA/2J yielded the largest number of differentially expressed (DE) probesets (1,394). ISS and ILS strains had only 174 probesets with expression levels that differed significantly between the two strains. There were similar quantities of DE probesets between HAP1 and LAP1 mice (n=753) and HAP2 and LAP2 mice (n=719). The HAP3 and LAP3 mice had only 18 probesets that reached statistical significance for differential expression. It is likely that selection is not complete within this pair (Grahame et al., personal communication), but the genes that are differentially expressed early in selection are likely to contribute substantially to the selected trait.
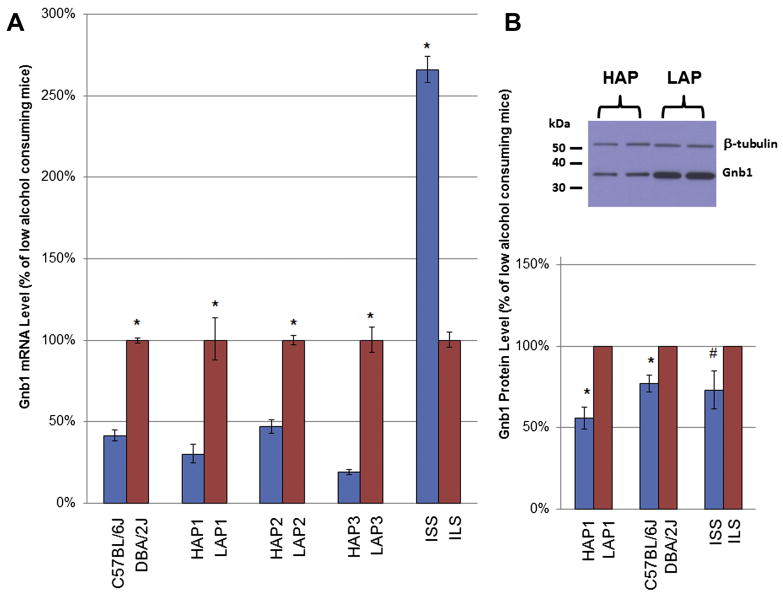
Only one probeset was differentially expressed in all five pairs, i.e., guanine nucleotide binding protein (G protein), beta 1 subunit (Gnb1) (FDR<0.01). Gnb1 is physically located on chromosome 4 near 155Mb and has a significant eQTL (cis eQTL) in the same region. In four of the five pairs, Gnb1 expression had an inverse relationship with alcohol consumption (i.e., higher mRNA levels in the group that consumed less alcohol). In contrast, in the ISS and ILS comparison, Gnb1 was expressed at a higher level in the ISS strain, which also had the higher alcohol consumption (Figure 3A). We examined expression measures for individual probes within the probeset for Gnb1 and all probes indicated a similar magnitude and direction of difference between the tested animals (data not shown).
Figure 3A and 3B. Comparison of Gnb1 Transcript Expression and Gβ1 Protein Expression Levels in Low- and High-Alcohol Consuming Mice.
A) Means and standard errors of expression levels of Gnb1 mRNA (1454696_at) in high-alcohol consuming mice (blue) and low-alcohol consuming mice (red) are shown as a percent of the group mean of the low alcohol–consuming mice in each comparison. B) The top image is a representative example of a western blot showing Gβ1 levels in brains of 1 HAP1 mouse and 1 LAP1 mouse (duplicate samples for each mouse). The bottom figure shows quantitation of Gβ1 protein levels in the brains of 4 HAP1 and 4 LAP1 mice, 5 C57BL/6 and 5 DBA/2 mice, and 2 ILS and 2 ISS mice (mean ± SEM). On each blot, tissue from one or two mice from each strain or line was assayed and data for the high alcohol consuming mice was calculated as a % of the value for the low alcohol consuming mice. Because the western blots were paired, no standard error is shown for the low alcohol consuming mice (i.e., in each case, the value for the low alcohol consuming mice was set to 100%). * FDR<0.01 (transcript expression); *p<0.01, #P=0.07 (protein expression).
3.3. Protein Expression Analysis
When we compared brain protein levels (Gβ1) between four pairs of low and high alcohol consuming mice, the greatest difference was between HAP1 and LAP1 mice (Figure 3B). The direction of difference in protein levels (higher in LAP1 mice) was the same as the direction of difference between transcript levels in these selectively bred animals. The same concordance in the direction of difference in transcript and protein levels was seen in inbred C57BL/6J vs DBA/2J mice, but, in our analysis, the difference in protein levels was more modest (30% higher in brains of DBA/2J mice, n=5 mice/strain). In our initial analysis of ILS and ISS mice, the difference in Gβ1 levels was similar to that seen in C57BL/6J and DBA/2J mice, but in this case, the direction of difference in protein levels (37% higher in ILS mice, n=2 mice/strain) was in the opposite direction to the difference in transcript levels, which was higher in ISS mice. Therefore, the transcript levels and the protein levels in brains of ISS and ILS strains contradict the simple hypothesis that a difference in transcript levels will be directly translated to a difference in protein levels. However, in all cases, there is an association between higher levels of Gβ1 protein and lower levels of alcohol consumption.
3.4. Gnb1 Transcript Isoforms
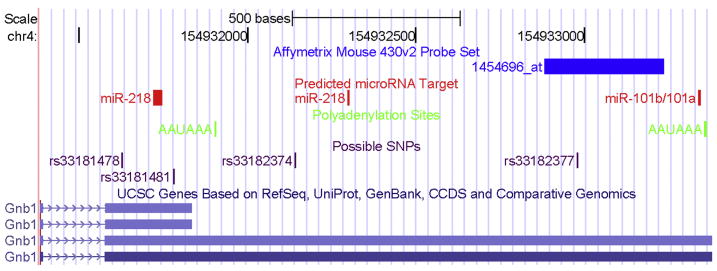
The bioinformatic analysis identified four mouse Gnb1 transcripts. In addition to 2 alternatively spliced exons, another feature that distinguishes transcripts is the presence of either a long or short version of the 3′-untranslated region (UTR). Two transcripts have the long 3′-UTR, and the other two have the short 3′-UTR (Figure 4). The Affymetrix probeset that was differentially expressed (1454696_at) interrogates an area of the 3′-UTR that is only present in the long form of the 3′-UTR (Figure 4). We also looked for a polyadenylation signal (AAUAAA) within the entire 3′-UTR, and found two alternative predicted polyadenylation sites that correspond to the ends of the two forms of the 3′-UTR (Figure 4).
Figure 4. Description of 3′-Untranslated Region of Gnb1.
Two versions of the 3′-UTR region of Gnb1 are predicted among the 4 different Gnb1 transcripts. The Affymetrix probeset that indicated differential expression between all groups of high- and low-alcohol consuming mice is shown in dark blue. Predicted polyadenylation sites (AAUAA) are shown in green, along with the area of the 3′-UTR predicted to bind to the microRNAs miR-101a/b and miR-218 (in red). Sites of SNPs predicted to differ between ISS and three other inbred strains (ILS, C57BL/6J, and DBA/2J) are also marked in purple. This graphic was generated using tools available with the UCSC Genome Browser (Kent et al., 2002).
To investigate the possibility of SNP(s) in the 3′-UTR that may impact the translation of specific transcripts of Gnb1, we retrieved data from the Imputed Genotype Resource at the Center for Genome Dynamics at The Jackson Laboratory (Szatkiewicz et al 2008). We specifically looked for genotyped SNPs in the long version 3′-UTR of Gnb1 (Chr 4:154,931,386 – 154,933,376 bp). Four SNPs were located in this region (rs33181478, rs33181481, rs33182374, and rs33182377). All four had been genotyped directly in the C57BL/6J and DBA/2J strains, but not in the ILS and ISS strains. The C57BL/6J and DBA/2J strains had identical genotypes for all four SNPs. The ILS imputed genotypes were identical to the C57BL/6J and DBA/2J strains (posterior probabilities from 32% to 45%), but the imputed genotypes for the ISS strain were different for all four SNPs (posterior probability of 97% for all four). Although this data set does not provide information on all possible SNPs, and therefore, cannot necessarily identify a functional SNP, it does indicate that ISS has a haplotype in this region that differs from not only ILS, but also from C57BL/6J and DBA/2J. If a SNP did fall within a microRNA binding site, or close enough to the binding site to prevent normal translational control by that microRNA, differences in stability or translation of that particular transcript may occur.
3.5. MicroRNA Binding Site Prediction
In the area that differed between the long and short versions (Chr 4:154,931,836-154,933,376 bp) of the 3′-UTR in Gnb1 mRNA, mmu-miR-218 had a predicted target in the same location across four of the seven software prediction algorithms (154,932,298-154,932,304 bp). In addition, mmu-miR-101a/mmu-miR-101b also had a predicted target in the Chromosome 4: 154,931,836-154,933-376 region across five of the seven software packages (154,933,336-154,933,342 bp). In the more proximal portion of the 3′-UTR of the Gnb1 transcript (i.e., the portion which would be present in all four transcript forms of Gnb1), mmu-miR-218 had an additional predicted target in the same location across three of the seven software packages (154,931,721-154,931,749 bp).
4. Discussion
Prior QTL analyses of the quantitative phenotypes of alcohol intake and alcohol preference in mice have used a number of substrates, including F2 populations derived from selectively bred animals, inbred mice or panels of recombinant inbred mice derived from two spontaneous mutation-derived inbred progenitor strains (Bice et al., 2009; Crabbe et al., 2010). The LXS recombinant inbred (RI) panel of animals is, on the other hand, originally derived from a heterogeneous stock of mice which were produced by purposeful cross-breeding of eight inbred strains (Williams et al., 2004). The LXS RI panel, compared to other currently available RI mouse panels, contains a much broader representation of polymorphisms which are an inherent part of the mouse genome and can contribute to the alcohol intake phenotype. We could expect to see evidence of the influence of these yet unexplored polymorphisms in an alcohol intake measure and a QTL analysis using the LXS RI panel of animals.
The phenotype of alcohol intake in mice has usually been measured using a procedure which allows for 24 hour access to two containers, where one container is filled with an ethanol solution and the other with water (“two-bottle choice paradigm”) (Crabbe et al., 2010). More recently, a robust measure of differences in alcohol intake has been introduced which takes advantage of a rodent's normal diurnal cycle and offers alcohol over a two-hour period at the early part of the daily dark cycle. This procedure has been termed “drinking in the dark” (DID) (Rhodes et al., 2005; Rhodes et al., 2007).
An initial study of the behavioral QTLs associated with DID used a cross between C57BL/6J and FVB/NJ mice (Phillips et al., 2010). The analysis of 600 F2 mice from this cross produced evidence for a QTL on mouse chromosome 11. Comparing these QTL results with the results obtained with the LXS mice underscores the importance of genotype and examined phenotype in the results one obtains in a QTL analysis. The DID phenotype measured in the studies of Phillips et al. (2010) was obtained with mice that had already consumed ethanol for 20 days, were withdrawn from access to ethanol for 24 hours and then tested for DID. In contrast, we were interested in a genetic predisposition to an inherent initial level of alcohol consumption, rather than a phenotype which includes periods of prior ethanol exposure and withdrawal. We did not find a suggestive or significant QTL on Chromosome 11 for the phenotype of DID as measured in our paradigm. When we compared the QTLs we calculated for DID in our studies (Table 1), with QTLs previously reported for alcohol drinking levels measured in paradigms that do not include an extensive period of prior ethanol exposure, we found overlaps with QTLs identified for quantities of alcohol consumed in the two bottle choice paradigm. The QTLs we identified on chromosomes 2 and 4 were similar to those noted by Belknap and Atkins (2001) to be significant and replicable across eight independent studies using the two bottle choice paradigm with populations of mice derived from C57BL/6J and DBA/2J strains. The QTL we identified on chromosome 1 was also noted in the meta-analysis performed by Belknap and Atkins (2001) to be significant, but this locus was not consistently found in all of the examined studies. However, a more recent QTL analysis of alcohol intake in the two bottle choice paradigm using an F2 population derived from HAP and LAP mouse progenitors again identified a significant QTL on chromosome 1 in the same region as identified by our work and in the meta-analysis by Belknap and Atkins (2001). The HAP and LAP mice were selectively bred from the same heterogeneous stock as the ISS and ILS mice, which are the progenitors of the LXS RIs used in our current study. The QTL that we identified on mouse chromosome 19 had not been previously identified in mouse studies of DID or the two bottle choice paradigm. However, we have recently completed a study of alcohol intake by rats (HXB/BXH RI panel) using the 24 hour access, two bottle choice paradigm, and found a QTL on chromosome 1 of the rat at 213-228 Mb (Tabakoff et al., 2009). Interestingly, this region is syntenic with the QTL region on chromosome 19 of the mouse (11-35 Mb).
As satisfying as it is to find regions of a genome that may be contributing to a particular phenotype, or what can be considered “genetically correlated” phenotypes, the question arises as to the identity of the “genes” within the QTL intervals which predispose to a particular trait. If one examines the four chromosomal locations which we identified as QTLs for DID with LXS RI mice, one finds that there are 4,020 protein coding regions in these intervals.
There are two major ways in which genotype can influence phenotype. One is a polymorphism which directly affects the biological function of a “gene” product, and the other is a polymorphism which affects the transcription of RNA from the DNA template, or the stability or translational efficiency of the mRNA. Thus, measures of RNA quantity or sequence can be used as intermediate phenotypes in determining the path between DNA sequence and a quantitative phenotype such as alcohol intake. In the current work, we adopted the premise that if a particular “gene” is contributing to the phenotype of interest, then we should find differences in the expression of mRNA for that “gene” between high alcohol drinking and low alcohol drinking mice. An important caveat would, of course, be that such a differentially expressed gene would have to be physically located within a QTL for the trait of alcohol drinking, and/or that the regulation of the quantity of transcript would have to be mediated by a genomic region located within the trait QTL (i.e., the transcript's eQTL would have to be within the bQTL region associated with alcohol drinking). We examined the progenitors of the populations that were used in the QTL meta-analysis for high and low alcohol intake performed by Belknap and Atkins (2001) (i.e., C57BL/6J and DBA/2 mice), the progenitors of the population used for alcohol intake QTL analysis by Bice et al. (2009) (i.e., the HAP1 and LAP1 mice) and the ISS and ILS mice that are the progenitors of the LXS RI panel used in the current studies, for levels of gene expression in brain. We also added the HAP2/LAP2 and HAP3/LAP3 animals, which are a result of replicate selection experiments using the identical selection paradigm (high and low alcohol intake in the two bottle choice paradigm) as was used for the selection of the HAP1 and LAP1 mice (Chester et al., 2003; Grahame et al., 1999). If a subset of genes is important in determining the trait for which the HAP1 and LAP1 animals were selected, one would expect that selective pressure would result in the segregation of at least some of the same alleles in the HAP2 and HAP3 versus the LAP2 and LAP3 mice.
We found only one transcript that was localized within a QTL for DID (and had an eQTL within this same region), and that was differentially expressed in brains of all five pairs of high and low alcohol consuming mice (C57BL/6J vs DBA/2J; HAP1 vs LAP1; HAP2 vs LAP2; HAP3 vs LAP3 and ISS vs ILS). That transcript (Gnb1) codes for the β1 subunit of the guanine nucleotide binding proteins, which transduce signals from G protein coupled receptors (GPCRs). A surprising observation was that although the transcript level for Gnb1 was higher in the brains of the low alcohol drinking mice in 4 of the pairs (DBA/2J, LAP1, LAP2, LAP3), in the ISS/ILS pair, higher levels of Gnb1 mRNA were found in the ISS mice, which consume more ethanol than the ILS mice. We therefore measured Gβ1 protein levels in brains of C57BL/6J, DBA/2J, HAP1, LAP1 and ISS, ILS mice. We found that Gβ1 protein levels were in all cases higher in the brains of the low alcohol consuming mice (DBA/2J, LAP1 and ILS).
To explain why the levels of mRNA for Gnb1 do not consistently reflect Gβ1 protein levels, we examined the DNA sequence of Gnb1. When one examines the genomic sequence in the region of the 3′-UTR of Gnb1 in the various pairs of mice, one finds that the ISS mice have a different haplotype in this region compared to other mice that we used for measures of brain RNA levels. The 3′-UTR of a transcript is an important region for control of stability and translational efficiency of that transcript (Bolognani and Perrone-Bizzozero, 2008; Danckwardt et al., 2008). A significant amount of processing of the 3′-UTR takes place prior to the addition of the polyA tail, and translocation of the polyA mRNA out of the nucleus (Danckwardt et al., 2008; Shi et al., 2009). We noted that the Gnb1 transcript contains two sites in the 3′-UTR that serve as binding sites for the protein complex that truncates the 3′-UTR and adds the polyA sequence (Shi et al., 2009). Our analysis is consistent with available data that two types of transcripts are generated from the Gnb1 gene (Figure 4). One type of transcript has a short 3′-UTR sequence prior to the polyA sequence, and one has a long 3′-UTR sequence prior to the polyA sequence. If one maps the location of the Affymetrix probes that we used to interrogate the Gnb1 transcript, one finds that the probes target the long form of the 3′-UTR (Figure 4). Therefore, the low alcohol consuming DBA/2J mice have more of this form of Gnb1 mRNA than the C57BL/6J mice, and the low alcohol consuming LAP mice have more of this form than the HAP mice. However, the ILS mice, which have more Gβ1 protein in their brain and drink less alcohol than the ISS mice, have less of the Gnb1 transcript with the long 3′-UTR.
A search for microRNA (miRNA) binding sites in the long version of the 3′-UTR region of Gnb1 revealed probable binding sites for two miRNAs (miR-101a/b and miR-218). These microRNAs have been shown to destabilize mRNA and reduce the translation of mRNA into protein (Fabian et al., 2010; Sempere et al., 2004; Simion et al., 2010; Vilardo et al., 2010). One can speculate that since the sequence of the 3′-UTR of the ISS mice is different from that of the other mice used in our studies, the ISS Gnb1 mRNA would bind the miRNAs with higher affinity and, even though there is more mRNA produced in the ISS mouse brain than the ILS brain, the poorer stability/translation of the ISS mRNA would generate less Gβ1 protein (consistent with our results). While this possible scenario does not directly involve an interaction of ethanol with miRNAs, it may be of interest to note that miRNAs have recently been implicated in several alcohol-related phenomena, including alcohol tolerance, alcohol-induced teratogenesis and alcohol-induced gastrointestinal disease (Miranda et al., 2010; Wang et al. 2009; Pietrzykonski et al. 2008).
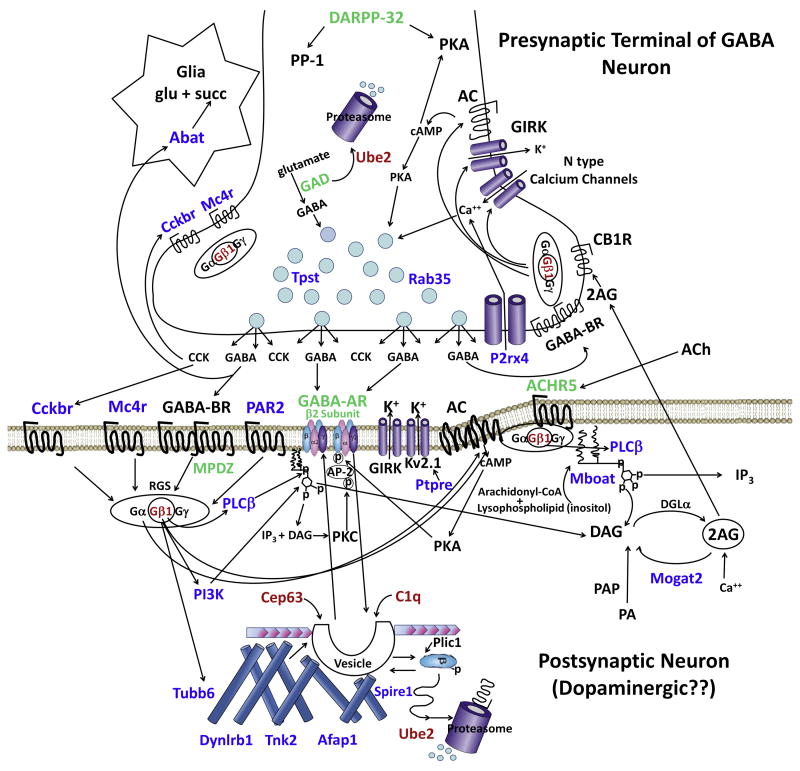
Is the Gβ1 protein the singular genetic determinant of alcohol drinking in male mice? Our own estimates of the contribution of Gnb1 to alcohol intake (Tabakoff et al., 2008) indicate that variation in transcript levels for the Gβ1 protein contribute between 13 and 20% to the overall genetic variance for the trait of alcohol drinking by mice derived from the C57BL/6J and DBA/2J progenitors (i.e., the BXD RI mice). Thus, there are more genetic determinants of alcohol intake than Gnb1. When we focused on pathways and used a systems approach to examine the relationship of differentially expressed genes that contribute to the trait of alcohol drinking in rats, as well as mice, and also examined genetic polymorphisms in the human genome that are associated with quantitative measures of alcohol intake (not alcohol dependence) (Tabakoff et al., 2009), we identified a neuro-signaling pathway that can accommodate most of the genes identified in mice, rats and men (Figure 5). This pathway, which was at the center of the work of Erminio Costa (1998), encompasses both presynaptic and postsynaptic elements of GABA signaling, and Gβ1 fits into this system as an important component (Dupre et al., 2009; Smrcka, 2008). For instance, our studies with humans identified a polymorphism in GABRB2 which was associated with levels of alcohol intake in men. Interestingly, associations between alcohol dependence and polymorphisms in the GABRB2 gene have been reported in Scottish, Finnish and Native American populations (Loh et al., 1999; Radel et al., 2005). Our results were obtained in a population of Australian subjects. GABRB2 is the β2 subunit of the GABAA receptor complex, which is composed of α, β and other (γ, δ, ε, θ, π) subunits in a pentameric structure. The complex always has two β subunits (β1-β3) and β2 is the most common of the β isoforms found in GABAA receptors in brain (α1/β2/γ2) (Whiting et al., 1999). A significant amount of work has been performed examining the effects of ethanol on GABAA receptor function (see review by Kumar et al., 2009), but in many of these studies, the specific role of the β2 subunit in the propensity for alcohol intake by an animal has not been explored. The β2 subunit of the GABAA receptor, and the β subunits in general, have been implicated in both pharmacologic and receptor trafficking components of GABAA receptor function. Chang et al. (2003) demonstrated that a change in a single amino acid in the transmembrane domain of the β2 subunit can alter GABAA receptor activation by GABA, and modulation of this activation by general anesthetics. The β subunits of the GABAA receptor have also been noted to contain a dileucine motif that is integral for the β subunits' interaction with AP-2 adaptin protein which is part of the clathrin-dependent endocytotic process (Herring et al., 2003; Stephenson, 2006). The coding region for this motif (Herring et al., 2003) is in the vicinity of the polymorphic SNP (rs0051667) which was used in our studies (Tabakoff et al., 2009) of the association of genetic polymorphisms and quantitative measures of alcohol consumption. Mutation of the dileucine motif on the GABAA β2 subunit significantly reduces receptor internalization and decreases GABAA receptor mediated Cl- currents in response to GABA (Herring et al., 2003). If one examines Figure 5, it becomes obvious that one of the general differentiating features between high alcohol and low alcohol consuming animals is a multicomponent difference in the microfilament and dynamin endocytotic machinery. Thus, it may be a kinetic issue related to GABAA receptor endocytotic machinery that influences alcohol consumption, rather than any singular component of the endocytotic system.
Figure 5. Proposed Interactions among the Protein Products of Candidate Genes Identified as Contributing to Predisposition to Alcohol Consumption in Mice, Rats and Men.
The figure depicts a GABAergic synapse in brain, with a presynaptic terminal that releases GABA onto a postsynaptic neuron (for example, a dopaminergic neuron in the VTA). The candidate genes and gene products associated with variations in non-dependent alcohol intake, that were identified using the genetic (human) and genetical genomic/phenomic approaches (rats and mice) described in the text, have been integrated into an overall system associated with modulation of presynaptic GABA synthesis and release, and postsynaptic GABA receptor responsiveness and trafficking. The function of each of the candidate gene products shown in this figure is described in Table 2. Candidate genes identified with mice are indicated in red; candidate genes identified with rats are indicated in blue; and candidate genes identified in humans are indicated in green (Tabakoff et al., 2008; Tabakoff et al., 2009).
How does the β subunit of the guanine nucleotide binding protein enter this picture of receptor endocytosis? As shown in Figure 5, activation of GPCRs, be they GABAB receptor, PAR2, CCK or muscarinic M5 receptors, can release Gβγ subunits from their cognate association with Gα. The Gβ1 subunit, in association with Gγ, interacts with a number of effectors, two of which, PLCβ and adenylyl cyclase (AC), generate the second messengers DAG, inositol tris-phosphate and cyclic AMP. These second messengers, in turn, control the activity of PKC and PKA isoforms. Both PKC and PKA can phosphorylate the GABAA receptor β subunit and alter the association of this subunit with the AP2 adaptin protein and, thus, the internalization of GABAA receptors (Song and Messing, 2005). There is some controversy as to whether it is the phosphorylation of the GABAA receptor β subunit that promotes endocytosis (McDonald and Moss, 1997) or the phosphorylation of the AP2 adaptin protein, but in either case, the guanine nucleotide binding protein Gβ1 can initiate the cascade which leads to the internalization of the GABAA receptor or other receptors.
4.1. Conclusions
What has struck us in our studies of the genetic regulation of the phenotype of alcohol consumption across species and strains (isogenic and selected), as well as different populations of humans (Tabakoff et al., 2008; Tabakoff et al., 2009), is that a multitude of seemingly disparate genes are associated with this phenotype. If one attempts to understand a complex phenotype such as alcohol consumption in different species by comparing individual candidate genes or gene products, one finds seemingly unrelated results. However, if one goes a step further and organizes the identified genes and their products from all species, into biologically relevant systems, a concerted picture emerges. What we see from our studies is that the systems that influence GABA release and metabolism and GABA receptor responsiveness and trafficking, are critical in the expression of the quantitative phenotype of alcohol consumption. The point at which this process is influenced in different species or individuals seems less important than the overall level of function of the GABA signaling system. This hypothesis is speculative at this juncture, but we hope that our data support a certain level of curiosity and that we and others can use the results to perform additional work to affirm or reject this hypothesis.
Table 2.
List of Candidate Genes which Influence Levels of Alcohol Consumption in Mice, Rats and Humans.
This list was compiled from the candidate genes derived from genetical/genomic/phenomic studies with mice and rats ((Tabakoff et al., 2008; Tabakoff et al., 2009) and current manuscript) and with humans in a genetic association study with levels of alcohol consumption as the phenotype (Tabakoff et al., 2009).
| Genes Identified in Mice (in Figure 5, the information in red denotes the genes whose protein products are included. | |||
|---|---|---|---|
| Correlation with Drinking | Gene Symbol | Gene Name | Description |
| - | Gnb1 | Guanine Nucleotide Binding Protein, β1 | A component of the trimeric G protein complex activated by GPCRs and can also act independently as an intracellular signaling molecule (Blackmer et al., 2005; Dupre et al., 2009; Smrcka, 2008) |
| + | C1qb | Complement component 1 q subcomponent, β polypeptide | The C1q family proteins have recently taken on a new role due to their location in the CNS: they have been shown to be involved in formation and stabilization of synaptic contacts and in regulation of endocytosis (Iijima et al., 2010; Matsuda et al., 2010; Yuzaki, 2008) |
| + | C1qc | Complement component 1 q subcomponent, C chain | The C1q family proteins have recently taken on a new role due to their location in the CNS: they have been shown to be involved in formation and stabilization of synaptic contacts and in regulation of endocytosis (Iijima et al., 2010; Matsuda et al., 2010; Yuzaki, 2008) |
| + | Scn 4b | Sodium channel, type 4, beta | This is one of the four β subunit proteins that interact with α subunits to form voltage gated sodium channels. This subunit is important for channel opening and closing at various stages of depolarization and repolarization (Bant and Raman, 2010). |
| + | Cep 63 | Centrosomal protein 63 | Centrosomal protein 63 is currently poorly characterized, but is a member of a class of proteins important for localization of centrosomal components and endomembrane compartments. Some evidence of importance in caveolar membrane traffic (King et al., 2003; Mundy et al., 2002) |
| + | Ube2j2 | Ubiquitin-conjugating enzyme E2 | This enzyme is responsible for ubiquitination of proteins targeted for proteasome-dependent degradation. The ubiquitination catalyzed by UBE2J2 can be on lysine or serine residues of the target protein (Wang et al., 2009). |
| Genes Identified in Rats (in Figure 5, the information in blue denotes the genes whose protein products are included). | |||
|---|---|---|---|
| Correlation with Drinking | Gene Symbol | Gene Name | Description |
| + | Afap 1l1 | Actin filament associated protein 1 like | The actin filament associated proteins interact with dynamin to promote endocytosis and also transduce mechanical stretch into c-src protein tyrosine kinase activation (Han et al., 2004) |
| + | Dynlrb1 | Dynein light chain road block type 1 | Part of the dynein complex which regulates protein transport, specifically interacts with Rab6 for transport from endosome via Golgi to ER (Lo et al., 2007; Wanschers et al., 2008) |
| + | Tubb6 | Tubulin B6 | A component of the tubulin complex which interacts with dynein. Tubulin components have also been shown to interact with Gβγ and PAR to mediate endocytosis. Tubulin-GTP can also act through Gq to activate PLCB1 (Popova and Rasenick, 2004; Swift et al., 2010) |
| + | Spire1 | Actin nucleation factor 1 | Important in formation of endosomes. Interacts with F-actin and annexin A2 to produce multivesicular intermediates for endosomes (Morel et al., 2009; Moss et al., 2009) |
| + | Rab 35 | RAS oncogene family member 35 | Rabs are GTPases that interact with a large array of proteins such as Ypt and SNARE to orchestrate membrane trafficking events (Pfeffer and Aivazian, 2004; Rivera-Molina and Novick, 2009; Schimmoller et al., 1998) |
| - | Tnk2 | Protein tyrosine kinase (ACK) | ACK2 possesses a highly conserved clathrin-binding motif and interacts with clathrin and Cdc42. ACK2 acts as an inhibitor of endocytosis (Yang et al., 2001) |
| + | F2rl1 | Protease-activated receptor (PAR2) | GPCR that is activated by trypsin-like serine proteases. PAR2 couples to Gq and Gi and modulates AC and PLC (Ricks and Trejo, 2009; Trejo, 2003) via G protein α and βγ |
| - | Ptpre | Protein tyrosine phosphatase epsilon | This protein tyrosine phosphatase can interact with microtubules (tubulin). This interaction reduces its phosphatase activity. One of the noted targets for PTP epsilon is the Kv2.1 potassium channel. The dephosphorylation of Kv2.1 downregulates its activity (Mohapatra et al., 2007; Tiran et al., 2003) |
| + | Mc4r | Melanocortin 4 receptor | This is a GPCR which has been considered to be coupled to Gs protein. The receptor is responsive to melanocortin and agouti-related peptide and this receptor exists on GABA-containing neurons which control feeding behavior (Fu and van den Pol, 2008; Meister, 2007) |
| + | Cckbr | Cholecystokinin 2 receptor | A GPCR which can exist both pre and postsynaptically. It is a Gq-coupled receptor. Cholecystokinin co-localizes with GABA and dopamine and the cholecystokinin 2 receptor has been shown to control both glutamate and GABA release (Ali and Todorova, 2010; Deng et al., 2010; Kombian et al., 2004) |
| + | Tpst 1 | Tyrosylprotein sulfotransferase 1 | Enzyme which produces the sulfated form of CCK which is necessary for high affinity binding to CCK receptors (Foucaud et al., 2008; Vargas et al., 1994) |
| + | Mboat2 | Membrane-bound D-acetyl transferase 2 | More recently referred to as lysophospholipid acetyltransferase (LPAT). Important in the synthesis and turnover of unsaturated fatty acids in glycerol phospholipids (e.g., phosphatidyl inositol, phosphatidyl choline, etc.). Uses arachidonoyl-CoA or oleoyl-CoA as donor to form the membrane glycerol phospholipids (Corda et al., 2009; Hishikawa et al., 2008) |
| - | Mogat 2 | Monoacylglycerol-O-acyltransferase 2 | Also referred to as Mgat2 catalyzes the conversion of monoacylglycerol to diacylglycerol (DAG). This path can produce DAG independently of phosphatidyl inositol and can also be hypothesized to convert the endogenous endocannabinoid, 2-AG, into DAG (Shi and Cheng, 2009) |
| - | Abat | 4-aminobutyrate (GABA) aminotransferase | Major enzyme for degradation of GABA. Present in both neurons and glia (De Biase et al., 1995) |
| - | P2rx4 | P2x purinoceptor 4 | Receptor gated ion channel responsive to ATP as the agonist. When activated is permeable to Ca++. Found presynaptically on GABA and DA neurons and can control release (enhance release) (Surprenant and North, 2009) |
| + | Fgfr2 | Fibroblast growth factor receptor 2 | Important in development of telencephalon and astrocyte proliferation. In adult, Fgfr2 is of substantial importance in maintaining the pool of dividing CNS progenitor cells in the sub-ventricular zone (Zheng et al., 2004) |
| Genes derived from genetic association studies with humans (phenotype of quantitative measure of alcohol consumption g/kg/day) (in Figure 5, the information in green denotes the genes whose protein products are included) | |||
|---|---|---|---|
| Gene Symbol | Gene Name | Description | |
| GAD1 | Glutamic acid decarboxylase 1 | This gene codes for the 67 kD form of glutamate decarboxylase which is the enzyme that synthesizes GABA from glutamate. There is also another gene that codes a 65 kD form of GAD. GAD67 is constitutively active and responsible for basal GABA production while GAD65 is activated in response to neurotransmission (Wei and Wu, 2008) | |
| MPDZ | Multiple PDZ domain protein | Also known as MUPP1, this is a scaffolding protein shown to promote activation of Gi by GPCRs. MUPP1 associates with the GABAB receptor subunit 2 and controls receptor stability at synapse (Balasubramanian et al., 2007) | |
| CHRM5 | Muscarinic cholinergic Receptor 5 | These are GPCRs coupled primarily to Gq proteins which act to generate depolarization through inhibition of K+ channels and activation of phospholipase C. Located on brainstem DA neurons (among others) (Brown, 2010) and considered as target for drug abuse medication (Raffa, 2009) | |
| GABRB2 | GABAA receptor subunit β2 | The β2 subunit of the GABAA receptor is a critical component of this pentameric receptor gated ion channel complex. The β subunits are part of the ion gating mechanism and are also critical for surface expression and endocytosis of the GABAA receptor. Membrane expression of GABAA receptors is in large part controlled by phosphorylation-dependent interactions of the β subunits with the AP2 clathrin adaptor protein complex (Kumar et al., 2009; Stephenson, 2006) | |
| TUBA8 | Tubulin alpha 8 | Cytoskeletal protein which participates in interactions with actin and dynamin. Recent evidence shows tubulin to also be part of the active zone for transmitter release (Poulain and Sobel, 2010) | |
| PPP1R1B | DARPP32 | Dopamine and cAMP regulated phosphoprotein of 32 KDa is expressed in medium spiny and aspiny neurons (GABA) juxtaposed to DA terminals in several brain areas. Considered as major modulator of DA signaling and NMDA glutamate signals. Recent work indicates involvement of DARPP-32 in excitatory drive of GABAergic neurons and cholinergic neurons (Onn et al., 2008) | |
Acknowledgments
This study was supported in part by grants from NIH, NIAAA: U01AA16649 (PH), U01AA016663 (BT), R24AA013162 (BT) and K01AA 026922 (KK) and by the Banbury Fund (BT). We thank Dr. Sanjiv Bhave, Adam Chapman, Catherine Salipante and Cristina Gonzalez for processing the microarrays; Dr. Tom Johnson, IBG, Boulder, CO for providing tissue from ILS/ISS mice; Drs. Tom Johnson and Gary Churchill, Jackson Laboratories, Bar Harbor, ME for providing SNP information on the LXS RI panel; and Dr. Nicholas Grahame, Indiana University, for providing HAP and LAP mice. We are grateful to Seija Hackl and Heather Daigle for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali AB, Todorova M. Asynchronous release of GABA via tonic cannabinoid receptor activation at identified interneuron synapses in rat CA1. Eur J Neurosci. 2010;31:1196–1207. doi: 10.1111/j.1460-9568.2010.07165.x. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Fourth. American Psychiatric Association; Washington DC: 1994. [Google Scholar]
- Balasubramanian S, Fam SR, Hall RA. GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J Biol Chem. 2007;282:4162–4171. doi: 10.1074/jbc.M607695200. [DOI] [PubMed] [Google Scholar]
- Bant JS, Raman IM. Control of transient, resurgent, and persistent current by open-channel block by Na channel beta4 in cultured cerebellar granule neurons. Proc Natl Acad Sci U S A. 2010;107:12357–12362. doi: 10.1073/pnas.1005633107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JM, Torregrossa MM, Arnold AP, Taylor JR. Dissociation of genetic and hormonal influences on sex differences in alcoholism-related behaviors. J Neurosci. 2010;30:9140–9144. doi: 10.1523/JNEUROSCI.0548-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hocberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Statist Soc B. 1995;57:289–300. [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database Issue):D154–158. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bice P, Valdar W, Zhang L, Liu L, Lai D, Grahame N, Flint J, Li TK, Lumeng L, Foroud T. Genomewide SNP screen to detect quantitative trait loci for alcohol preference in the high alcohol preferring and low alcohol preferring mice. Alcohol Clin Exp Res. 2009;33:531–537. doi: 10.1111/j.1530-0277.2008.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmer T, Larsen EC, Bartleson C, Kowalchyk JA, Yoon EJ, Preininger AM, Alford S, Hamm HE, Martin TF. G protein betagamma directly regulates SNARE protein fusion machinery for secretory granule exocytosis. Nat Neurosci. 2005;8:421–425. doi: 10.1038/nn1423. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Metten P, Finn DA, Rhodes JS, Bergeson SE, Harris RA, Crabbe JC. Hybrid C57BL/6J × FVB/NJ mice drink more alcohol than do C57BL/6J mice. Alcohol Clin Exp Res. 2005;29:1949–1958. doi: 10.1097/01.alc.0000187605.91468.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Brown DA. Muscarinic acetylcholine receptors (mAChRs) in the nervous system: some functions and mechanisms. J Mol Neurosci. 2010;41:340–346. doi: 10.1007/s12031-010-9377-2. [DOI] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, Lu L, Chesler EJ, Alberts R, Jansen RC, Williams RW, Cooke MP, de Haan G. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Carlborg O, De Koning DJ, Manly KF, Chesler E, Williams RW, Haley CS. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21:2383–2393. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- Chang CS, Olcese R, Olsen RW. A single M1 residue in the beta2 subunit alters channel gating of GABAA receptor in anesthetic modulation and direct activation. J Biol Chem. 2003;278:42821–42828. doi: 10.1074/jbc.M306978200. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chester JA, Lumeng L, Li TK, Grahame NJ. High- and low-alcohol-preferring mice show differences in conditioned taste aversion to alcohol. Alcohol Clin Exp Res. 2003;27:12–18. doi: 10.1097/01.ALC.0000046340.06154.9F. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D, Zizza P, Varone A, Filippi BM, Mariggio S. The glycerophosphoinositols: cellular metabolism and biological functions. Cell Mol Life Sci. 2009;66:3449–3467. doi: 10.1007/s00018-009-0113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E. From GABAA receptor diversity emerges a unified vision of GABAergic inhibition. Annu Rev Pharmacol Toxicol. 1998;38:321–350. doi: 10.1146/annurev.pharmtox.38.1.321. [DOI] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, Tsaih SW, Churchill GA, Broman KW. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Belknap JK. The Complexity of Alcohol Drinking: Studies in Rodent Genetic Models. Behav Genet. 2010 doi: 10.1007/s10519-010-9371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DA, Li TK, Grant BF. A prospective study of risk drinking: at risk for what? Drug Alcohol Depend. 2008;95:62–72. doi: 10.1016/j.drugalcdep.2007.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D, Barra D, Simmaco M, John RA, Bossa F. Primary structure and tissue distribution of human 4-aminobutyrate aminotransferase. Eur J Biochem. 1995;227:476–480. doi: 10.1111/j.1432-1033.1995.tb20412.x. [DOI] [PubMed] [Google Scholar]
- Deng PY, Xiao Z, Jha A, Ramonet D, Matsui T, Leitges M, Shin HS, Porter JE, Geiger JD, Lei S. Cholecystokinin facilitates glutamate release by increasing the number of readily releasable vesicles and releasing probability. J Neurosci. 2010;30:5136–5148. doi: 10.1523/JNEUROSCI.5711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaud LL, Risinger FO, Selvage D. Impact of the hormonal milieu on the neurobiology of alcohol dependence and withdrawal. J Gen Psychol. 2006;133:337–356. doi: 10.3200/GENP.133.4.337-356. [DOI] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Rebois RV, Hebert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagon PK. Alcoholic liver injury: influence of gender and hormones. World J Gastroenterol. 2010;16:1377–1384. doi: 10.3748/wjg.v16.i11.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Walter NA, Dick DM, Buck KJ, Crabbe JC. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addict Biol. 2010;15:185–199. doi: 10.1111/j.1369-1600.2009.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Foucaud M, Archer-Lahlou E, Marco E, Tikhonova IG, Maigret B, Escrieut C, Langer I, Fourmy D. Insights into the binding and activation sites of the receptors for cholecystokinin and gastrin. Regul Pept. 2008;145:17–23. doi: 10.1016/j.regpep.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LY, van den Pol AN. Agouti-related peptide and MC3/4 receptor agonists both inhibit excitatory hypothalamic ventromedial nucleus neurons. J Neurosci. 2008;28:5433–5449. doi: 10.1523/JNEUROSCI.0749-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, Lumeng L. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29:47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dufour MC, Harford TC. Epidemiology of alcoholic liver disease. Semin Liver Dis. 1988;8:12–25. doi: 10.1055/s-2008-1040525. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Bai XH, Lodyga M, Xu J, Yang BB, Keshavjee S, Post M, Liu M. Conversion of mechanical force into biochemical signaling. J Biol Chem. 2004;279:54793–54801. doi: 10.1074/jbc.M406880200. [DOI] [PubMed] [Google Scholar]
- Hasin D, Hatzenbuehler ML, Keyes K, Ogburn E. Substance use disorders: Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) and International Classification of Diseases, tenth edition (ICD-10) Addiction. 2006;101 1:59–75. doi: 10.1111/j.1360-0443.2006.01584.x. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Herring D, Huang R, Singh M, Robinson LC, Dillon GH, Leidenheimer NJ. Constitutive GABAA receptor endocytosis is dynamin-mediated and dependent on a dileucine AP2 adaptin-binding motif within the beta 2 subunit of the receptor. J Biol Chem. 2003;278:24046–24052. doi: 10.1074/jbc.M301420200. [DOI] [PubMed] [Google Scholar]
- Hishikawa D, Shindou H, Kobayashi S, Nakanishi H, Taguchi R, Shimizu T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc Natl Acad Sci U S A. 2008;105:2830–2835. doi: 10.1073/pnas.0712245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PL, Bennett B, Saba LM, Bhave SV, Carosone-Link PJ, Hornbaker CK, Kechris KJ, Tabakoff B. Using the Phenogen website for “in silico” analysis of morphine-induced analgesia: identifying candidate genes. Addict Biol. 2010 doi: 10.1111/j.1369-1600.2010.00254.x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326:792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- Iijima T, Miura E, Watanabe M, Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombian SB, Ananthalakshmi KV, Parvathy SS, Matowe WC. Cholecystokinin activates CCKB receptors to excite cells and depress EPSCs in the rat rostral nucleus accumbens in vitro. J Physiol. 2004;555:71–84. doi: 10.1113/jphysiol.2003.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Drug abuse: hedonic homeostatic dysregulation. Science. 1997;278:52–58. doi: 10.1126/science.278.5335.52. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kril JJ, Halliday GM, Svoboda MD, Cartwright H. The cerebral cortex is damaged in chronic alcoholics. Neuroscience. 1997;79:983–998. doi: 10.1016/s0306-4522(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Kumar S, Porcu P, Werner DF, Matthews DB, Diaz-Granados JL, Helfand RS, Morrow AL. The role of GABA(A) receptors in the acute and chronic effects of ethanol: a decade of progress. Psychopharmacology (Berl) 2009;205:529–564. doi: 10.1007/s00213-009-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits, guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Leeman RF, Heilig M, Cunningham CL, Stephens DN, Duka T, O'Malley SS. Ethanol consumption: how should we measure it? Achieving consilience between human and animal phenotypes. Addict Biol. 2010;15:109–124. doi: 10.1111/j.1369-1600.2009.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KW, Kogoy JM, Rasoul BA, King SM, Pfister KK. Interaction of the DYNLT (TCTEX1/RP3) light chains and the intermediate chains reveals novel intersubunit regulation during assembly of the dynein complex. J Biol Chem. 2007;282:36871–36878. doi: 10.1074/jbc.M705991200. [DOI] [PubMed] [Google Scholar]
- Loh EW, Smith I, Murray R, McLaughlin M, McNulty S, Ball D. Association between variants at the GABAAbeta2, GABAAalpha6 and GABAAgamma2 gene cluster and alcohol dependence in a Scottish population. Mol Psychiatry. 1999;4:539–544. doi: 10.1038/sj.mp.4000554. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M, Yuzaki M. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- McDonald BJ, Moss SJ. Conserved phosphorylation of the intracellular domains of GABA(A) receptor beta2 and beta3 subunits by cAMP-dependent protein kinase, cGMP-dependent protein kinase protein kinase C and Ca2+/calmodulin type II-dependent protein kinase. Neuropharmacology. 1997;36:1377–1385. doi: 10.1016/s0028-3908(97)00111-1. [DOI] [PubMed] [Google Scholar]
- McLellan AT, Lewis DC, O'Brien CP, Kleber HD. Drug dependence, a chronic medical illness: implications for treatment, insurance, and outcomes evaluation. JAMA. 2000;284:1689–1695. doi: 10.1001/jama.284.13.1689. [DOI] [PubMed] [Google Scholar]
- Meister B. Neurotransmitters in key neurons of the hypothalamus that regulate feeding behavior and body weight. Physiol Behav. 2007;92:263–271. doi: 10.1016/j.physbeh.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Mohapatra DP, Park KS, Trimmer JS. Dynamic regulation of the voltage-gated Kv2.1 potassium channel by multisite phosphorylation. Biochem Soc Trans. 2007;35:1064–1068. doi: 10.1042/BST0351064. [DOI] [PubMed] [Google Scholar]
- Morel E, Parton RG, Gruenberg J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev Cell. 2009;16:445–457. doi: 10.1016/j.devcel.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Moss FJ, Imoukhuede PI, Scott K, Hu J, Jankowsky JL, Quick MW, Lester HA. GABA transporter function, oligomerization state, and anchoring: correlates with subcellularly resolved FRET. J Gen Physiol. 2009;134:489–521. doi: 10.1085/jgp.200910314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy DI, Machleidt T, Ying YS, Anderson RG, Bloom GS. Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J Cell Sci. 2002;115:4327–4339. doi: 10.1242/jcs.00117. [DOI] [PubMed] [Google Scholar]
- Onn SP, Lin M, Liu JJ, Grace AA. Dopamine and cyclic-AMP regulated phosphoprotein-32-dependent modulation of prefrontal cortical input and intercellular coupling in mouse accumbens spiny and aspiny neurons. Neuroscience. 2008;151:802–816. doi: 10.1016/j.neuroscience.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Reed C, Burkhart-Kasch S, Li N, Hitzemann R, Yu CH, Brown LL, Helms ML, Crabbe JC, Belknap JK. A method for mapping intralocus interactions influencing excessive alcohol drinking. Mamm Genome. 2010;21:39–51. doi: 10.1007/s00335-009-9239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova JS, Rasenick MM. Clathrin-mediated endocytosis of m3 muscarinic receptors. Roles for Gbetagamma and tubulin. J Biol Chem. 2004;279:30410–30418. doi: 10.1074/jbc.M402871200. [DOI] [PubMed] [Google Scholar]
- Poulain FE, Sobel A. The microtubule network and neuronal morphogenesis: Dynamic and coordinated orchestration through multiple players. Mol Cell Neurosci. 2010;43:15–32. doi: 10.1016/j.mcn.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Raffa RB. The M5 muscarinic receptor as possible target for treatment of drug abuse. J Clin Pharm Ther. 2009;34:623–629. doi: 10.1111/j.1365-2710.2009.01059.x. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Ford MM, Yu CH, Brown LL, Finn DA, Garland T, Jr, Crabbe JC. Mouse inbred strain differences in ethanol drinking to intoxication. Genes Brain Behav. 2007;6:1–18. doi: 10.1111/j.1601-183X.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Ricks TK, Trejo J. Phosphorylation of protease-activated receptor-2 differentially regulates desensitization and internalization. J Biol Chem. 2009;284:34444–34457. doi: 10.1074/jbc.M109.048942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci U S A. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L, Bhave SV, Grahame N, Bice P, Lapadat R, Belknap J, Hoffman PL, Tabakoff B. Candidate genes and their regulatory elements: alcohol preference and tolerance. Mamm Genome. 2006;17:669–688. doi: 10.1007/s00335-005-0190-0. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Simon I, Pfeffer SR. Rab GTPases, directors of vesicle docking. J Biol Chem. 1998;273:22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- Schulte MT, Ramo D, Brown SA. Gender differences in factors influencing alcohol use and drinking progression among adolescents. Clin Psychol Rev. 2009;29:535–547. doi: 10.1016/j.cpr.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Cheng D. Beyond triglyceride synthesis: the dynamic functional roles of MGAT and DGAT enzymes in energy metabolism. Am J Physiol Endocrinol Metab. 2009;297:E10–18. doi: 10.1152/ajpendo.90949.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion A, Laudadio I, Prevot PP, Raynaud P, Lemaigre FP, Jacquemin P. MiR-495 and miR-218 regulate the expression of the Onecut transcription factors HNF-6 and OC-2. Biochem Biophys Res Commun. 2010;391:293–298. doi: 10.1016/j.bbrc.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Smrcka AV. G protein betagamma subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 2008;65:2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Snell LD, Nunley KR, Lickteig RL, Browning MD, Tabakoff B, Hoffman PL. Regional and subunit specific changes in NMDA receptor mRNA and immunoreactivity in mouse brain following chronic ethanol ingestion. Brain Res Mol Brain Res. 1996;40:71–78. doi: 10.1016/0169-328x(96)00038-1. [DOI] [PubMed] [Google Scholar]
- Song M, Messing RO. Protein kinase C regulation of GABAA receptors. Cell Mol Life Sci. 2005;62:119–127. doi: 10.1007/s00018-004-4339-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson FA. Structure and trafficking of NMDA and GABAA receptors. Biochem Soc Trans. 2006;34:877–881. doi: 10.1042/BST0340877. [DOI] [PubMed] [Google Scholar]
- Strong MN, Yoneyama N, Fretwell AM, Snelling C, Tanchuck MA, Finn DA. “Binge” drinking experience in adolescent mice shows sex differences and elevated ethanol intake in adulthood. Horm Behav. 2010;58:82–90. doi: 10.1016/j.yhbeh.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surprenant A, North RA. Signaling at purinergic P2X receptors. Annu Rev Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- Swift S, Xu J, Trivedi V, Austin KM, Tressel SL, Zhang L, Covic L, Kuliopulos A. A novel protease-activated receptor-1 interactor, Bicaudal D1, regulates G protein signaling and internalization. J Biol Chem. 2010;285:11402–11410. doi: 10.1074/jbc.M110.105403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Cornell N, Hoffman PL. Alcohol tolerance. Ann Emerg Med. 1986;15:1005–1012. doi: 10.1016/s0196-0644(86)80119-6. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Hoffman PL. Animal models in alcohol research. Alcohol Res Health. 2000;24:77–84. [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19:352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiran Z, Peretz A, Attali B, Elson A. Phosphorylation-dependent regulation of Kv2.1 Channel activity at tyrosine 124 by Src and by protein-tyrosine phosphatase epsilon. J Biol Chem. 2003;278:17509–17514. doi: 10.1074/jbc.M212766200. [DOI] [PubMed] [Google Scholar]
- Trejo J. Protease-activated receptors: new concepts in regulation of G protein-coupled receptor signaling and trafficking. J Pharmacol Exp Ther. 2003;307:437–442. doi: 10.1124/jpet.103.052100. [DOI] [PubMed] [Google Scholar]
- US Department of Health and Human Services . 10th Special Report to the US Congress on Alcohol and Health. National Instutes of Health, National Institute on Alcohol Abuse and Alcoholism; Bethesda, MD: 2000. [Google Scholar]
- Vargas F, Frerot O, Brion F, Trung Tuong MD, Lafitte A, Gulat-Marnay C. 3′-Phosphoadenosine 5′-phosphosulfate biosynthesis and the sulfation of cholecystokinin by the tyrosylprotein-sulfotransferase in rat brain tissue. Chem Biol Interact. 1994;92:281–291. doi: 10.1016/0009-2797(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, Hansen TH. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol. 2009;187:655–668. doi: 10.1083/jcb.200908036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanschers B, van de Vorstenbosch R, Wijers M, Wieringa B, King SM, Fransen J. Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton. 2008;65:183–196. doi: 10.1002/cm.20254. [DOI] [PubMed] [Google Scholar]
- Wei J, Wu JY. Post-translational regulation of L-glutamic acid decarboxylase in the brain. Neurochem Res. 2008;33:1459–1465. doi: 10.1007/s11064-008-9600-5. [DOI] [PubMed] [Google Scholar]
- Whiting PJ, Bonnert TP, McKernan RM, Farrar S, Le Bourdelles B, Heavens RP, Smith DW, Hewson L, Rigby MR, Sirinathsinghji DJ, Thompson SA, Wafford KA. Molecular and functional diversity of the expanding GABA-A receptor gene family. Ann N Y Acad Sci. 1999;868:645–653. doi: 10.1111/j.1749-6632.1999.tb11341.x. [DOI] [PubMed] [Google Scholar]
- Williams RW, Bennett B, Lu L, Gu J, DeFries JC, Carosone-Link PJ, Rikke BA, Belknap JK, Johnson TE. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- Yang W, Lo CG, Dispenza T, Cerione RA. The Cdc42 target ACK2 directly interacts with clathrin and influences clathrin assembly. J Biol Chem. 2001;276:17468–17473. doi: 10.1074/jbc.M010893200. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. Cbln and C1q family proteins: new transneuronal cytokines. Cell Mol Life Sci. 2008;65:1698–1705. doi: 10.1007/s00018-008-7550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Nowakowski RS, Vaccarino FM. Fibroblast growth factor 2 is required for maintaining the neural stem cell pool in the mouse brain subventricular zone. Dev Neurosci. 2004;26:181–196. doi: 10.1159/000082136. [DOI] [PubMed] [Google Scholar]