Abstract
S. cerevisiae grown on plastic surfaces formed organized structures, termed minicolonies, that consisted of a core of round (yeast-like) cells surrounded by chains of filamentous cells (pseudohyphae). Minicolonies had much higher affinity for plastic than unstructured yeast communities growing on the same surface. Pseudohyphae at the surface of these colonies developed further into chains of asci. These structures suggest that pseudohyphal differentiation and sporulation are sequential processes in minicolonies. Consistent with this idea, minicolonies grown under conditions that stimulated pseudohyphal differentiation contained higher frequencies of asci. Furthermore, a flo11Δ mutant, which fails to form pseudohyphae, yielded normal sporulation in cultures but was defective for minicolony sporulation. When minicolonies were dispersed in water and cells then allowed to settle on the plastic surface, these cells sporulated very efficiently. Taken together, our results suggest that sporulation in minicolonies is stimulated by pseudohyphal differentiation because these pseudohyphae are dispersed from the core of the colony.
Keywords: pseudohyphae, sporulation, Flo11, meiosis, minicolony
Introduction
Biofilms are bacterial or fungal communities that tightly bind plastic or other surfaces. For example, the yeast C. albicans, a common pathogen in humans, forms an organized bi-layer on plastic surfaces consisting of a basal layer of ovoid cells and an upper layer of chains of filamentous cells (Baillie & Douglas, 1999, Chandra, et al., 2001, Khot, et al., 2006). Biofilms that form on medical devices like catheters and heart valves are a major contributor to the high mortality rate associated with hospital-acquired fungal infections (Puzniak, et al., 2004, Pfaller & Diekema, 2007, Tumbarello, et al., 2007). S. cerevisiae is the most extensively studied yeast species, but the extent that this non-pathogenic yeast can form biofilms is uncertain. S. cerevisiae can adhere tightly to plastic surfaces (Reynolds & Fink, 2001), but a clinical isolate of S. cerevisiae growing on plastic did not generate the extracellular material and increased drug resistance characteristic of C. albicans biofilms (Chandra, et al., 2001). One limitation to using S. cerevisiae as a model for biofilms is that the structure of S. cerevisiae communities growing on plastic is not known. Thus, the degree to which S. cerevisiae can serve as a model for pathogenic biofilms remains an open question.
C. albicans biofilms undergo a dimorphic transition from ovoid to mycelial cells. This transition stimulates two key aspects of pathogenesis: adhesion to plastic and ability to invade host tissue (Hube, 2006, Verstrepen & Klis, 2006). Diploid S. cerevisiae also can undergo a dimorphic switch, in this case from ovoid to pseudohyphal cells, i.e. elongated cells containing one nucleus per cell. This dimorphic transition occurs during conditions of nutrient limitation, especially limitation for a nitrogen source (Gancedo, 2001, Dickinson, 2008). Nutrient limitation also promotes sporulation, i.e. meiosis and spore formation, in this yeast (Honigberg & Purnapatre, 2003, Kassir, et al., 2003). The connection between pseudohyphal differentiation and sporulation is unclear, but a number of regulators independently activate the dimorphic switch and repress sporulation. These regulators include the transcription factor Rme1p, the G1 cyclin Cln2p, the trimeric G-protein subunit Gpa2p, and the monomeric G-protein, Ras2 (Matsuura, et al., 1990, Gimeno, et al., 1992, Colomina, et al., 1999, Donzeau & Bandlow, 1999, Purnapatre, et al., 2002, van Dyk, et al., 2003). These results suggest that sporulation and the dimorphic switch may be mutually exclusive differentiation programs. However, in some cases pseudohyphal differentiation and spore formation may occur sequentially; for example, colonies of the SK1 strain background form high frequencies of pseudohyphae at their bottom surface, and these elongated cells can eventually sporulate to form dyad and 3–4 spore linear asci (Piccirillo & Honigberg, 2010).
Different strain backgrounds of laboratory yeast show considerable variation both in their ability to form pseudohyphae and in their ability to sporulate. Much of the variation in pseudohyphae can be traced to the FLO11 gene. FLO11 (also known as MUC1) encodes a cell surface protein, termed an adhesin, required not only for pseudohyphal differentiation but also for the affinity of cells to plastic and to other cells (Lambrechts, et al., 1996, Lo & Dranginis, 1998). Interestingly, although required for diploid pseudohyphal growth, FLO11 is not required for the most aspects of haploid filamentous growth (Cullen & Sprague, 2002, Vyas, et al., 2003, Vopalenska, et al., 2010). Allelic variation between strains is common both in FLO11 itself (Kron, 1997, Fidalgo, et al., 2006) and in regulators of FLO11 expression such as the Flo8p transcriptional activator (Liu, et al., 1996, Kobayashi, et al., 1999). Expression of other adhesin proteins (e.g. Flo1p and Flo10p) can also affect pseudohyphal formation (Fichtner, et al., 2007, Van Mulders, et al., 2009). Variability between laboratory strain backgrounds in their sporulation efficiency is also common. This variability can be attributed in part to allelic differences in key transcription factors regulating the initiation of sporulation such as IME1 and RME1 (Deutschbauer & Davis, 2005, Ben-Ari, et al., 2006, Gerke, et al., 2009).
The current study reports that a common laboratory strain of S. cerevisiae (SK1) formed organized structures when grown on plastic surfaces, and these structures were absent in other commonly used laboratory strains. We term these structures “minicolonies” to distinguish them from microcolonies, i.e. early stages of colony development on agar plates. We investigated the organization of ovoid, pseudohyphal and sporulated cells within minicolonies and also examined the connection between pseudohyphal differentiation and sporulation within these colonies.
Materials and methods
Strains, media, and growth conditions
Strains are shown in Table 1. Except where noted, 400 cells/250 μl YPA-1 medium/well were inoculated in 96-well polystyrene microtiter plates (Nunclon) and grown at 24° C without shaking for six days. YPA-1 medium contains 0.25% yeast extract, 0.5% peptone and 1% potassium acetate, pH 5.8. YPA-2 medium contained 0.5% yeast extract, 1% peptone and 1% potassium acetate, pH 5.8; YPA-3 medium contained 1% yeast extract, 2% peptone and 1% potassium acetate, pH 5.8.
Table 1.
Yeast strains used in this study
| Strain | Background | Genotype |
|---|---|---|
| SH3881 | W303 |
MATa/MATα ade2/ade2 can1:ADE2:CAN1/can1:ADE2:CAN1 his3-11,15/his3-11,15 lys2(3′Δ):HIS3:lys2(5′Δ)/LYS2 URA3/ura3-1 |
| SH2533 | Σ1278b |
MATa/MATα ura3-52/ura3-52 his3::hisG/HIS3 leu2::hisG/leu2::hisG YEp24 |
| SH2081 | S288C |
MATa/MATα his3Δ/his3Δ leu2Δ/leu2Δ met15Δ/MET15 LYS2/lys2Δ ura3Δ/ura3Δ |
| SH4329 | SK1 |
MATa/MATα ade3/ADE3 ARG6/arg6 gal80::LEU2/gal80::LEU2 his4-G/his4-N ho::LYS2/ho::LYS2 leu2::hisG/leu2::hisG lys2/lys2 trp1::hisG/trp1::hisG ura3/URA3 |
| SH4325 | SK1 | Same as SH4329 except ura3/ura3 flo11Δ:URA3/flo11Δ:URA3 |
Minicolony growth and differentiation assays
Before assaying minicolony diameter, cell growth, sporulation, or viability, the microtiter wells containing the minicolonies were first examined using a dissecting microscope. Rare wells containing “flors”-aggregations of cells growing on top of the medium- or otherwise containing many cells not associated with minicolonies were not analyzed further. Minicolony diameter was measured using AnalySIS software (SIS) on light micrographs; for each minicolony image, we drew the smallest circle possible that contained all cells (including pseudohyphae), then measured the diameter of this circle.
To measure cell number in minicolonies, 200 μl of the medium was removed from the well without disturbing the minicolonies. Cells removed at this stage amounted to less than 5% of the cells remaining in the well. Next, minicolonies from a single well were suspended in the remaining approximately 50 μl of spent medium by vigorous pipetting, then sonicated for three 6-second pulses with a GE 50 Ultrasonic Processor. To measure sporulation, the same procedure was followed except that all of the medium was removed from the well, and then cells resuspended from the central portion of the well using 25 μl of sterile dH2O. Dilutions of the cell suspensions were examined by light microscopy using a hemacytometer to calculate both the total number of cells in the well and the fraction of these cells that had sporulated (sporulation efficiency).
To measure viability and sporulation capability in minicolonies, we resuspended cells as above, transferred them to a 1.6 ml Eppendorf tube, and then washed the cells 3× in 1 ml sterile dH2O. After washing, cells were suspended in 200 μl sterile dH2O, sonicated as above, and examined in the microscope to ensure that >90% of cells were not attached to other cells. To measure viability, approximately 500 cells from a serial dilution were plated on YPD medium, and the fraction of cells that developed into colonies determined. To measure sporulation capability, cells were diluted ten-fold from their original concentration in sterile dH2O, and 250 ul of this dilution (containing ~ 1 × 105 cells) was placed into a microtiter well and incubated as before at 24° C. After three days, cells were resuspended and examined by microscope to determine the fraction that had sporulated.
Plastic-affinity assays
Affinity of minicolonies for polystyrene microtiter plates was measured after 6 days of incubation, except where noted. At this time, the spent medium was removed from each well, and the wells washed four times. Washes were performed by placing 200 μl of dH2O into the well, incubating at room temperature for 3 minutes, and then removing the water by touching a P-200 pipetman tip to the edge of the well. The total number of cells present in the spent medium and the washes was compared to the number resuspended in a final resuspension of cells from the plastic surface. This final resuspension was achieved by vigorously pipetting 200 μl of dH2O up and down in the well approximately eight times.
Scanning electron microscopy
SK1 at the same concentration as above in 400 μl YPA-1 medium was inoculated in a single well of an 8-chamber Permanox slide (Lab-Tek). After incubation at 24°C, spent medium was removed from the well, and the well was washed 4× with 200 μl cacodylate buffer (0.2M sodium cacodylate [pH 7.2]). The sample was then incubated for 90 minutes at room temperature in 2.5 % glutaraldehyde. The fixative was removed from the wells, and the wells then washed 2× with sodium cacodylate buffer and 2× with O buffer (100 mM KH2PO4, 10 mM MgCl2, pH 6.0). Wells were then incubated for 1 hr at room temperature in a fume hood in 1% OsO4 in O buffer, then washed 2× in O buffer and 3× in dH2O. After removing the last wash, cells were progressively incubated for 5 minutes each in 33%, 67%, 85%, 95% and 2× in 100% ethanol. Finally, ethanol was replaced with hexamethyldisilazane, the sample dried in a fume hood overnight, and coated with gold/palladium. Samples were visualized in a FEG ESEM XL30 scanning electron microscope (FEI/Philips, Hillsboro, OR).
Light microscopy and time-lapse photography
Light microscopy images in Figures 1, 2 and 4 were captured using a Nikon Eclipse TE2000-U inverted microscope, a Colorview II camera, and AnalySIS software (SIS). For time-lapse photography, 14 hrs after inoculation, a Nikon TEM inverted microscope was focused on a single minicolony with a 20× objective. Images were captured every 10 minutes using a Canon Powershot G5 digital camera controlled by GBTimelapse software (Granite Bay Software). Video was created in Adobe® Premiere Pro CS4 and After Effects CS4. After the video was assembled, variation in luminescence between frames was reduced using VirtualDub (1.3).
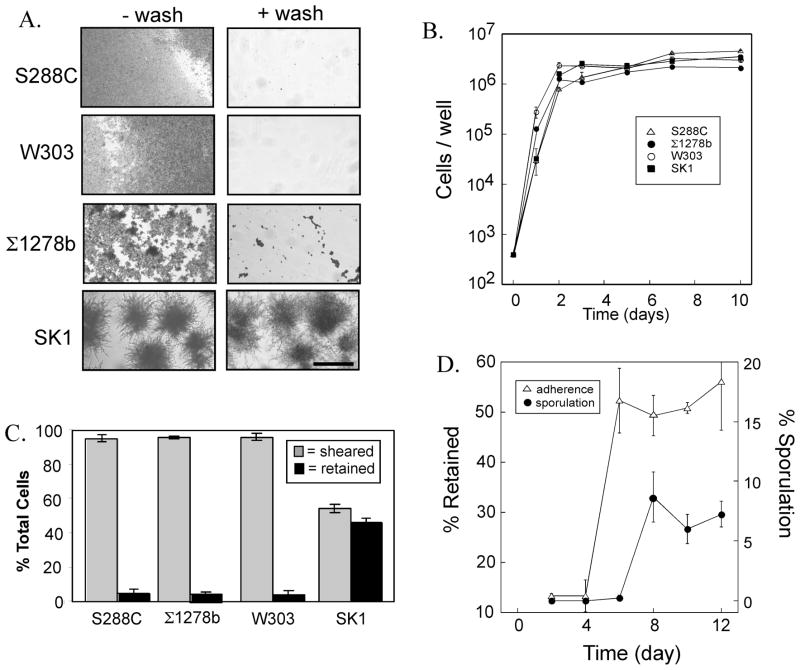
Fig. 1. Comparison of yeast strain backgrounds for minicolony sporulation and affinity to plastic.
A) 6-day minicolonies of the indicated strain backgrounds before washing (left column of images) and after washing (right column). Strains used: W303 (SH3881), ©1278b (SH2533), S288C (SH2081), SK1 (SH4329). Scale bar = 200 μ. B) Growth rates of the same strains as in (A) on a plastic surfaces, growth measured as the total number of cells/well at the indicated times, data expressed as mean ± S.E.M., n=3. C) Affinity to plastic for the indicated strains after 6-day incubation, “sheared cells” are cells removed in the supernatant or in any of four sequential washes, “retained cells” are cells remaining after these washes. D) Affinity to plastic and sporulation of SK1 strain. At the indicated times, the fraction of cells retained on the plastic after washing (open triangles) and the fraction of asci in minicolonies (filled circles) were assayed.
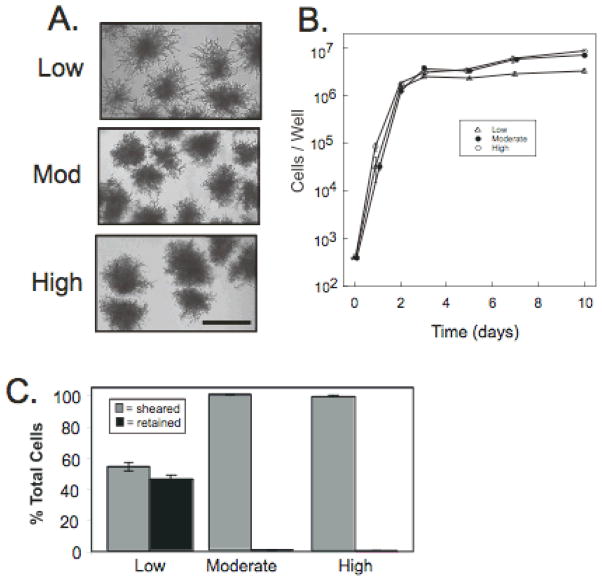
Fig. 2. Effect of nutrient concentration on pseudohyphae formation and cell-surface affinity.
A) Morphology of SK1 (SH4329) minicolonies grown for 6 days in YPA-1 (low concentration nitrogen source), YPA-2 (intermediate concentration nitrogen source), and YPA-3 (high concentration nitrogen source). B) Growth rate of SK1 in the indicated media as in (A). C) Affinity to plastic for 6 day SK1 minicolonies grown in the indicated media as in (A), and assayed as in Fig. 1.
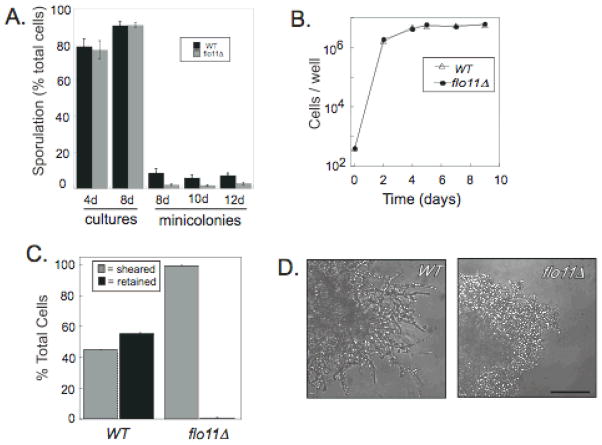
Fig. 4. Roles of FLO11 in minicolony development.
A) Sporulation levels of wild type (WT, SH4329) and flo11Δ (SH4325) in suspension cultures and minicolonies. Strains were grown in suspension for 4 or 8 days in YPA-1 (n=3) or grown as minicolonies for 8, 10, or 12 days in the same medium (n=6), and the percentage asci in the population determined B) Growth rate of WT and flo11Δ minicolonies. C) Affinity of 6-day WT and flo11Δ minicolonies to well of microtiter plate assayed as in Fig. 1. D) Morphology of WT and flo11Δ minicolonies after 6 days incubation. Scale bar = 50 μ.
Results
Comparison of four laboratory strains of S. cerevisiae for growth and sporulation on plastic surfaces
To begin an investigation of the growth, morphology and sporulation capacity of S. cerevisiae on plastic surfaces, we examined four common laboratory strains of S. cerevisiae (SK1, W303, Σ1278b, and S228C) for their growth on polystyrene microtiter plates. Both sequence analysis and historical evidence indicates that among these four strains, S288C and W303 are the most closely related, whereas SK1 is the most distant strain (Mortimer & Johnston, 1986, Schacherer, et al., 2007, Liti, et al., 2009). We grew these strains in rich acetate medium (YPA-1) with the idea that this medium would allow growth, pseudohyphal differentiation and sporulation.
Cells were inoculated in 96-well format microtiter plates and incubated at 24°C without agitation. After six days of incubation, the structure of the cell community growing on the plastic surface was much different in the four strains (Fig. 1A). In the SK1 culture, and to some extent in the Σ1278b culture, discreet structures were formed on the plastic surface, whereas in the other two strains, growth resulted in a thin layer of cells that covered the entire plastic surface. Despite these differences in appearance, the rate of growth of the four strains was similar, and growth was largely complete by 48 hrs in all four strains (Fig. 1B).
We term the compact structures growing in SK1 and Σ1278b cultures “minicolonies”. SK1 minicolonies reached a diameter of approximately 261 ± 8 μ (n=24) after 6 days and did not increase significantly after a further 3 days of incubation (260 ± 8 μ, n=24). These SK1 minicolonies uniformly contained chains of pseudohyphal (elongated) cells extending out from a dense central core. Σ1278b minicolonies contained fewer and shorter pseudohyphal chains than SK1 minicolonies. S288C and W303 minicolonies did not yield visible pseudohyphae.
We next determined the efficiency of sporulation in each of the four strain backgrounds grown as above. Consistent with the relative efficiencies of these laboratory strains in both colonies and cultures (Piccirillo & Honigberg), we found that after 10 days of incubation, 5.8 ± 1.1 % of SK1 cells formed asci, whereas only 2.9 ± 0.4% of W303, 0.8 ± 0.3% of Σ1278b, and 0 % of S288C formed asci (n=3).
Relative affinity of four laboratory strains to plastic
To measure the affinity of the four strains to the surface of microtiter wells, minicolonies were grown for 6 days as above, then the wells washed repeatedly with water. We compared the fraction of cells that were removed from the wells to those that were retained on the plastic surface after washing (see Materials and Methods). We found that approximately 45% of SK1 cells were retained on the plates after four washes, whereas <5% of any of the three other strains were retained under these conditions (Fig. 1A and 1C). Because SK1 minicolonies efficiently formed pseudohyphae, sporulated, and adhered to plastic, we chose this strain background for subsequent experiments.
Timing of sporulation and increase in affinity in SK1
We next determined the timing of adherence and sporulation in minicolonies. After 2 or 4 days of incubation, the affinity of minicolonies to plastic remained low (<5% of cells adhered) but by 6 days approximately 50% of the cells adhered and this fraction did not change even after 12 days of incubation (Fig. 1D). Sporulation increased in minicolonies between 6 and 8 days; thus, the increase in affinity preceded spore formation (Fig 1D).
Effect of nutrient concentration on minicolony growth, morphology, affinity to plastic, and sporulation
Because conditions containing plentiful nutrients, and especially nitrogen, inhibit pseudohyphal differentiation, we next investigated whether pseudohyphal differentiation, affinity to the underlying surface, and sporulation in SK1 minicolonies depended on the concentration of nutrients in the rich medium. For this purpose, we compared cultures grown as above and containing the same concentration of acetate but varying concentrations of the other components of the medium (yeast extract and peptone). These two components provide nitrogen and other essential nutrients such as vitamins, salts and minerals. We found that minicolonies formed at all three nutrient media, termed YPA-1 (lowest nutrient concentration), YPA-2 (intermediate nutrient concentration), and YPA-3 (highest nutrient concentration). However, pseudohyphae were most prominent at the lowest nutrient concentrations (YPA-1, Fig. 2A), which was the medium used for the experiments shown in Fig. 1. Not surprisingly, cell number reached a slightly lower plateau value in this medium than in medium containing higher concentrations of nutrients (Fig. 2B). In line with the correlation between pseudohyphae and cell affinity observed in different strain backgrounds, minicolonies grown on medium that yielded the highest level of pseudohyphae also had the highest affinity to plastic (Fig. 2C). In addition, as the concentration of nitrogen in the media was increased, the efficiency of sporulation in minicolonies progressively decreased (8.7±2.1 in YPA-1, 3.7±0.7 in YPA-2, and undetectable in YPA-3, n=6). Because YPA-1 medium led to the highest level of pseudohyphal differentiation, adherence to plastic, and sporulation in minicolonies, this medium was employed for all subsequent experiments.
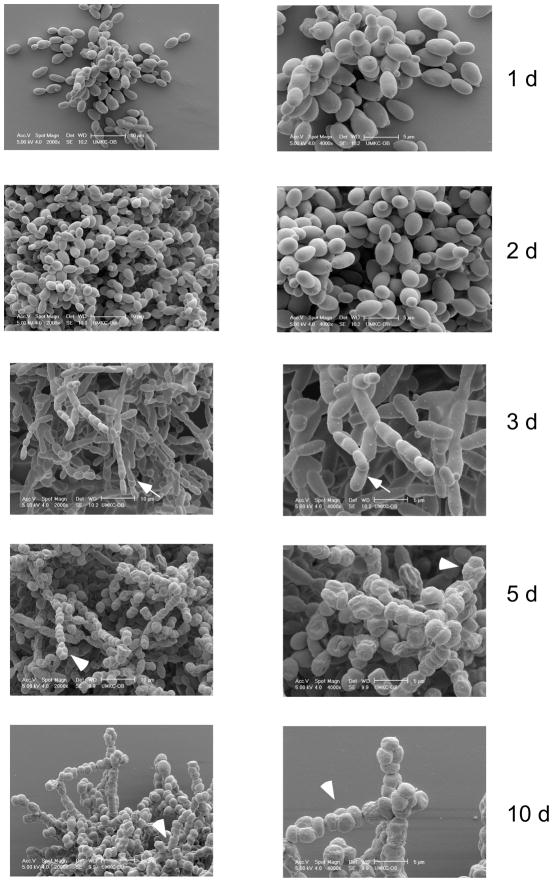
Three phases of minicolony development
To further investigate the structure of minicolonies containing pseudohyphal cells, we examined the structures of these colonies at various stages of development using scanning electron microscopy (Fig. 3). After one or two days, minicolonies consisted of mostly ovoid (i.e. yeast-like) cells. After three days, many pseudohyphal cells were visible extending from the body of the colony. After five days, many of these pseudohyphal cells had sporulated to form linear asci, and by ten days most of the cells on the surface of the colony had sporulated. Thus the development of minicolonies proceeded through a phase of growth as ovoid cells, then a phase of growth as pseudohyphae, and finally a phase in which the pseudohyphae formed asci. The development of pseudohyphae occurred at the same time or slightly before the increase in affinity we noted previously (compare Fig. 3 to Fig. 1D).
Fig. 3. Yeast minicolony development.
Scanning electron micrographs of minicolonies incubated for the indicated times. The left column of images are magnified 1000×, 10 μ bar. The right column are magnified 2000×, 5 μ bar. Arrows indicate representative chains of elongated cells. Arrowheads indicate representative sporulated cells.
As an independent test of minicolony development, we monitored minicolony development over time using time-lapse light microscopy. Except at the earliest times, this method only discriminates cells at the edge of the minicolony. Nevertheless, this experiment confirmed that wild-type yeast at the minicolony’s edge transition from round to pseudohyphal cells at 2–3 days, and that these pseudohyphae begin to sporulate by 5–6 days (Supplementary Information, Video 1).
Increased frequencies of dyads and linear tetrads among minicolony asci
The experiments in the previous section suggest that sporulation in minicolonies occurs preferentially in the pseudohyphal cells extending outward from the colony’s core. This interpretation is consistent with our finding that the medium that best promoted pseudohyphal differentiation (YNA-1) also yielded the highest rates of sporulation. Also consistent with this interpretation is the observation that many of the asci visible in scanning electron microscopy and time-lapse microscopy were dyads or linear 3–4 spore asci rather than tetrahedral asci. Indeed, earlier studies indicate that pseudohyphae and other elongated cells often sporulate to form two-spored asci (dyads) and linear tetrads (Hawthorne, 1955, Thomas & Botstein, 1987, Piccirillo & Honigberg, 2010). To further test the idea that pseudohyphae in minicolonies sporulate preferentially to other cells in the colony, we compared the structure of asci formed in minicolonies to that in liquid cultures grown in the same cultures. We found that 73 ± 7 % (n=4) of the asci observed after resuspending 8 day minicolonies were dyads as compared to only 11 ± 1 % (n=3) dyads in 4 day liquid cultures. Furthermore, 55 ± 9% (n=4) of the 3–4 spored asci in 8 day minicolonies were linear rather than tetrahedral as compared to only 0.2 ± 0.2 % (n=3) in 4 day liquid cultures. These results are consistent with the idea that pseudohyphal differentiation is a prerequisite for efficient sporulation in minicolonies.
Role of Flo11p adhesin in minicolony development
To further test the idea that sporulation in minicolonies occurs primarily in pseudohyphae extending from the core of the colony, we measured sporulation in a flo11Δ mutant, which is defective in forming pseudohyphae. Indeed, the ability of different strain backgrounds to form pseudohyphae in minicolonies correlates with their reported FLO11 expression-levels. That is, FLO11 is expressed at much higher levels in SK1 than in Σ1278b (Strudwick, et al.), whereas this gene is expressed in much lower levels in W303 and S288C than in Σ1278b (Trachtulcova, et al., 2000, Fichtner, et al., 2007). The latter two strains express only low levels of FLO11 because they both contain non-functional alleles of FLO8, a transcriptional activator required for efficient FLO11 transcription (Liu, et al., 1996).
We first compared suspended cultures of the flo11Δ mutant to the wild type for ability to sporulate in suspended cultures. The mutant sporulated to approximately the same high levels as the wild type in these cultures (Fig. 4A, bars 1–4). In contrast, flo11Δ minicolonies sporulated significantly less efficiently than wild-type minicolonies (P<0.05) at all three time points examined (Fig. 4A, bars 5–10). The flo11Δ minicolonies grew at approximately the same rate as the wild type (Fig. 4B), and as reported previously (Reynolds & Fink, 2001), had much lower affinity for plastic (Fig. 4C). As a result of this lower affinity, we were not able to prepare mutant minicolonies for SEM; however, we could perform time-lapse light microscopy without disturbing the sample. As expected, light microscopy revealed that the flo11Δ mutant failed to undergo the dimorphic transition observed for the wild type (Fig. 4D, and Video 2 in Supplementary Information). Sporulation in flo11Δ minicolonies was also significantly delayed relative to the wild type, and even after extended incubation, the majority of asci visible in the flo11Δ samples were either detached from the minicolonies or in chains of ovoid cells extending outward from the minicolonies (Video 2 in Supplementary Information). The efficient sporulation of rare dispersed cells around flo11Δ minicolonies suggests that sporulation occurs efficiently in the minicolony pseudohyphae because these pseudohyphae are dispersed from the core of the minicolony.
To determine whether at least a part of the sporulation defect in flo11Δ minicolonies resulted from a loss in viability in this mutant, cells from wild-type or flo11Δ minicolonies were suspended after 6 days of incubation and plated on rich glucose medium (n=3). In this experiment, we found that 63±2 % of cells in flo11Δ minicolonies were able to re-enter growth and form colonies, as compared to only 34±2 % of the cells from wild-type minicolonies. Thus the viability in the mutant minicolonies was actually higher than in the wild type.
Cells from dispersed minicolonies sporulate efficiently
As a test of the idea that pseudohyphae sporulate efficiently in minicolonies because they are dispersed from the core of the colony, cells from 3-day minicolonies were resuspended, washed, dispersed by sonication, and then diluted ten-fold in water. The cultures were then allowed to settle in a microtiter well, incubated for an additional 3 days, and examined through the microscope. We observed very high levels of sporulation in the dispersed cells from both wild-type minicolonies (69 ± 10 % of cells, n=3) and flo11Δ minicolonies (74 ± 7 % of cells, n=3) cultures relative to the levels in undisturbed minicolonies (Fig. 4A). This result is consistent with the idea that pseudohyphae sporulate efficiently in minicolonies because these pseudohyphae are dispersed from the colony.
To summarize our results regarding the relationship between pseudohyphal differentiation and sporulation in minicolonies: 1) sporulation in minicolonies occurred primarily in pseudohyphae extending outward from the colony’s core, 2) media conditions that prevented pseudohyphae formation in minicolonies also inhibited sporulation in these colonies, 3) flo11Δ minicolonies, which do not form pseudohyphae, failed to sporulate efficiently though they maintained high cell viability, 4) A high percentage of the asci were dyads or linear 3–4 spored asci. 5) Cells dispersed mechanically from minicolonies sporulate much more efficiently than cells in the core of undisturbed minicolonies. Taken together, these results indicate that sporulation in minicolonies is predominately limited to the pseudohyphal cells extending from the periphery of the colony, perhaps simply because these cells are dispersed from the core of the colony.
Discussion
The key result from this study is that a common laboratory strain of S. cerevisiae (SK1) forms organized structures (minicolonies) when grown on plastic surfaces. The organization of three cell types-- ovoid-shaped yeast cells, pseudohyphal cells and asci— within a minicolony reflects two sequential rounds of cell differentiation during colony development. First, “yeast-like” ovoid cells in the minicolony’s core differentiate into pseudohyphal chains that extend outward from this core. Second, these pseudohyphae differentiate further to yield asci. As a result, most sporulation in minicolonies occurs specifically at the surface of the minicolony.
Minicolony development provides a clear example of the relationship between pseudohyphal differentiation (the dimorphic switch) and sporulation. Because a number of cellular regulators stimulate pseudohyphal growth through one set of targets while inhibiting sporulation through a second set of targets (see Introduction), these two differentiation programs can be considered alternative, mutually-exclusive fates. Indeed, pseudohyphal differentiation is likely a foraging response triggered when nutrients are limiting but not absent, whereas sporulation is a stress response triggered (in part) by the absence of essential nutrients (Zaman, et al., 2008).
Examining minicolony development indicates that sporulation and the dimorphic switch are not only alternative fates, but more precisely, they are also sequential fates, at least with respect to minicolony development. Specifically, several lines of evidence suggest that formation of pseudohyphae in minicolonies leads to the sporulation of the pseudohyphal cells. First, asci in minicolonies were mostly observed in the pseudohyphal cells extending from the colony’s core. Second, nutrient conditions that increased pseudohyphal growth in minicolonies also increased sporulation. Third, most of the asci in minicolonies are dyads or linear asci, as expected if these asci derive from pseudohyphal cells. Finally, a flo11Δ mutant, which is defective in pseudohyphal differentiation, was also defective in sporulation. Since FLO11 is not required for sporulation in cultures, only in minicolonies, it is likely that the pseudohyphae extending outward from surface of minicolonies are more competent for sporulation than are the ovoid cells that compose the core of the minicolony. Consistent with sporulation following pseudohyphal differentiation during colony development, large colonies that form on top of an agar surface can form pseudohyphal chains that efficiently invade the agar surface, and these pseudohyphae also sporulate quite efficiently below the agar surface (Piccirillo & Honigberg).
The reason sporulation in minicolonies occurs most efficiently in the pseudohyphae may be that cells need to be dispersed from the minicolony’s core before they can sporulate. In agreement with this interpretation, although flo11Δ minicolonies sporulate to low levels, the relatively few cells that are dispersed from these minicolonies sporulate relatively efficiently. Also in agreement with this view, when wild-type minicolonies are mechanically dispersed, most of the dispersed cells go on to sporulate. The widely different sporulation efficiency in the core vs. the periphery of colonies may reflect differences in microenvironment of the two regions or differences in the number of cell-cell contacts.
Recently, a second link between pseudohyphae and sporulation was identified in SK1 microcolonies grown on agar medium containing nonfermentable carbon sources (Strudwick, et al.). These authors demonstrate that two key regulators of meiosis and sporulation, the Ime1p transcription factor and the Ime2p protein kinase, are required for pseudohyphal growth in SK1 colonies. This result suggests that metabolic or other changes induced as part of the sporulation program may, at least in some strains and conditions, also stimulate pseudohyphal differentiation.
The SK1 strain background may prove particularly useful for investigating the growth of yeast on plastic surfaces. Different laboratory strain backgrounds vary dramatically with respect to their ability to form pseudohyphae, invade agar, and adhere to plastic. For example, the commonly used laboratory S288C and W303 strains proliferated on a submerged plastic surface, but these strains failed to form minicolonies or adhere tightly to this surface. In contrast, both Σ1278b and SK1 formed minicolonies that adhered tightly to the plastic surface. SK1 minicolonies formed a more extensive pseudohyphal network than did Σ1278b and also had higher affinity to the underlying surface. It is not known whether SK1 minicolonies can serve as a tractable model for the biofilms formed by pathogenic yeast such as C. albicans, but it is promising that both S. cerevisiae minicolonies and C. albicans biofilms involves a dimorphic switch from ovoid to filamentous cells, and this dimorphic switch correlates with adherence to the underlying plastic surface for both species.
Supplementary Material
Acknowledgments
This work was funded by NIH R15GM094770 (to S.M.H.).
References
- 1.Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Ari G, Zenvirth D, Sherman A, et al. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2:e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colomina N, Gari E, Gallego C, Herrero E, Aldea M. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. Embo J. 1999;18:320–329. doi: 10.1093/emboj/18.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen PJ, Sprague GF., Jr The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell. 2002;13:2990–3004. doi: 10.1091/mbc.E02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 7.Dickinson JR. Filament formation in Saccharomyces cerevisiae--a review. Folia Microbiol (Praha) 2008;53:3–14. doi: 10.1007/s12223-008-0001-6. [DOI] [PubMed] [Google Scholar]
- 8.Donzeau M, Bandlow W. The yeast trimeric guanine nucleotide-binding protein alpha subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol Cell Biol. 1999;19:6110–6119. doi: 10.1128/mcb.19.9.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288C. Mol Microbiol. 2007;66:1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A. 2006;103:11228–11233. doi: 10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gancedo JM. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 12.Gerke J, Lorenz K, Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 14.Hawthorne DC. The Use of Linear Asci for Chromosome Mapping in Saccharomyces. Genetics. 1955;40:511–518. doi: 10.1093/genetics/40.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116:2137–2147. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 16.Hube B. Infection-associated genes of Candida albicans. Future Microbiol. 2006;1:209–218. doi: 10.2217/17460913.1.2.209. [DOI] [PubMed] [Google Scholar]
- 17.Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 18.Khot PD, Suci PA, Miller RL, Nelson RD, Tyler BJ. A small subpopulation of blastospores in candida albicans biofilms exhibit resistance to amphotericin B associated with differential regulation of ergosterol and beta-1,6-glucan pathway genes. Antimicrob Agents Chemother. 2006;50:3708–3716. doi: 10.1128/AAC.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi O, Yoshimoto H, Sone H. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr Genet. 1999;36:256–261. doi: 10.1007/s002940050498. [DOI] [PubMed] [Google Scholar]
- 20.Kron SJ. Filamentous growth in budding yeast. Trends Microbiol. 1997;5:450–454. doi: 10.1016/S0966-842X(97)01131-1. [DOI] [PubMed] [Google Scholar]
- 21.Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci U S A. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuura A, Treinin M, Mitsuzawa H, Kassir Y, Uno I, Simchen G. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. Embo J. 1990;9:3225–3232. doi: 10.1002/j.1460-2075.1990.tb07521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccirillo S, Honigberg SM. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res Microbiol. 2010;161:390–398. doi: 10.1016/j.resmic.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purnapatre K, Piccirillo S, Schneider BL, Honigberg SM. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells. 2002;7:675–691. doi: 10.1046/j.1365-2443.2002.00551.x. [DOI] [PubMed] [Google Scholar]
- 30.Puzniak L, Teutsch S, Powderly W, Polish L. Has the epidemiology of nosocomial candidemia changed? Infect Control Hosp Epidemiol. 2004;25:628–633. doi: 10.1086/502452. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 32.Schacherer J, Ruderfer DM, Gresham D, Dolinski K, Botstein D, Kruglyak L. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE. 2007;2:e322. doi: 10.1371/journal.pone.0000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strudwick N, Brown M, Parmar VM, Schroder M. Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on non-fermentable carbon sources. Mol Cell Biol. 2010;30:5514–5530. doi: 10.1128/MCB.00390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas JH, Botstein D. Ordered Linear Tetrads Are Produced by the Sporulation of Newly Formed Zygotes of Saccharomyces cerevisiae. Genetics. 1987;115:229–232. doi: 10.1093/genetics/115.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trachtulcova P, Janatova I, Kohlwein SD, Hasek J. Saccharomyces cerevisiae gene ISW2 encodes a microtubule-interacting protein required for premeiotic DNA replication. Yeast. 2000;16:35–47. doi: 10.1002/(SICI)1097-0061(20000115)16:1<35::AID-YEA504>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Tumbarello M, Posteraro B, Trecarichi EM, et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol. 2007;45:1843–1850. doi: 10.1128/JCM.00131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dyk D, Hansson G, Pretorius IS, Bauer FF. Cellular differentiation in response to nutrient availability: The repressor of meiosis, Rme1p, positively regulates invasive growth in Saccharomyces cerevisiae. Genetics. 2003;165:1045–1058. doi: 10.1093/genetics/165.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Mulders SE, Christianen E, Saerens SM, et al. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:178–190. doi: 10.1111/j.1567-1364.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 39.Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 40.Vopalenska I, St’ovicek V, Janderova B, Vachova L, Palkova Z. Role of distinct dimorphic transitions in territory colonizing and formation of yeast colony architecture. Environ Microbiol. 2010;12:264–277. doi: 10.1111/j.1462-2920.2009.02067.x. [DOI] [PubMed] [Google Scholar]
- 41.Vyas VK, Kuchin S, Berkey CD, Carlson M. Snf1 kinases with different beta-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol Cell Biol. 2003;23:1341–1348. doi: 10.1128/MCB.23.4.1341-1348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.