Abstract
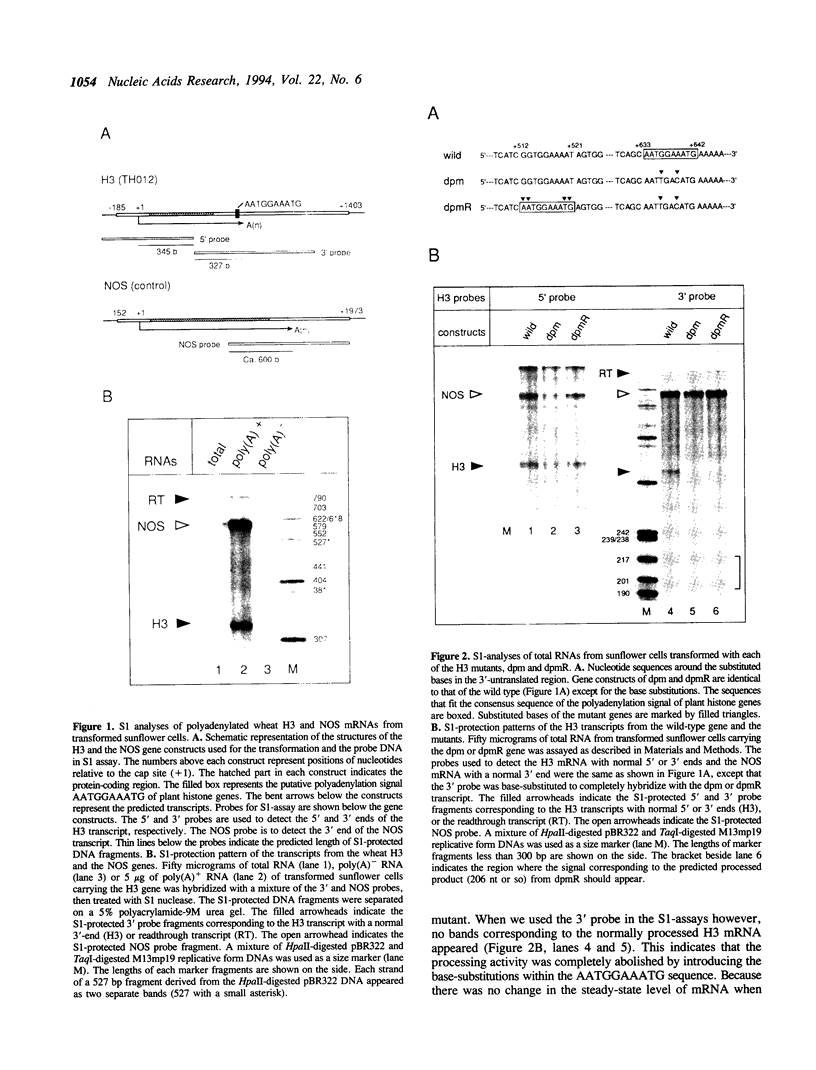
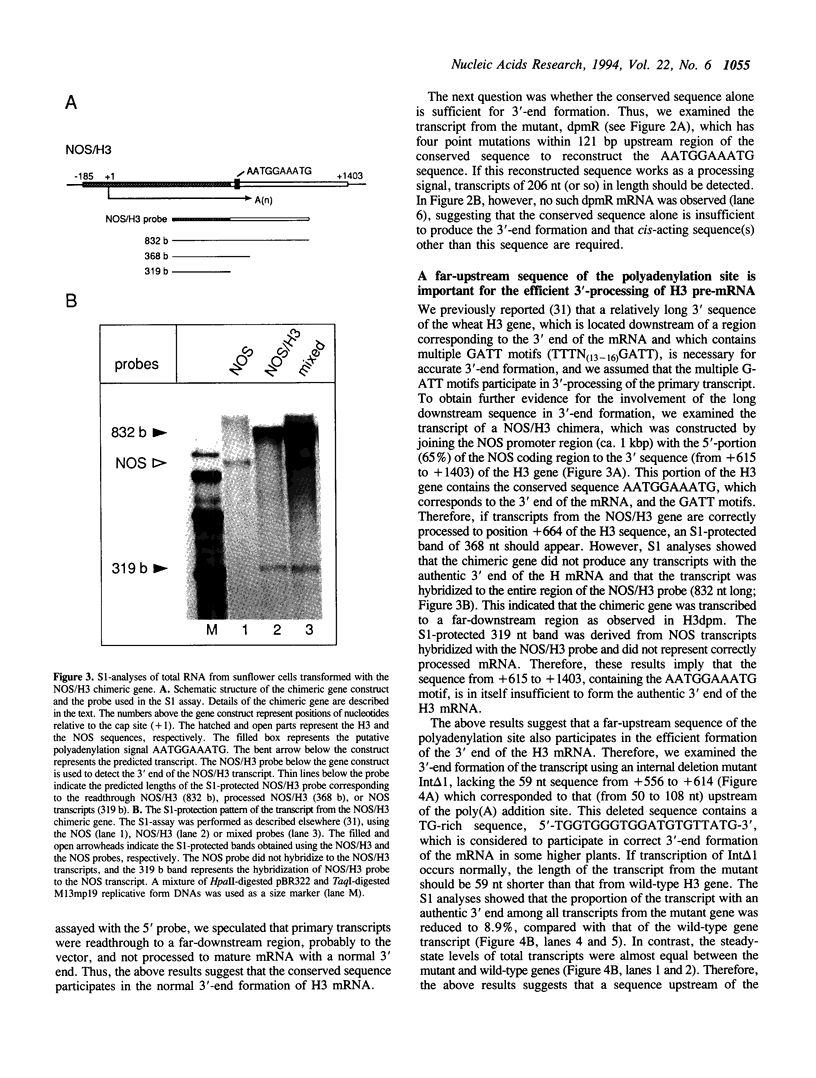
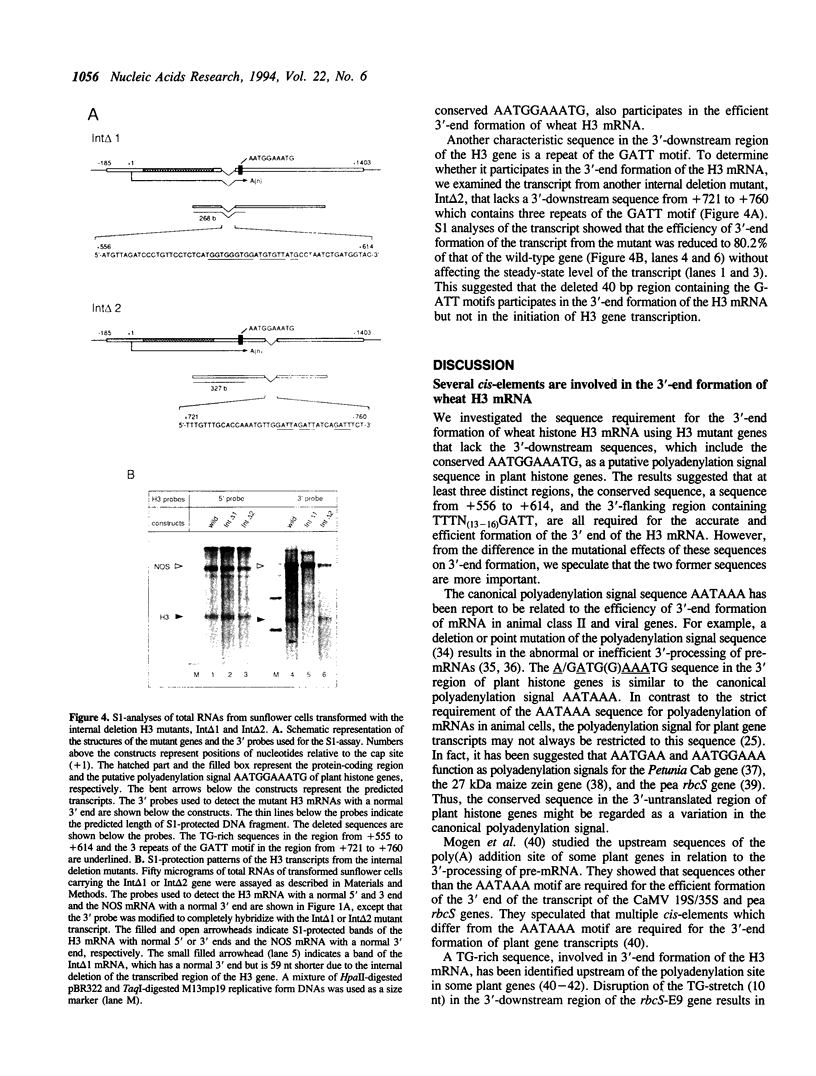
We examined the 3'-flanking regions required for accurate 3'-end formation of wheat histone H3 mRNA using gene expression in transformed sunflower cells. The introduction of mutations into the conserved sequence AATGGAAATG in the 3'-flanking region of plant histone genes, located 22 bp upstream from the polyadenylation site of the wheat H3 gene (TH012), completely abolished the 3'-end formation of mRNA at the authentic 3' end without affecting the transcription efficiency. However, a 0.8 kbp sequence containing this motif could not produce a normal 3' end when joined to the 3' end of the nopaline synthase (NOS) gene instead of its 3' sequence. The results indicated that this conserved sequence is necessary but not sufficient for the 3'-end formation of H3 or NOS mRNA. Deletion of a 59 bp sequence, located 19 bp upstream from the AATGGAAATG sequence, also reduced the 3'-end formation efficiency by a factor of 10, compared with the efficiency in wild-type gene. We concluded that 3'-end formation of wheat histone H3 mRNA is regulated by multiple sequences including the AATGGAAATG motif.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach L. L., Marashi F., Plumb M., Stein G., Stein J. Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry. 1984 Apr 10;23(8):1618–1625. doi: 10.1021/bi00303a006. [DOI] [PubMed] [Google Scholar]
- Bevan M., Barnes W. M., Chilton M. D. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 1983 Jan 25;11(2):369–385. doi: 10.1093/nar/11.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chaboute M. E., Chaubet N., Clement B., Gigot C., Philipps G. Polyadenylation of histone H3 and H4 mRNAs in dicotyledonous plants. Gene. 1988 Nov 15;71(1):217–223. doi: 10.1016/0378-1119(88)90095-9. [DOI] [PubMed] [Google Scholar]
- Chaubet N., Chaboute M. E., Clément B., Ehling M., Philipps G., Gigot C. The histone H3 and H4 mRNAs are polyadenylated in maize. Nucleic Acids Res. 1988 Feb 25;16(4):1295–1304. doi: 10.1093/nar/16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Broglie R., Edwards C., Chua N. H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J. 1984 Aug;3(8):1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Tamaki S., Dunsmuir P., Favreau M., Katayama C., Dooner H., Bedbrook J. mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 1986 Mar 11;14(5):2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N. J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991 Jul;10(7):1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gick O., Krämer A., Vasserot A., Birnstiel M. L. Heat-labile regulatory factor is required for 3' processing of histone precursor mRNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J. Control of mRNA Stability in Higher Plants. Plant Physiol. 1993 Aug;102(4):1065–1070. doi: 10.1104/pp.102.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991 Mar 26;1088(3):327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Ingelbrecht I. L., Herman L. M., Dekeyser R. A., Van Montagu M. C., Depicker A. G. Different 3' end regions strongly influence the level of gene expression in plant cells. Plant Cell. 1989 Jul;1(7):671–680. doi: 10.1105/tpc.1.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C. P. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987 Dec 10;15(23):9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T., Nakayama T., Ohtsubo N., Tabata T., Iwabuchi M. Cell cycle-regulated gene expression in transgenic plant cells. Dev Genet. 1990;11(3):205–213. doi: 10.1002/dvg.1020110306. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M. H., Mogen B. D., Hunt A. G. Characterization of the polyadenylation signal from the T-DNA-encoded octopine synthase gene. Nucleic Acids Res. 1991 Oct 25;19(20):5575–5581. doi: 10.1093/nar/19.20.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- Melin L., Soldati D., Mital R., Streit A., Schümperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992 Feb;11(2):691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B. D., MacDonald M. H., Graybosch R., Hunt A. G. Upstream sequences other than AAUAAA are required for efficient messenger RNA 3'-end formation in plants. Plant Cell. 1990 Dec;2(12):1261–1272. doi: 10.1105/tpc.2.12.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B. D., MacDonald M. H., Leggewie G., Hunt A. G. Several distinct types of sequence elements are required for efficient mRNA 3' end formation in a pea rbcS gene. Mol Cell Biol. 1992 Dec;12(12):5406–5414. doi: 10.1128/mcb.12.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T. D., Weber L. A., Hickey E., Stein G. S., Stein J. L. Changes in the stability of a human H3 histone mRNA during the HeLa cell cycle. Mol Cell Biol. 1991 Jan;11(1):544–553. doi: 10.1128/mcb.11.1.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3' end formation in vitro. Mol Cell Biol. 1987 May;7(5):1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Identification of the human U7 snRNP as one of several factors involved in the 3' end maturation of histone premessenger RNA's. Science. 1987 Dec 18;238(4834):1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. snRNP mediators of 3' end processing: functional fossils? Trends Biochem Sci. 1988 Nov;13(11):447–451. doi: 10.1016/0968-0004(88)90220-4. [DOI] [PubMed] [Google Scholar]
- Newman T. C., Ohme-Takagi M., Taylor C. B., Green P. J. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993 Jun;5(6):701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G., Chaubet N., Chaboute M. E., Ehling M., Gigot C. Genomic organization and nucleotide sequences of two corn histone H4 genes. Gene. 1986;42(2):225–229. doi: 10.1016/0378-1119(86)90301-x. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Sanfaçon H., Brodmann P., Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991 Jan;5(1):141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- Schümperli D. Cell-cycle regulation of histone gene expression. Cell. 1986 May 23;45(4):471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Pilch D. R., Marzluff W. F. The histone mRNA 3' end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992 Nov 25;20(22):6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Wu L., Ueda T., Messing J. 3'-end processing of the maize 27 kDa zein mRNA. Plant J. 1993 Sep;4(3):535–544. doi: 10.1046/j.1365-313x.1993.04030535.x. [DOI] [PubMed] [Google Scholar]
- Wu S. C., Györgyey J., Dudits D. Polyadenylated H3 histone transcripts and H3 histone variants in alfalfa. Nucleic Acids Res. 1989 Apr 25;17(8):3057–3063. doi: 10.1093/nar/17.8.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. C., Végh Z., Wang X. M., Tan C. C., Dudits D. The nucleotide sequences of two rice histone H3 genes. Nucleic Acids Res. 1989 Apr 25;17(8):3297–3297. doi: 10.1093/nar/17.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]