Abstract
KIR2DL4 (2DL4, CD158d) is a unique killer cell Ig-like receptor (KIR) expressed on human NK cells, which stimulates cytokine production, but mechanisms regulating its expression and function are poorly understood. By yeast two-hybrid screening, we identified the E3 ubiquitin ligase, Triad3A, as an interaction partner for the 2DL4 cytoplasmic domain. The protein interaction was confirmed in vivo, and Triad3A expression induced polyubiquitylation and degradation of 2DL4. Overexpression of Triad3A selectively abrogated cytokine-producing function of 2DL4, while Triad3A shRNA reversed ubiquitylation and restored cytokine production. Expression of Triad3A in an NK cell line did not affect receptor surface expression, internalization, or early signaling, but significantly reduced receptor turnover and suppressed sustained NF-κB activation. 2DL4 endocytosis was found to be vital to stimulate cytokine production, and Triad3A expression diminished localization of internalized receptor in early endosomes. Our results reveal a critical role for endocytosed 2DL4 receptor to generate sustained NF-κB signaling and drive cytokine production. We conclude that Triad3A is a key negative regulator of sustained 2DL4-mediated NF-κB signaling from internalized 2DL4, which functions by promoting ubiquitylation and degradation of endocytosed receptor from early endosomes. “This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.”
Keywords: natural killer cells, human, signal transduction, cell surface molecules, cell activation
Introduction
KIR2DL4 (2DL4, CD158d) is a structurally and functionally unique activating member of the human killer cell Ig-like receptor (KIR) family and the most highly conserved KIR among primates (1). The receptor is normally only expressed on the surface of a minor subset of CD56high NK cells that elicit mostly cytokine production (2, 3). In contrast to other KIR, surface expression of 2DL4 is up-regulated on NK cells by IL-2 stimulation (2, 3). Structurally, 2DL4 contains features of both activating and inhibitory KIR family members. The receptor has a basic arginine residue in the transmembrane domain, which allows association with the FcεRI-γ transmembrane adaptor protein (4). The cytoplasmic domain of 2DL4 also contains a single ITIM, which has the capacity to recruit SHP-2 protein tyrosine phosphatase, at least in the context of a chimeric receptor construct (5). The role of SHP-2 recruitment on function of the full receptor is currently unclear, however.
The only known ligand of 2DL4 is a soluble form of the non-classical MHC-class I molecule, HLA-G, which is normally produced by fetal-derived trophoblast cells in pregnant women (6–8). Since 2DL4 is expressed on uterine NK cells during pregnancy (9), it has been proposed that the interaction of 2DL4 with HLA-G may benefit pregnancy (8). Uterine NK cells have limited cytolytic capacity, but produce cytokines that may promote remodeling of the maternal vasculature to support fetal development in mice and humans (10, 11). Interestingly, 2DL4 ligation stimulates production of several pro-angiogenic cytokines, suggesting that it may contribute to this process in the decidual tissue during pregnancy (8, 12).
2DL4 transduces activating signals through at least two different modules. Association with the transmembrane adaptor, FcεRI-γ (4), provides a cytoplasmic ITAM that recruits Syk/ZAP-70 tyrosine kinases. FcεRI-γ supports 2DL4-mediated Ca2+ mobilization, weak target cell killing, and production of numerous cytokines (4, 12). Surprisingly, 2DL4-mediated stimulation of MAP kinases, NF-κB, and MIP1α production are largely independent of FcεRI-γ association (12). Furthermore, 2DL4 can stimulate production of IL-8 in FcεRI-γ-deficient 293T cells, which requires the receptor cytoplasmic domain (8), suggesting that the cytoplasmic domain contains a second signaling module to directly recruit signaling interacting partners. Recent evidence indicates that the cytoplasmic domain of 2DL4 interacts with the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) to stimulate the phosphorylation and activation of Akt (13), which is likely to contribute to FcεRI-γ-independent activating function.
2DL4 has further been shown to internalize and accumulate in Rab5-containing early endosomes, where it has been suggested to transduce intracellular signals (8) through the association with DNA-PKcs at this intracellular site (13). It has been recently recognized that endocytosis of several receptor types can promote sustained intracellular signaling processes, including the activation of MAP kinases (14, 15). On the other hand, roles for receptor-mediated endocytosis in downregulating receptor function have been widely appreciated, whereby internalized receptors can be sorted to degradation pathways mediated by lysosomes or the proteasome.
Conjugation with ubiquitin (Ub) is a common mechanism for tagging proteins for trafficking, degradation, and signaling (16, 17). Ubiquitylation of transmembrane proteins can promote either the initial step of internalization and/or subsequent sorting from the early endosome to degradation in lysosomes or the proteasome. Ubiquitin peptides are covalently attached to lysine residues on target proteins through substrate-specific recognition by distinct ubiquitin-protein ligases, known as E3 subunits.
Using a yeast two-hybrid approach, we have identified the RING finger-containing E3 ubiquitin ligase, Triad3A, as a 2DL4 cytoplasmic binding partner. We show that Triad3A interacts with 2DL4 and induces ubiquitylation and degradation of the receptor, although surface receptor expression is unchanged. Our data also show that Triad3A suppresses 2DL4-mediated cytokine production, accumulation in early endosomes, and sustained NF-κB activation, but does not affect early signaling events, suggesting that Triad3A is a critical regulator of late-term 2DL4 signaling at an intracellular site.
Materials and Methods
Cells and culture
The KHYG-1 cell line [obtained from Health Science Research Resources Bank, Japan Health Sciences Foundation; JCRB0156 (18)] was cultured in IL-2-containing α-MEM medium as previously described (5, 12). HEK-293T cells (kindly provided by Dr. Shao Cong-Sun, M.D. Anderson Cancer Center) were cultured in DMEM + 10% fetal bovine serum, 100U/ml penicillin and 100mg/ml streptomycin (Invitrogen). The retrovirus producing Phoenix-Ampho cells were cultured as described (19). Sorted primary CD56+CD3−DX9+ NK cells were cultured in RPMI 1640 with 5% (v/v) autologous serum and IL-2 (2% (v/v) supernatant from cultures of J558L myeloma cells transfected with human IL-2 gene) (20). Volunteer blood donors were recruited by informed consent as approved by the FCCC Institutional Review Board.
Antibodies and reagents
Antibodies included: anti-FLAG mAb (M2) from Sigma-Aldrich (St. Louis, MO); anti-2DL4 mAb (53.1; mouse IgG1) and anti-NKp44 mAb (3.43.13) from Dr. M. Colonna (Washington University, St. Louis, MO); anti-phospho-p42/44 ERK (pThr202/pTyr204), anti-phospho-JNK (pThr183/pTyr185), anti-phospho-c-Jun (pSer63), anti-phospho-p38 MAP kinase (pThr180/pTyr182), and anti-phospho-IKKα (pSer180)/IKKβ (pSer181) antibodies from Cell Signaling Technology; rabbit polyclonal anti-Triad3A antibody was from Abcam or kindly provided by Dr. T.-H. Chuang (Scripps, La Jolla, CA); HRP-conjugated goat anti-mouse IgG and goat anti-rabbit IgG from Jackson Immunoresearch; mouse monoclonal antibody to GAPDH from Chemicon (Temecula, CA); anti-p65 and anti-myc (9E10) antibodies from Santa Cruz Biotech. Lipofectamine™ reagent, Plus™ reagent and Lipofectamine 2000 were from Invitrogen; PMA and ionomycin were from FisherBiotech.
cDNA and shRNA constructs and cell transfection/transduction
Retroviral expression constructs for FLAG- or myc-tagged WT-2DL4 in the bicistronic pBMN-IRES-EGFP vector (from Dr. G. Nolan, Stanford University, Stanford, CA) were previously described (5). The cDNA of human FLAG-Triad3A in pRK5 vector from Dr. T.-H. Chuang (Scripps) was cloned into pBMN-IRES-EGFP. Retroviral transduction of KHYG-1 cells was previously described (19). Transduced cells were sorted in a FACS VantageSE flow cytometer (BD) for expression of EGFP or surface FLAG- or myc-tag. HA-Ub in pcDNA vector was from Dr. K. Tanaka (Tokyo Metropolitan Institute of Medical Science, Japan). The GST-c-Jun (1–79 aa) construct was from Dr. L. Heasley (University of Colorado, Denver, CO). NF-κB luc-pRL-TK Renilla Luciferase Reporter Vector was from Dr. K. Sada (Kobe University, Japan). 293T cells were transiently transfected using Lipofectamine 2000, and cells were used 48 h later. Triad3A (Genbank #AF513717.1) shRNA sequences were designed to target all Triad3 isoforms using software from Oligoengine (www.oligoengine.com, Seattle WA): shRNA #1 (targeting 1243–1263, aagtgctcagtagtcaggaca; N-terminal of RING domain #1), #2 (targeting 1527–1546, gagcaggagttctatgagca; N-terminal of RING domain #1), #3 (targeting 2290–2309, tctggaccgatcccactgaa; C-terminal to RING domain #2). Sense and antisense oligos (3μg of each) incorporating an intervening hairpin and terminal Bgl II and Hind III restriction site overhangs were annealed together and ligated into pSuperior.retro.neo or .puro vectors (Oligoengine).
Yeast two-hybrid screen
A cDNA encoding the cytoplasmic domain of human 2DL4 (amino acids 858–1032) was subcloned into the pEG202 vector (a gift of Dr. Roger Brent) to generate bait for yeast two-hybrid screening. The yeast strain EGY48 was transformed sequentially with the 2DL4(Cyto)/pEG202 vector and a human Jurkat cDNA library in pJG4-5 vector (kindly provide by Dr. Brent Passerin, formerly at NIAID, Rockville, MD) using a yeast transformation kit (Zymogen). Positive clones were selected by growth in leucine-, histidine-, tryptophan- and uracil-deficient medium and the specificity of their interaction was verified by testing for β-galactosidase production as previously described (21, 22). The cDNA from positive clones was purified, sequenced, and identified by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Immunoprecipitation and immunoblotting
Cell lysate preparation in 1% Triton X-100 buffer, immunoprecipitation, SDS-PAGE, and immunoblotting were performed as previously described (12). For 2D gel electrophoresis, proteins were solubilized with sample buffer (7 M urea, 2 M thiourea, 4% CHAPS) and 100 μg was applied to each pH (3–10) NL isoelectric focusing (IEF) strip, focused on a PROTEAN IEF CELL (Bio-Rad, Hercules, CA) (23), separated on 10% SDS-PAGE, transferred to PVDF membrane, and immunoblotted.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated with the RNeasy Kit (Qiagen; Valencia, CA), and qRT-PCR was performed using TaqMan PCR reagents and Triad3- or GAPDH-specific primers and probes on a 7500 Real-Time PCR system [all from Applied Biosystems (Foster City, CA)]. The mRNA copy numbers of a particular gene in each sample were normalized to the mRNA copy number of the housekeeping gene GAPDH in that sample.
Assay of JNK activity
JNK was assayed using GST-c-Jun as a substrate to measure the phosphorylation by JNK in an in vitro kinase assay (24). Briefly, GST-c-Jun (aa 1–79) was isolated on glutathione-agarose (Pierce) and was used to precipitate JNK from lysates of stimulated NK cells. After immunoprecipitation JNK-GST-c-Jun complexes were suspended in 40 μl JNK assay buffer (50 mM β-glycerophosphate, pH 7.2, 0.1 mM sodium orthovanadate, 10 mM MgCl2, 1 mM EGTA, and 20 μM ATP), incubated for 20 min at 30°C while mixing the assay components by frequent vortexing, denatured with 3X Laemmli buffer, separated by SDS-PAGE, transferred to PVDF membrane, and immunoblotted.
Cellular assays
For cytokine/chemokine assays, KHYG-1 cells transduced with FLAG-2DL4 ± Triad3A (2×105 cells/well) were cultured for 24 h in a 96-well plate containing plate-bound mAb, and culture supernatants were assayed using ELISA kits for IL-8, MIP1α (Pierce Biotechnology), or IFN-γ (BD Pharmingen) as previously described (12).
KIR2DL4 internalization assay
The internalization assay was adopted from previously established assays (20, 25, 26).Cells were stained with 10 μg/ml biotinylated anti-FLAG mAb for 1 h on ice, washed twice, kept at 37°C for the indicated times, stained with PE-conjugated streptavidin for 1 h on ice, washed and analyzed by FACS. The percent (%) internalization was calculated from mean fluorescence intensity (MFI) of staining at the indicated time-point (t) compared to time zero (0) using the calculation: % internalization = 100−[(MFI ‘t’ min/MFI ‘0’ min)/100].
KIR2DL4 turnover assay
2DL4 surface turnover was assayed as previously described (20, 27)). Briefly, cells were blocked with 50 μg/ml unlabeled anti-FLAG polyclonal Ab (Sigma) in HBSS plus1% FBS for 40 min at 4°C, washed, and incubated in α-MEM at37°C. Cell aliquots were stained on ice with biotinylated anti-FLAG mAb and PE-streptavidin, analyzed on a FACScan (BD Biosciences), and evaluated withFlowJo software. The % turnover of 2DL4 staining = 100 × (MFIexp − MFIblocked)/(MFItotal − MFIblocked), where MFIexp is 2DL4 staining at the indicated time, MFItotal is staining of untreated cells and MFIblocked is staining cells blocked with unlabeled Ab and kept at 4°C. In some experiments, cells were treated with 50 μg/ml cycloheximide during blocking and throughout the assay (20).
EMSA analysis
Nuclear extracts were prepared by using a nuclear extraction kit (Panomics). For EMSA, the NF-κB consensus oligonucleotide 5′-GGGGACTTTCCC-3′ (Santa Cruz Biotechnology) was end labeled with [γ32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (Promega) and purified using Microspin G-50 columns (GE Healthcare). Nuclear extracts were mixed with the labeled probe in 20μl of buffer [3.75 mM HEPES pH 7.6, 1.5 % vol/vol glycerol, 0.5 mM DTT, 0.015 mM EDTA, 0.5μg poly (dI-dC) (Roche)] and incubated at RT for 30 min. For supershifts, antibodies to the p65 subunit of NF-κB (Santa Cruz Biotechnology) were added to extracts for 30 min prior to addition of radiolabeled oligonucleotide. DNA-protein complexes were separated on 5% non-denaturing PAGE and autoradiographed.
Immunostaining and confocal microscopy
Cells were surface stained with primary anti-myc antibody (9E10) and secondary AlexaFluor 647-conjugated anti-mouse Ig antibody (Invitrogen) on ice, incubated at 37°C for 1–2 h to allow receptor internalization, settled onto poly-L-lysine coated culture slides (Lab Scientific, BD Pharmingen) for 10–30 min at RT, fixed and permeabilized in 3–4% paraformaldehyde/PBS + 0.1% Triton X-100 for 15 minutes, stained with anti-Rab5 or anti-EEA1 antibody for 15 minutes at room temperature or overnight at 4°C, respectively, and AlexaFluor 350-conjugated secondary antibody for 30 min at room temperature in PBSS (0.1% saponin, 1% BSA and 0.04% azide in PBS), washed, and mounted using Fluoromount-G (Southern Biotechnology, Birmingham, AL). Cells were imaged on a Nikon TE300 inverted microscope fitted for phase contrast and epifluorescence, including a ProScan II filter/shutter/objective z-step controller (Prior Scientific) and CoolSnap HQ CCD camera (Photometrics), as previously described (28). For 3D analysis of 2DL4 and Rab5 early endosomal localization, a minimum of 13 images in 1 μm increments through z-space were collected for each cell. The resulting image stack for each fluorescence channel was max projected into a single plane image, and then each fluorescence channel was overlayed into a single image. Co-localization was measured using Meta Vue software Version 7.0 (Molecular Devices, Sunnyvale, CA). For analysis of EEA1 with 2DL4, 15 randomly selected cells from each condition (± Triad3A expression) were imaged at 0.5 μm z-step intervals. Background/non-specific staining was defined by staining myc-2DL4 negative cells. Image thresholds and area of intersection between 2DL4 and EEA1 were determined within each z-section using NIS-Elements software (version AR 3.10, Nikon). The percentages of total 2DL4 staining intersecting with EEA1 staining were then calculated for each z-step interval from each cell. The 2-tailed Mann-Whitney test was used to determine the significance of percentages of 2DL4/Rab5 or 2DL4/EEA1 co-localization between the two conditions (± Triad3A expression) using the InStat program (version 3.06, Graphpad Software Inc.).
NF-κB luciferase reporter assay
The NF-κB reporter gene construct NF-κB luc-pRL-TK (5 ng) were transiently transfected into 293T cells in combination with 2DL4 and/or Triad3A with Lipofectamine 2000. After 48 h, cells were lysed and luciferase activities were measured by the Steady-Glo Luciferase Assay (Promega, Madison, WI) using a GloMax® 20/20 Luminometer (Promega). Reporter gene activity was expressed as fold increase compared with that in cells transfected with the NF-κB reporter only.
Results
Identification of Triad3A as a 2DL4-interacting protein by yeast two-hybrid screening
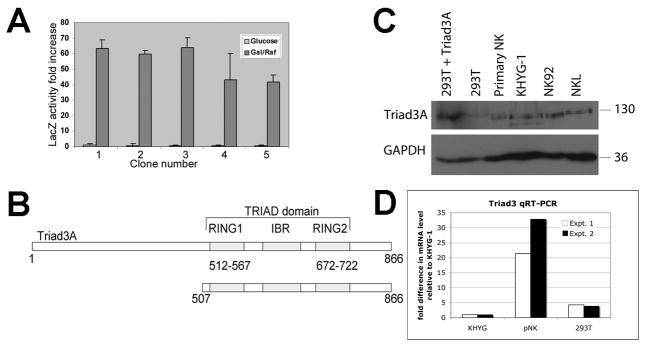
Previous work from our lab and others (8, 12) suggested that the cytoplasmic domain of 2DL4 might interact directly with other proteins to mediate intracellular signaling. Therefore, we utilized the yeast two-hybrid system to identify proteins that interact with the 2DL4 cytoplasmic domain. Five positive clones isolated from the library screen growth assay were also found to interact with the cytoplasmic domain in a secondary assay for β-galactosidase production, as shown in Fig. 1A. All five clones contained cDNA encoding the C-terminal sequence of human Triad3 (also known as ZIN or RNF216), which exhibits E3 ubiquitin ligase activity (29, 30). The 2DL4-interacting sequence (amino acids 507–866) contained the two TRIAD (Two RING finger and DRIL) domains and the IBR (in between RING) region (Fig. 1B). Triad3 is one of six mRNA splice variants (designated Triad3 and Triad3A-E), all of which include the common C-terminal domain isolated in our screen. Subsequently, we focused our studies on Triad3A, which is one of the largest and most abundantly expressed forms (29). Immunoblot and qRT-PCR analysis (Figs. 1C, D) confirmed that Triad3A is expressed in human NK cell lines (KHYG-1, NKL, and NK-92), primary human NK cells, and 293T cells, and the size of the major endogenous protein in these cells (~130 kDa) is similar to that of transfected Triad3A.
FIGURE 1. Identification of Triad3A as a 2DL4 binding partner by yeast two hybrid screening.
The 2DL4 cytoplasmic domain (aa 858–1032) was used to identify interacting proteins in a yeast two-hybrid screen. A, Five positive clones identified in a growth assay, which all encoded the C-terminus of Triad3A, also tested positive for β-galactosidase reporter activity. Results are mean fold increase LacZ activity ±S.D. comparing inducing conditions (galactose/raffinose; Gal/Raf) to uninduced conditions (glucose; activity = 1) from 3 assays. B, The 2DL4-interacting portion of Triad3A protein identified in all 5 clones (aa 507–866; bottom) includes the Triad domain [two RING domains and IBR domain] from the full protein (top). C, Immunoblotting for expression of Triad3 and GAPDH in total lysates of primary NK cells, KHYG-1 NK cell line, NK-92 NK cell line, NKL NK cell line, 293T cells, and 293T cells transfected with Triad3A cDNA. Results are representative of at least 3 experiments. D, qRT-PCR analysis of Triad3 mRNA expression in KHYG-1, primary NK, and 293T cells. Results from two experiments were normalized to GAPDH mRNA levels in each cell type and expressed as fold difference as compared to the amounts detected in KHYG-1 cells, which were set at a relative value of 1.
Previous work has shown that Triad3A interacts with several Toll like receptors (TLRs), promotes their degradation by ubiquitylation, and inhibits their function (29). Triad3A also acts as a negative regulator in TNF-α receptor signaling by interacting with and inducing ubiquitylation and degradation of RIP-1 kinase, as well as degradation of the TIRAP and TRIF adaptors, all of which are important for initiating NF-κB signaling (30). Recently, it has been shown that Triad3A also targets TRAF3 for ubiquitylation and degradation and negatively regulates the RIG-I/MAVS pathway (31). These results suggested that Triad3A might similarly suppress 2DL4 functions.
Triad3A interacts with 2DL4 in 293T cells to promote polyubiquitylation and degradation
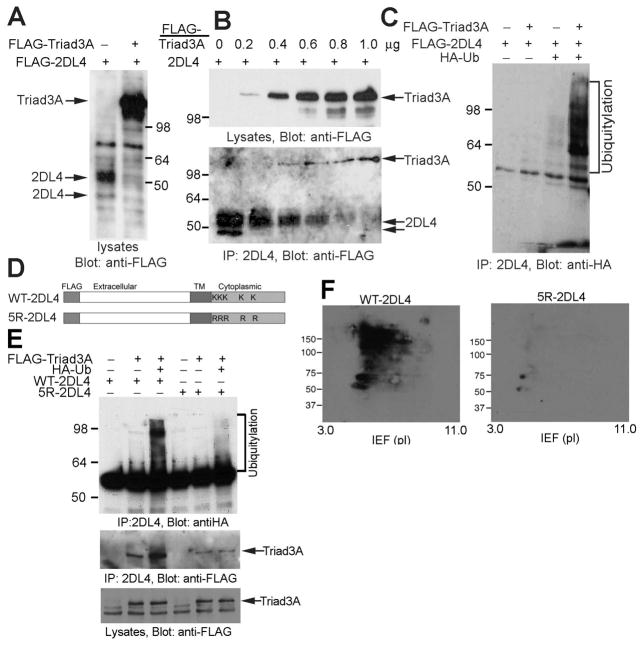
We tested whether Triad3A interacts with 2DL4 and promotes its degradation by co-transfecting cDNAs encoding both proteins into 293T cells. Co-transfection of Triad3A dramatically diminished the total level of 2DL4 protein detected in 293T cell lysates (Fig. 2A), which is consistent with the previously reported impacts on TLRs (29). To test whether Triad3A associates with 2DL4, we co-transfected 293T cells with increasing amounts of Triad3A cDNA and a constant amount of 2DL4 cDNA. Immunoprecipitation of the receptor showed progressively decreased 2DL4 protein recovery that correlated with increasing amounts of Triad3A protein expression and coordinate increases in the amount of Triad3A co-immunoprecipitating with the receptor (Fig. 2B).
FIGURE 2. Triad3A promotes polyubiquitylation and degradation of 2DL4 in 293T cells.
A, Co-expression of Triad3A reduces 2DL4 expression in 293T cells. 293T cells were transfected with FLAG-2DL4 ± FLAG-Triad3A, lysed 48 h later, separated by SDS-PAGE, and immunoblotted with anti-FLAG mAb. B, Triad3A associates with and induces degradation of 2DL4 protein. 293T cells were co-transfected with indicated amount of FLAG-Triad3A- and 1 μg of FLAG-2DL4-encoding plasmids, lysed 48 h later (lysates in top panel), and 2DL4 immunoprecipitates (IP in bottom panel) were immunoblotted with anti-FLAG mAb. C, Triad3A induces ubiquitylation of 2DL4. 293T cells were transfected with indicated cDNAs, lysed 48 h later, immunoprecipitated with anti-2DL4 mAb, and analyzed for Ub by anti-HA immunoblotting. D. Structure of WT-2DL4 and the 5R-2DL4 constructs (TM = transmembrane, K = Lys, R = Arg). E. Direct polyubiquitylation of lysines in the cytoplasmic domain of 2DL4 by Triad3A. 293T cells were transfected with indicated combinations of cDNAs, lysed 48 h later, immunoprecipitated with anti-2DL4 mAb, and immunoblotted for Ub with anti-HA Ab. Immunoprecipitates and lysates were also probed with anti-FLAG mAb to detect Triad3A (bottom). F. Analysis of lysine-dependent ubiquitylation of 2DL4 by 2D gel electrophoresis. 293T cells were transfected with FLAG-Triad3A, HA-Ub, and FLAG-WT-2DL4 (left) or FLAG-5R-2DL4 (right), lysed 48 h later, immunoprecipitated with anti-2DL4 mAb, separated by 2D gel electrophoresis (IEF/SDS-PAGE), and immunoblotted for Ub with anti-HA Ab.
We next tested whether Triad3A also induces the ubiquitylation of 2DL4 by transfecting FLAG-tagged Triad3A, FLAG-2DL4 and hemagglutinin-tagged ubiquitin (HA-Ub) cDNAs into 293T cells. As shown in Fig. 2C, Triad3A induced strong polyubiquitylation of 2DL4 when all three cDNAs were co-expressed. Mutation of the five cytoplasmic lysines in 2DL4 (Fig. 2D) to arginine (5R-2DL4 mutant) abrogated the polyubiquitylation of 2DL4 (Fig. 2E), indicating direct polyubiquitylation of at least one of these lysines by Triad3A. Two-dimensional gel electrophoresis demonstrated the extensive degree of direct 2DL4 polyubiquitylation that was disrupted by lysine mutation (Fig. 2F).
Triad3A suppresses KIR2DL4 function in 293T cells
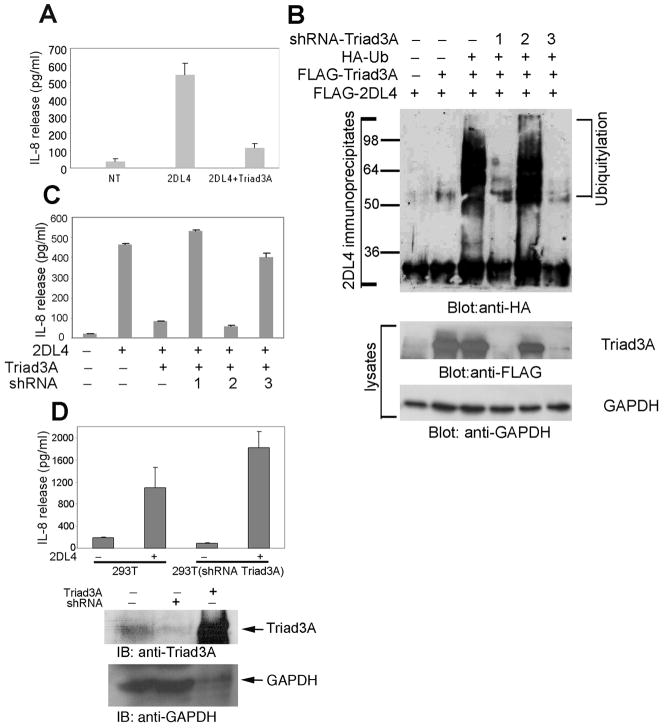
A previous report showed that 2DL4 transfection into 293T cells resulted in spontaneous IL-8 production (8), suggesting that these cells may contain a ligand that engages the receptor to result in productive signal transduction. Thus, we speculated that 2DL4-mediated IL-8 production might be impacted by co-transfection with Triad3A. As shown in Fig. 3A, 2DL4 transfection indeed promoted spontaneous IL-8 production in 293T cells, and Triad3A co-transfection essentially eliminated this IL-8 production, suggesting that Triad3A-induced ubiquitylation and degradation of 2DL4 profoundly inhibits receptor function.
FIGURE 3. Triad3A suppresses 2DL4-mediated production of IL-8 in 293T cells.
A, Triad3A co-expression suppresses 2DL4-mediated IL-8 production. 293T cells were transfected with FLAG-2DL4 ± FLAG-Triad3A cDNAs and tested 48 h later for IL-8 secretion by ELISA. Results are mean ±S.D. of single assays from 8 experiments. B, Triad3A knockdown reduces 2DL4 ubiquitylation in 293T cells. 293T cells were transfected with cDNAs encoding combinations of FLAG-2DL4, FLAG-Triad3A, HA-Ub, and one of three shRNAs targeting Triad3A. After 48 h, cells were lysed and 2DL4 immunoprecipitates (top panel) or cell lysates (bottom panels) were immunoblotted as indicated. C, 2DL4-mediated IL-8 production was restored by Triad3A knockdown. 293T cells were transfected with indicated combinations of FLAG-2DL4, FLAG-Triad3A, and one of the shRNA constructs, and tested 48 h later for IL-8 production by ELISA. Results are mean ±S.D. of single determinations from 3 experiments. D, Knockdown of endogenous Triad3A significantly enhanced 2DL4-mediated IL-8 production in 293T cells. 293T cells were transfected with shRNA3 and stably transfected cells were selected by treating with puromycin (2 μg/ml) for 7 days. The shRNA transfectants and 293T parent cells were then transfected with 2DL4 and assayed 48 hrs later for IL-8 secretion by ELISA (top panel). Results are mean ±S.D. of a single assay from 3 experiments. Lysates from 293T transfectants were immunoblotted with anti-Triad3A and anti-GAPDH to assess knockdown efficiency (bottom). Note that 10-fold less lysate was added for the Triad3A transfected sample (lane 3).
To demonstrate the direct involvement of Triad3A in regulating 2DL4 function we used shRNA to knockdown expression in 293T cells. We prepared three shRNA expression constructs targeting different regions of Triad3A. Both shRNA1 and shRNA3 efficiently knocked down the cDNA-driven expression of Triad3A protein by more than 90% (Fig. 3B), while shRNA2 was ineffective. We then tested whether knockdown of Triad3A will reverse Triad3A-mediated 2DL4 ubiquitylation and restore receptor function. As expected, co-transfection of shRNA1 or shRNA3 nearly abolished 2DL4 ubiquitylation (Fig. 3B), whereas shRNA2 had no effect. Furthermore, shRNA1 and shRNA3 efficiently reversed the inhibition of 2DL4-mediated IL-8 production by Triad3A in 293T cells (Fig. 3C), demonstrating direct involvement of Triad3A in regulating 2DL4 function. To further confirm that endogenous levels of Triad3A impact 2DL4 function, we found that knocking down expression levels of endogenous Triad3A in 293T cells with shRNA3 significantly enhanced 2DL4-mediated IL-8 production (Fig. 3D), demonstrating the importance of physiological levels of Triad3A in controlling 2DL4 function.
2DL4-mediated cytokine production is suppressed by Triad3A in an NK cell line
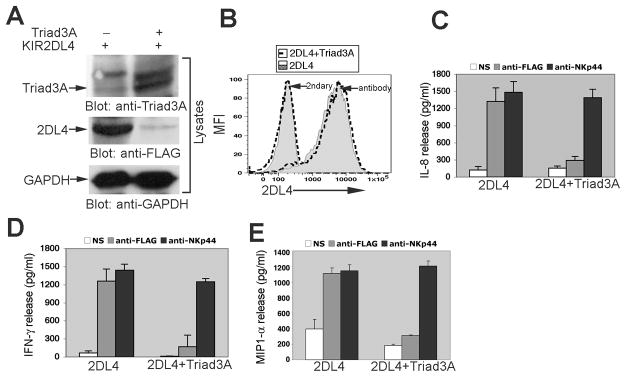
We and others have previously reported that engagement of 2DL4 with plate bound antibody stimulates the production of cytokines and chemokines in NK cell culture supernatants (2, 9, 12). To test the effect of Triad3A expression on the 2DL4-mediated induction of cytokine production, we engineered the KHYG-1 NK cell line to stably express FLAG-2DL4 alone or in combination with Triad3A by retroviral transduction. As in 293T cells, total 2DL4 protein expression in NK cell lysates was significantly suppressed by Triad3A co-expression, indicating that 2DL4 is degraded by ubiquitylation (Fig. 4A). We were surprised to find, however, that 2DL4 surface expression on the NK cell line was unaffected by Triad3A co-expression (Fig. 4B), indicating that Triad3A decreases the intracellular pool of 2DL4, while not impacting the level of surface receptor. Next, we tested the effect of Triad3A co-expression on the 2DL4-mediated activation of cytokine production in NK cells. KHYG-1 cells expressing FLAG-2DL4 alone or in combination with Triad3A were either left unstimulated or stimulated with plate-bound anti-FLAG mAb or anti-NKp44 mAb to ligate another NK cell activating receptor. Consistent with results in 293T cells, Triad3A co-expression significantly suppressed 2DL4-mediated production of IL-8, MIP-1α, and IFN-γ by the NK cell line, while NKp44-mediated cytokine production was unaffected (Figs. 4C-E). Our data demonstrate that expression of Triad3A selectively and efficiently suppresses 2DL4 function in NK cells, and suppression correlates with substantially decreased total levels of intracellular 2DL4, while surface receptor expression levels are unaffected.
FIGURE 4. Triad3A significantly reduces total 2DL4 levels and suppresses 2DL4-mediated cytokine production without impacting surface receptor expression in the KHYG-1 NK cell line.
A, Triad3A expression reduced the total level of 2DL4 protein in KHYG-1 cell lysates. KHYG-1 cells were transduced with FLAG-2DL4 ± FLAG-Triad3A and cell lysates were analyzed by immunoblotting with anti-Triad3A (top panel; top band is non-specific), anti-FLAG (middle) and anti-GAPDH (bottom). B, Surface expression of 2DL4 was not affected after co-transduction of Triad3A in KHYG-1 cells. KHYG-1 cells (assayed 2 days after IL-2 stimulation) that had been stably transduced with FLAG-2DL4 alone (shaded histograms) or in combination with FLAG-Triad3A (dashed line histograms) were stained for surface expression with anti-FLAG mAb plus PE-conjugated anti-κ secondary (antibody) or secondary alone (2ndary) by FACS. (C-E). KHYG-1 cells stably transduced with FLAG-2DL4 ± FLAG-Triad3A were harvested on day 2 after IL-2 stimulation, cultured for 24 hr (without IL-2) either with or without (NS) plate-bound anti-FLAG mAb (M2) or anti-NKp44 mAb (3.43.13), and analyzed by ELISA for secretion of IL-8 (C), IFN-γ (D), and MIP1-α (E). Results are representative mean ±S.D. from at least 3 experiments.
Triad3A suppresses 2DL4-mediated late-term NF-κB activation without affecting early signaling
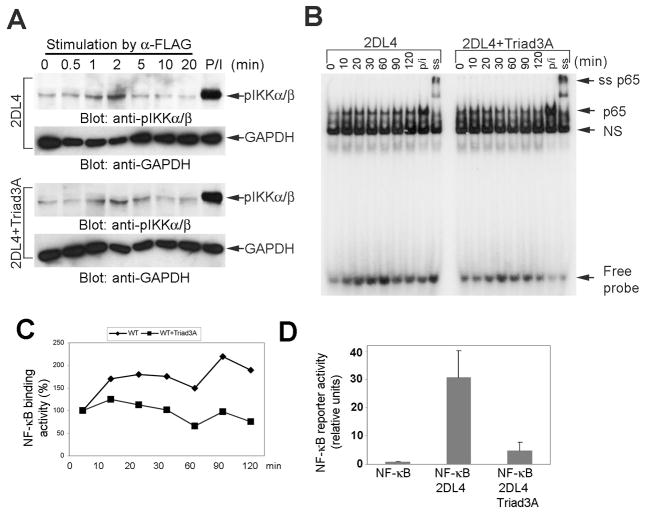
In addition to stimulating cytokine production, 2DL4 crosslinking activates MAP kinases (phosphorylation of JNK, ERK and p38) and NF-κB (phosphorylation of IKKβ and IκBα, and degradation of IκBα) (12). We also previously showed that all of these pathways are critical for downstream cytokine production in NK cells, and stimulation of p38 and ERK are upstream of NF-κB activation (12). Therefore, we tested whether exogenous Triad3A expression in KHYG-1 cells regulates 2DL4-mediated activation of MAP kinases and NF-κB. To our surprise, Triad3A expression did not suppress 2DL4-mediated phosphorylation of ERK or p38 (Fig. 5A), and the activation of JNK was unaffected (Fig. 5B). In addition, Triad3A expression did not affect early phosphorylation of IKKα/β, which is an upstream kinase of the NF-κB activation cascade (Fig. 6A). The canonical NF-κB pathway culminates in translocation of the p65 subunit to the nucleus, where it binds to specific DNA sequences to regulate gene transcription. Therefore, we further tested the effect of Triad3A co-expression on 2DL4-mediated activation of NF-κB by EMSA. Our results show that expression of Triad3A in KHYG-1 cells consistently diminished the maintenance of p65 DNA binding activity in response to 2DL4 engagement, especially at later time points (Figs. 6B and 6C). To further confirm the negative impact of Triad3A on sustained NF-κB activation, we measured activity in 293T cells 48 h after transfection with 2DL4 ±Triad3A using a luciferase reporter assay. Our data demonstrated that Triad3A expression nearly abolishes 2DL4-mediated activation of NF-κB (Fig. 6D). In conclusion, Triad3A does not affect the 2DL4-mediated early activation of MAP kinases and IKKα/β, but suppresses sustained activation of NF-κB, demonstrating that only late-term signaling is disrupted by Triad3A.
FIGURE 5. Activation of MAP kinases upon 2DL4 cross-linking in an NK cell line was unaffected by Triad3A expression.
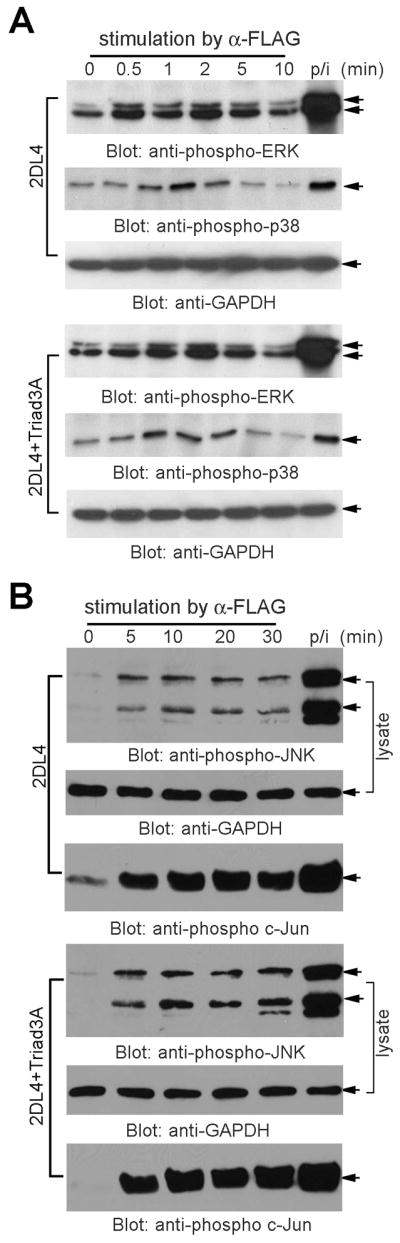
KHYG-1 cells (harvested 2 days after fresh IL-2 culture) transduced to express FLAG-2DL4 ± FLAG-Triad3A were stimulated with soluble anti-FLAG (M2) mAb (10 μg/ml) for the indicated minutes or PMA and ionomycin (p/i) for 10 min. A, Cells were then lysed and aliquots of precleared lysates were separated by SDS-PAGE and immunoblotted with anti-phospho-ERK (pERK), anti-phospho-p38, and anti-GAPDH. The results are representative of at least three independent experiments. B, JNK was assayed from the same cell lysates with an in vitro kinase assay using GST-c-Jun (1–79 aa) as a JNK-binding matrix and substrate. Stimulated lysates were adsorbed with GST-c-Jun on glutathione-sepharose and suspended with JNK assay buffer containing cold ATP at 30°C for 20 min. Reactions were terminated with 3X Laemmli buffer and analyzed by immunoblotting with anti-phospho-c-Jun. Unmanipulated lysates from same samples were immunoblotted with anti-pJNK and anti-GAPDH.
FIGURE 6. Triad3A expression suppresses sustained activation of NF-κB in response to 2DL4 ligation.
A, KHYG-1 cells transduced with FLAG-2DL4 ± FLAG-Triad3A (day 2 after IL-2 culture) were stimulated with soluble anti-FLAG mAb (M2; 10 μg/ml) for the indicated time or PMA (P; 10 nM)) + ionomycin (I; 1 μM) (10 min). Cell lysates were immunoblotted with anti-phospho-IKKα/β (pIKKα/β) and anti-GAPDH. B, 2DL4 ligation-induced nuclear DNA binding activity of NF-κB p65 complexes. KHYG-1 cells expressing FLAG-2DL4 ± FLAG-Triad3A were stimulated with soluble anti-FLAG mAb (10 μg/ml) for indicated time or PMA and ionomycin (P/I) for 120 min. Nuclear extracts were preincubated with radiolabeled ds NF-κB oligonucleotide probe, and analyzed on non-denaturing PAGE. The last lane shows anti-p65 Ab supershift (ss) analysis of a PMA/ionomycin-stimulated extract. NS = non-specific band. Results are representative of at least 3 experiments. C, The p65 DNA binding band in panel B was quantified with ImageJ software and plot shows % binding activity compared to unstimulated control at time zero (= 100%) from panel B. D, Triad3A expression suppressed 2DL4-stimulated NF-κB luciferase reporter activity. 293T cells were transfected with NF-κB renilla luciferase reporter plasmid in combination with FLAG-2DL4 ±FLAG-Triad3A cDNAs and luciferase activity in lysates was assayed 48 h later. Results are mean ± S.D. of relative units NF-κB activity from 6 experiments.
Triad3A inhibits 2DL4 turnover without affecting receptor internalization
Endocytosis is a critical event influencing the turnover, signal transduction, and biological functions of cell surface receptors. Internalized receptors generally enter Rab5+ early endosomes, where some receptors are sorted to lysosome and proteasome compartments for degradation, while others can be recycled back to the cell surface (15, 32).
Previous work showed that vesicle-mediated endocytosis is required to transport 2DL4 to the Rab5+ early endosomal compartment, since expression of a dominant-negative (DN) form of dynamin inhibited 2DL4 internalization to that location (8). Dynamins are large GTPases that facilitate vesicle invagination and scission during endocytic processes, especially involving clathrin-coated vesicles or caveolae (33). In view of the DN-dynamin impact and our observations that Triad3A expression in KHYG-1 cells selectively abolished 2DL4-mediated cytokine production without altering surface expression or early signaling events, we speculated that the E3 ubiquitin ligase might be disrupting intracellular receptor trafficking to prevent sustained NF-κB signaling and achieve cytokine production. Therefore, we examined whether Triad3A expression affects 2DL4 endocytosis, turnover, and intracellular localization in response to receptor ligation.
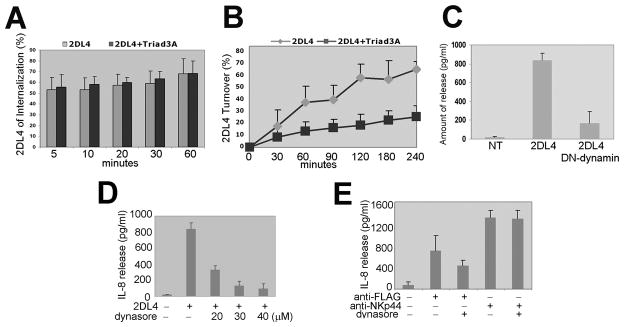
First, we found that Triad3A expression does not affect receptor endocytosis over time in KHYG-1 cells (Fig. 7A). Consistent with our previous findings, these data further demonstrate that Triad3A does not impact 2DL4 at the cell surface. Next, we tested whether 2DL4 receptor turnover back to the cell surface was affected by Triad3A expression. While 2DL4 was efficiently turning over in KHYG-1 cells expressing the receptor alone, Triad3A expression significantly suppressed receptor turnover (Fig. 7B). This result was consistent when performed in the presence of the protein synthesis inhibitor, cycloheximide (data not shown), indicating that Triad3A expression is not suppressing 2DL4 surface expression derived from de novo synthesis, but is instead suppressing the recycling of internalized 2DL4 back to the cell surface. Therefore, we hypothesize that Triad3A expression promotes the ubiquitylation of the majority of internalized 2DL4, which is shuttled to the proteasome or lysosome for degradation, thereby preventing recycling back to the cell surface.
FIGURE 7. Triad3A alters 2DL4 turnover and vesicle-mediated internalization is required for cytokine production.
A, 2DL4 internalization was not affected in KHYG-1 cells co-transduced with Triad3A. KHYG-1 cells transduced with FLAG-2DL4 ± Triad3A were assayed for 2DL4 internalization as in Materials and Methods. Results are mean ±S.D. of 8 experiments. B, 2DL4 turnover was suppressed in KHYG-1 cells co-transduced with Triad3A. KHYG-1 cells expressing FLAG-2DL4 ± Triad3A were assayed for 2DL4 turnover as in Materials and Methods. Results are mean ±S.D. from 3 experiments. C, Dominant negative (DN) dynamin blocks 2DL4-mediated IL-8 production in 293T cells. 293T cells were not transfected (NT) or transfected with FLAG-2DL4 ± DN-dynamin and tested 48 hr later for IL-8 production by ELISA. D, Dynasore inhibits 2DL4-mediated IL-8 production in 293T cells. 293T cells were transfected with FLAG-2DL4 and cultured with indicated concentrations of dynasore for 48 h and tested for IL-8 secretion by ELISA. E, 2DL4-mediated IL-8 production in KHYG-1 cells was suppressed by dynasore. KHYG-1 cells pretreated with 40 μM dynasore for 30 min were stimulated with either plate-bound anti-FLAG mAb (M2) or anti-NKp44 (3.43.13) and tested 24 h later for IL-8 secretion by ELISA.
Triad3A disrupts 2DL4 localization with Rab5+ early endosomes
We next determined whether vesicle-mediated endocytosis is necessary for 2DL4 to stimulate the production of IL-8 in 293T cells. As shown in Fig. 7C, co-transfection of 2DL4 in combination with DN-dynamin nearly abolished 2DL4-mediated IL-8 production, indicating a requirement for receptor internalization to trigger cytokine production. We further observed that the pharmacological dynamin inhibitor, dynasore (34), similarly blocked 2DL4-mediated IL-8 production in 293T cells in a dose-dependent manner (Fig. 7D). Although DN-dynamin expression was toxic to KHYG-1 cells, we found that dynasore also suppressed IL-8 production in response to engaging 2DL4 in the NK cell line without affecting production of the cytokine upon engaging NKp44 (Fig. 7E). Our results demonstrate that 2DL4-mediated cytokine production requires receptor internalization, which is consistent with recent work by Rajagopalan et al. (13).
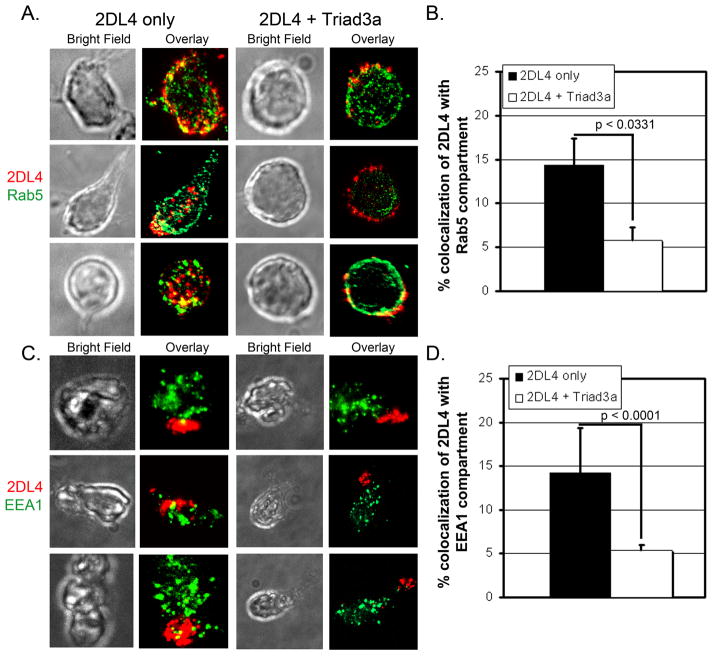
Previous evidence indicates that 2DL4 localization into the Rab5+ early endosomes is a critical step to induce cytokine production (8, 13), so we tested whether Triad3A expression alters the localization of 2DL4 to early endosomes after 2DL4 ligation in KHYG-1 cells. Cells expressing myc-tagged 2DL4 either with or without Triad3A were surface stained with anti-myc antibody and fluorophore-conjugated secondary antibody, incubated at 37°C for 1–2 hours to allow for receptor internalization, fixed, permeabilized, and stained for the early endosomal markers, Rab5 (Figs. 8A, B, and Supplemental Figure 1) or EEA1 (Figs. 8C, D, and Supplemental Figure 2). When 2DL4 was expressed alone, about 15% of the receptor entered the Rab5/EEA1 compartment under these crosslinking conditions (Figs. 8A-D and Supplemental Figures 1 and 2), although the fraction accumulating in the early endosomes with our cellular system was significantly lower than shown in previous work by Rajagopalan et al. (8, 13). Nonetheless, when cells co-expressed Triad3A, the 2DL4 appeared to be distributed more at the periphery of the cells or in cellular extensions and only about 5% of the receptor co-localized with Rab5 or EEA1 (Figs. 8A-D and Supplemental Figures 1 and 2). Taken together, our data demonstrate that antibody-engaged 2DL4 is endocytosed and some localizes to Rab5+/EEA1+ early endosomes. However, this endosomal localization is significantly diminished by co-expression of Triad3A (Figs. 8C, D), which according to our previous evidence, promotes polyubiquitylation of 2DL4 and trafficking to the proteasome or lysosome for degradation.
Figure 8. Intracellular trafficking of engaged 2DL4 to early endosomes is reduced by Triad3A.
KHYG-1 cells transduced to express myc-2DL4 ± Triad3A were stained with anti-myc Ab + AlexaFluor 647-labelled secondary Ab on ice, cultured at 37°C for 2 h (A,B) or 1 h (C,D), settled onto poly-L-lysine coated slides, fixed, permeabilized, and stained with polyclonal antibody to early endosomal markers (Rab5, A,B or EEA1, C,D) + secondary Ab conjugated with AlexaFlour 350. Cells were imaged by bright field (BF) and immunofluorescence microscopy for myc (2DL4; red) and Rab5 or EEA1 (green). A, Bright field and overlayed max-projected fluorescence images of all z-stack sections from three representative cells stained for Rab5 from each condition (± Triad3A expression). B, Mean ± S.E. percentage area of 2DL4 compartmentalized with Rab5 was quantified using Meta Vue software from max-projected images of z-stacks of 12–15 cells for each bar. C, Bright field and overlayed max-projected fluorescence images of all z-stack sections stained with EEA1 from three representative cells from each condition. D, Mean ±S.E. percentage area of 2DL4 compartmentalized with EEA1 was quantified using NIS-Elements software from all of the individual z-sections from 15 cells for each bar (>200 data points/bar). Percentage values were compared using the 2-tailed Mann-Whitney test and resulting p values are listed in panels B and D.
Discussion
We have identified the E3 ubiquitin ligase, Triad3A (29), as an important negative regulator of 2DL4 function. This is the first report describing the regulation of KIR by ubiquitylation. Our data demonstrate that Triad3A recruitment promotes the direct polyubiquitylation and degradation of 2DL4, and expression of the E3 ligase dramatically reduces total expression of the receptor in cell lysates. However, Triad3A expression did not influence 2DL4 surface expression, endocytosis, or early signaling events triggered through the receptor. Nonetheless, Triad3A expression significantly suppressed cytokine production and blunted sustained NF-κB signaling in response to 2DL4 engagement. Furthermore, expression of Triad3A suppressed 2DL4 recycling to the cell surface and promoted trafficking of internalized 2DL4 away from the early endosome, which was previously shown to be a site of intracellular 2DL4 accumulation and signaling (8, 13). Importantly, we also showed that function of another NK cell activating receptor, NKp44, was not impacted by expression of Triad3A in the same NK cells. Therefore, we conclude that Triad3A selectively controls intracellular 2DL4 function.
It is well established that polyubiquitylation of lysine residues can tag transmembrane, cytosolic and nuclear proteins for sorting to multivesicular bodies and subsequent transport to proteasomal or lysosomal degradation (16, 35). Several shuttle factors, such as RAD23, Dsk2/ubiquilin, and DDI1, contain both a ubiquitin binding domain and a proteasome-targeting motif, which bind to polyubiquitylated cargo proteins and direct their transport to the proteasome, respectively (36). Evidence is accumulating that attachment with different ubiquitylation branching arrays to proteins results in distinct functional outcomes: 1) polyubiquitylation chains linked at the lysine-48 (K48) residue of the ubiquitin molecules generally targets proteins to the proteasome for degradation, 2) K63-linked polyubiquitylation initiates intracellular signaling or trafficking, and 3) monoubiquitylation can alter protein activity, promote endocytosis, or target to lysosomal degradation (16). Our observation that polyubiquitylation of 2DL4 by Triad3A target only internalized receptor away from the early endosomes to a degradation pathway is consistent with K48-linked branching. Furthermore, a recent report demonstrated that Triad3A can incorporate K48-linked polyubiquitin chains (31). Other examples of ubiquitylation-mediated trafficking of proteins away from early endosomes include the diubiquitylation of Ig-β of the B cell antigen receptor complex by Itch (37) and the monoubiquitylation of Lst2 protein (38), although neither of these proteins are targeted to the proteasome.
We previously reported that 2DL4-mediated cytokine production requires activation of MAP kinases and downstream activation of NF-κB (12). Here, we have found that Triad3A overexpression in the NK cell line, KHYG-1, does not affect 2DL4-mediated early activation profiles of ERK, JNK, p38, and IKK-α/β. We conclude that these 2DL4-mediated signaling events occur at the cell surface and the surface expression and membrane-proximal function of 2DL4 was unchanged in Triad3A-expressing cells. The reduction of total 2DL4 protein levels by Triad3A was most likely due to ubiquitylation-mediated degradation of the intracellular pool of receptor, and we observed a coordinate reduction in sustained NF-κB activation, which we conclude to be critical for transducing intracellular signals leading to cytokine release. In this model, 2DL4 signals sequentially, first from the cell surface and subsequently from an intracellular endosomal compartment. Taken together, our data demonstrate that both early MAP kinase stimulation and sustained activation of NF-κB are necessary for the induction of 2DL4-mediated cytokine production and Triad3A disrupts the latter signal.
The Triad3A-mediated reduction in sustained activation of NF-κB is consistent with growing evidence that the E3 ligase targets the NF-κB pathway, particularly in innate immune responses. Chuang et al. reported that Triad3A interacts with distinct toll-like receptors (TLR4, 5, and 9, but not TLR2) and down-regulates their function (29). Importantly, the same report demonstrated that Triad3A inhibited the capacity of these TLRs to stimulate NF-κB reporters by promoting receptor ubiquitylation and degradation via the proteasome. Triad3A has also been shown to act as a negative regulator of TNF-α- and IL-1-mediated activation of NF-κB by associating with and promoting the degradation of the NF-κB signaling proteins RIP-1, TIRAP, and TRIF (30, 39). Recently, Triad3A was also shown to interact with and induce ubiquitylation and degradation of TRAF3 (31). Triad3A was further shown to negatively regulate RIG-I/MAVS-mediated NF-κB activation through targeting TRAF3, thereby modulating innate immune receptor responses toward several invading pathogens (31). In view of the impacts of Triad3A on the receptor-proximal NF-κB signaling pathway, we cannot rule out that direct recruitment of the E3 ligase to 2DL4 may similarly promote the ubiquitylation and degradation of receptor-associated signaling proteins within the NF-κB pathway. Our data, however, are the first to demonstrate that Triad3A specifically influences late-term NF-κB signaling at an intracellular site, but does not disrupt membrane proximal events.
Six different alternative spliced forms of Triad3 have been reported, named Triad3 and Triad3A-E (29), and all of these contain the C-terminal domain found to interact with 2DL4 in our yeast screen. The major protein form that we have identified in NK cells migrates nearly identical to that of Triad3A in SDS-PAGE. Although our studies have therefore focused on the impacts of the common Triad3A form, it is possible that other differentially spliced variants also influence 2DL4 function.
Our data indicate that 2DL4 ubiquitylation by Triad3A is occurring subsequent to internalization and this prevents accumulation of the receptor in Rab5+/EEA1+ early endosomes to disrupt function. It has been shown that 2DL4 is internalized in a dynamin-dependent manner to early endosomes after receptor ligation in NK cells or 293T cells (8). Consistent with a recent report (13), we show that DN-dynamin and the dynamin inhibitor, dynasore, suppress 2DL4-induced production of IL-8 in both 293T cells and NK cells, indicating a major role for internalized receptor in cytokine production. Although it is believed that ligand-induced endocytosis is a mechanism of receptor inactivation, numerous studies have shown that receptors can remain active within the endosomal compartment (14, 15), and the temporal dynamics of receptor signaling can dramatically influence functional outcomes (40). Our results indicate that the cytokine-producing function of 2DL4 requires intracellular shuttling of the receptor to early endosomes to stimulate prolonged signaling, which is suppressed by Triad3A. In this way, polyubiquitylation of 2DL4 by Triad3A prevents surface recycling and shuttles the receptor away from the early endosomes, thereby negatively regulating 2DL4 function by limiting the lifetime of its active signaling state at this intracellular site.
Supplementary Material
Acknowledgments
We thank Drs. Dietmar Kappes and Siddharth Balachandran for constructive advice on the manuscript, Dr. Suresh Basagoudanavar for technical assistance, Dr. Edna Cukierman for assistance in analyzing the immunofluorescence microscopy data, Dr. Xin-Ming Li for 2D gel electrophoresis, and Drs. Erica Golemis, Roger Brent, Brent Passerin, and Ilya J. Serebriiskii for advice and reagents during the yeast two-hybrid screening. We also thank Drs. Marco Colonna, Garry Nolan, Lynn Heasley, Tung-Hsein Chuang, Keiji Tanaka, Shao Cong-Sun, Kiyonao Sada and Masato Yagita for reagents. Special thanks also to the DNA Synthesis, DNA Sequencing, Flow Cytometry, Biochemistry and Biotechnology, and Cell Culture Facilities at FCCC.
Abbreviations
- KIR
killer cell Ig-like receptor
- 2DL4
KIR2DL4 (KIR with two Ig-like domains and a long cytoplasmic domain 4)
- ITIM
Immunoreceptor tyrosine-based inhibitory motif
- ITAM
Immunoreceptor tyrosine-based activation motif
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor α
- shRNA
short hairpin RNA
- RIG-I
Retinoic acid inducible gene-I
- MAVS
mitochondrial antiviral signaling
Footnotes
This work was supported by grant R01-CA100226 (K.S.C.), training grant T32-CA009035 (S.M.S.M., A.K.P. and D.A.A.A.), Cancer Center Support Grant CA06927 (FCCC) from the NIH, and an appropriation from the Commonwealth of Pennsylvania.
Conflict of Interest Disclosure:
The authors declare no conflict of interest or financial interests.
References
- 1.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 2.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 3.Goodridge JP, Witt CS, Christiansen FT, Warren HS. KIR2DL4 (CD158d) genotype influences expression and function in NK cells. J Immunol. 2003;171:1768–1774. doi: 10.4049/jimmunol.171.4.1768. [DOI] [PubMed] [Google Scholar]
- 4.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 5.Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168:5047–5057. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- 6.Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, Bertone S, Moretta A, Moretta L, Mingari MC. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodridge JP, Lathbury LJ, John E, Charles AK, Christiansen FT, Witt CS. The genotype of the NK cell receptor, KIR2DL4, influences INFgamma secretion by decidual natural killer cells. Mol Hum Reprod. 2009;15:489–497. doi: 10.1093/molehr/gap039. [DOI] [PubMed] [Google Scholar]
- 10.Trundley A, Moffett A. Human uterine leukocytes and pregnancy. Tissue Antigens. 2004;63:1–12. doi: 10.1111/j.1399-0039.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 11.Ashkar AA, Croy BA. Interferon-gamma contributes to the normalcy of murine pregnancy. Biol Reprod. 1999;61:493–502. doi: 10.1095/biolreprod61.2.493. [DOI] [PubMed] [Google Scholar]
- 12.Miah SM, Hughes TL, Campbell KS. KIR2DL4 differentially signals downstream functions in human NK cells through distinct structural modules. J Immunol. 2008;180:2922–2932. doi: 10.4049/jimmunol.180.5.2922. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan S, Moyle MW, Joosten I, Long EO. DNA-PKcs controls an endosomal signaling pathway for a proinflammatory response by natural killer cells. Sci Signal. 2010;3:ra14. doi: 10.1126/scisignal.2000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiley HS, Burke PM. Regulation of receptor tyrosine kinase signaling by endocytic trafficking. Traffic. 2001;2:12–18. doi: 10.1034/j.1600-0854.2001.020103.x. [DOI] [PubMed] [Google Scholar]
- 15.Sadowski L, Pilecka I, Miaczynska M. Signaling from endosomes: location makes a difference. Exp Cell Res. 2009;315:1601–1609. doi: 10.1016/j.yexcr.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 17.Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 18.Yagita M, Huang CL, Umehara H, Matsuo Y, Tabata R, Miyake M, Konaka Y, Takatsuki K. A novel natural killer cell line (KHYG-1) from a patient with aggressive natural killer cell leukemia carrying a p53 point mutation. Leukemia. 2000;14:922–930. doi: 10.1038/sj.leu.2401769. [DOI] [PubMed] [Google Scholar]
- 19.Miah SM, Campbell KS. Expression of cDNAs in Human Natural Killer Cell Lines by Retroviral Transduction. In: Campbell KS, editor. Meth in Mol Biol. 2. Springer; 2010. pp. 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Arias DA, Campbell KS. Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. J Immunol. 2007;179:5281–5290. doi: 10.4049/jimmunol.179.8.5281. [DOI] [PubMed] [Google Scholar]
- 21.Campbell KS, Cooper S, Dessing M, Yates S, Buder A. Interaction of p59fyn kinase with the dynein light chain, Tctex-1, and colocalization during cytokinesis. J Immunol. 1998;161:1728–1737. [PubMed] [Google Scholar]
- 22.Campbell KS, Giorda R. The cytoplasmic domain of rat NKR-P1 receptor interacts with the N-terminal domain of p56(lck) via cysteine residues. Eur J Immunol. 1997;27:72–77. doi: 10.1002/eji.1830270111. [DOI] [PubMed] [Google Scholar]
- 23.Li XM, Patel BB, Blagoi EL, Patterson MD, Seeholzer SH, Zhang T, Damle S, Gao Z, Boman B, Yeung AT. Analyzing alkaline proteins in human colon crypt proteome. J Proteome Res. 2004;3:821–833. doi: 10.1021/pr049942j. [DOI] [PubMed] [Google Scholar]
- 24.Heasley LE, Winn RA. Analysis of Wnt7a-stimulated JNK activity and cJun phosphorylation in non-small cell lung cancer cells. Methods Mol Biol. 2008;468:187–196. doi: 10.1007/978-1-59745-249-6_14. [DOI] [PubMed] [Google Scholar]
- 25.Hou P, Araujo E, Zhao T, Zhang M, Massenburg D, Veselits M, Doyle C, Dinner AR, Clark MR. B cell antigen receptor signaling and internalization are mutually exclusive events. PLoS Biol. 2006;4:e200. doi: 10.1371/journal.pbio.0040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masilamani M, Narayanan S, Prieto M, Borrego F, Coligan JE. Uncommon endocytic and trafficking pathway of the natural killer cell CD94/NKG2A inhibitory receptor. Traffic. 2008;9:1019–1034. doi: 10.1111/j.1600-0854.2008.00738.x. [DOI] [PubMed] [Google Scholar]
- 27.Dietrich J, Menne C, Lauritsen JP, von Essen M, Rasmussen AB, Odum N, Geisler C. Ligand-induced TCR down-regulation is not dependent on constitutive TCR cycling. J Immunol. 2002;168:5434–5440. doi: 10.4049/jimmunol.168.11.5434. [DOI] [PubMed] [Google Scholar]
- 28.Purdy AK, Campbell KS. SHP-2 expression negatively regulates NK cell function. J Immunol. 2009;183:7234–7243. doi: 10.4049/jimmunol.0900088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 30.Fearns C, Pan Q, Mathison JC, Chuang TH. Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem. 2006;281:34592–34600. doi: 10.1074/jbc.M604019200. [DOI] [PubMed] [Google Scholar]
- 31.Nakhaei P, Mesplede T, Solis M, Sun Q, Zhao T, Yang L, Chuang TH, Ware CF, Lin R, Hiscott J. The E3 Ubiquitin Ligase Triad3A Negatively Regulates the RIG-I/MAVS Signaling Pathway by Targeting TRAF3 for Degradation. PLoS Pathog. 2009;5:e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 34.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 36.Grabbe C, Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem Rev. 2009;109:1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M, Veselits M, O’Neill S, Hou P, Reddi AL, Berlin I, Ikeda M, Nash PD, Longnecker R, Band H, Clark MR. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J Immunol. 2007;179:4435–4443. doi: 10.4049/jimmunol.179.7.4435. [DOI] [PubMed] [Google Scholar]
- 38.Mosesson Y, Chetrit D, Schley L, Berghoff J, Ziv T, Carvalho S, Milanezi F, Admon A, Schmitt F, Ehrlich M, Yarden Y. Monoubiquitinylation regulates endosomal localization of Lst2, a negative regulator of EGF receptor signaling. Dev Cell. 2009;16:687–698. doi: 10.1016/j.devcel.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D, Li X, Zhai Z, Shu HB. A novel zinc finger protein interacts with receptor-interacting protein (RIP) and inhibits tumor necrosis factor (TNF)- and IL1-induced NF-kappa B activation. J Biol Chem. 2002;277:15985–15991. doi: 10.1074/jbc.M108675200. [DOI] [PubMed] [Google Scholar]
- 40.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.