The ability to study proteins in live cells using genetically encoded fluorescent proteins (FPs) has revolutionized cell biology(1), (2), (3). Researchers have created numerous FP biosensors and optimized FPs for specific organisms and subcellular environments in a rainbow of colors(4), (5). However, expressing FPs in oxidizing environments such as the eukaryotic endoplasmic reticulum (ER) or the bacterial periplasm can impair folding, thereby preventing fluorescence(6), (7). A substantial fraction of EGFP oligomerizes to form non-fluorescent mixed disulfides in the ER(6) and EGFP does not fluoresce in the periplasm when targeted via the SecYEG translocon(7). To overcome these obstacles, we exploited the highly efficient folding capability of superfolder GFP (sfGFP)(8). Here, we report sfGFP does not form disulfide-linked oligomers in the ER and maltose-binding protein signal sequence (peri)-sfGFP(9) is brightly fluorescent in the periplasm of E. coli. Thus, sfGFP represents an important research tool for studying resident proteins of oxidizing environments.
When properly implemented(10), FPs are a valuable asset to many experimental systems. However, the unintended and unknown effects of using suboptimal FPs represent a significant caveat for FP experiments. For example, EGFP can form weak non-covalent dimers. When fused to integral membrane proteins and overexpressed, the dimeric tendency of EGFP leads to inappropriate interactions, resulting in false-positive FRET signals(11) and dysmorphic organelles(12). A single point mutation, A206K, can prevent dimerization and resolved these issues.
Similarly, it has been reported that EGFP can form covalent oligomers via interchain disulfide bonds in the oxidizing environment of the secretory pathway in endocrine cells(6, 13), (14). The structure of correctly folded GFP consists of an internal fluorophore surrounded by a tight β-barrel and does not require cellular chaperones for folding(3), (15). GFP contains two cysteine residues, C49 and C71, which are both located in the interior of the barrel and flank the chromophore (Ser65-Tyr66-Gly67). Since endogenous GFP is a cytoplasmic protein, disulfide bond formation cannot adversely affect GFP formation in jellyfish. However, when GFP is expressed in oxidizing environments (i.e. the ER lumen or the periplasm of gram negative bacteria), C49 and C71 are exposed during folding and can potentially bind other folding GFP molecules or cysteine-containing proteins to form mixed-disulfides. To form a fluorophore and produce a fluorescent signal, GFP must form and maintain the tight b barrel structure(15). The cysteines are separated by 2.4 nm, too far apart to form an intramolecular disulfide bond(3). Therefore, intermolecular disulfide-bonded EGFP must be inherently misfolded and thus, non-fluorescent. While GFP can clearly fold and form fluorescent molecules in the ER(16), (17, 18), anti-GFP immunoblots of non-reducing SDS-PAGE gels reveal up to 50% of total ER GFP is incorporated into disulfide bonded oligomers(6). Such effects confound quantitation of total levels of GFP in the secretory pathway (19). To minimize perturbation of the secretory pathway, maximize fluorescent GFP signal, and maintain functionality of FP-fusion secretory proteins, errant disulfide bond formation must be addressed.
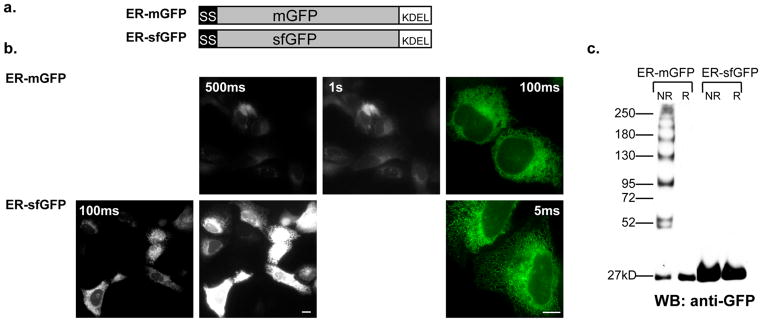
As inappropriate disulfide bond formation must be occurring during nascent GFP folding in the ER, we hypothesized that a rapidly folding and robustly stable mutant of EGFP could potentially fold before disulfide bonds can form. SfGFP has both of these properties(8). However, sfGFP also contains the C49 and C71 cysteines (at positions C48 and C72, respectively). To test the hypothesis that sfGFP would remain truly monomeric in the ER, we replaced mGFP with sfGFP in our previously described fluorescent ER marker, ER-mGFP(20), which contains a prolactin signal-sequence and a KDEL retrieval motif (known from here on as ER-sfGFP) (Fig. 1a). We expressed both ER markers in human osteosarcoma epithelial (U2OS) cells, noting ER-sfGFP is significantly brighter than ER-mGFP (Fig. 1b), treated the cells with N-ethylmaleimide (NEM) and collected the cell lysates in SDS-PAGE buffer with or without DTT. We immunoblotted the samples with anti-GFP and detected both the ER-mGFP and ER-sfGFP reduced samples as single bands at 27kD as expected. The non-reduced ER-mGFP resembled a protein ladder with bands ranging from 27kD to greater than 250kD. As reported in Jain et al., this is consistent with the presence of oligomeric mixed disulfides of ER-mGFP(6). In contrast, even at longer exposures, the non-reduced ER-sfGFP only produced a single 27kD band (Fig. 1c). The misfolded, and thus non-fluorescent, mixed-disulfide GFP oligomers are likely oligomerizing fusion protein, as well. This result signifies a fundamental shift towards using sfGFP rather mGFP for the creation of secretory fusion proteins is necessary for markers and fusion proteins localized to the secretory pathway.
Figure 1.
(a) The ER-mGFP construct was created for expression in mammalian cells to fluorescently label the ER and has been previously described(20). To make ER-sfGFP we replaced mGFP with sfGFP while all other elements of the constructs remain identical. (b) In the epifluorescent micrographs of fields of U2OS cells at 20X or 63X (color) magnification expressing either ER-mGFP at 500ms or 1s, or ER-sfGFP at 100ms or 500ms exposures, we show ER-sfGFP is much brighter than mGFP in the ER and can be visualized at much lower exposures. Scale bars are 10μm. (c) We evaluated the tendencies of the constructs to form mixed disulfide complexes in the ER by western blot analysis. Both samples were treated with 20mM NEM for 15 minutes and collected in either reducing (100mM DTT) or non-reducing conditions. The data shows ER-mGFP forms many disulfide complexes of varying sizes up to a few hundred kilodaltons, whereas ER-sfGFP does not form any mixed disulfides. None are detectable even at longer exposures.
We hypothesized sfGFP might also fold robustly enough to fold and fluoresce in the highly oxidizing periplasm of gram-negative bacteria when targeted cotranslationally via the bacterial Sec61 homolog, the SecYEG translocon. The formation of disulfide-bonded GFP oligomers is not easily observed in the periplasm of gram-negative bacteria and has remained unreported to date. However, structural disulfide folding intermediates have been detected at the site of translocation and indicate the presence of cysteine residues not only affects the folding reaction, but also the type of translocation used as well (post or cotranslational)(21). The presence of cysteines in FPs is strongly implicated in the complete absence of EGFP fluorescence following SecYEG translocation(7). In support of this hypothesis, mCherry, which does not contain any cysteine residues, correctly folds and fluoresces in the periplasm(22). Cytoplasmic membrane proteins are targeted to the SecYEG translocon via the signal recognition particle (SRP) and then cotranslationally translocated. The vast majority of periplasmic proteins are posttranslationally targeted to and translocated through the SecYEG translocon, dependent on either SRP or SecA/B(23). For both pathways, proteins are maintained in an unfolded state by cytoplasmic chaperones, threaded through the translocon, and then fold in the periplasm(24). Alternatively, a protein targeted to the periplasm via the twin-arginine translocation system (Tat), can first fold in the cytoplasm and translocate in a folded state into the periplasm(25). Proteins targeted using this pathway employ a –RR motif rather than an N-terminal signal sequence for localization. Although EGFP did not fluoresce when targeted to the periplasm via the SecYEG pathway(7), Thomas et al. demonstrated an EGFP fusion protein, TorA-GFP, could be exported to the periplasm by the Tat pathway in a fully active state and remain fluorescent(25). More recently, Cava et al. used a PhoA-sfGFP construct to demonstrate sfGFP-fusion proteins fluoresce in the periplasm when targeted via the Tat system, in Thermus thermophilus(26).
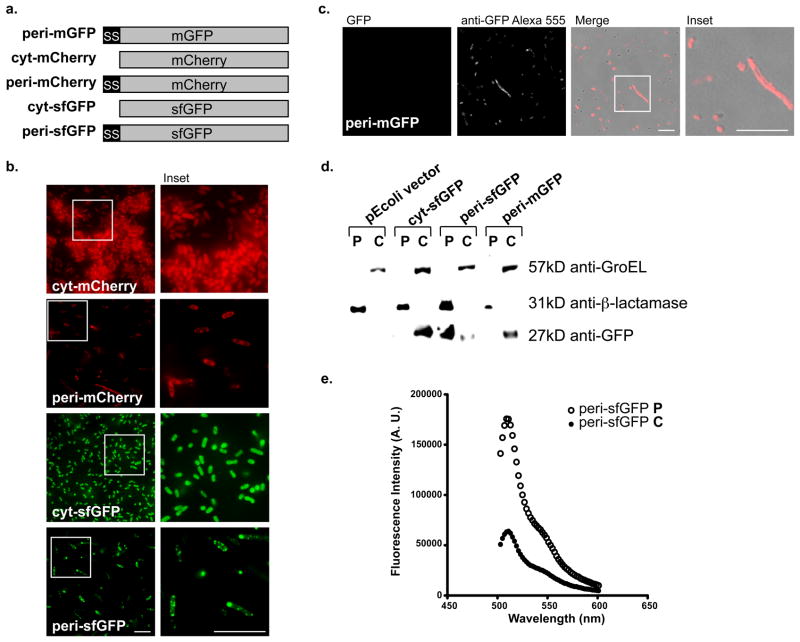
In a recent attempt to form a green fluorescent fluorophore in the periplasm via the SecYEG translocon, Fisher et al., created a ssMBP-sfGFP bacterial expression construct, which accumulated primarily in the cytoplasm and remained inactive in the periplasm(27). The authors concluded sfGFP, like EGFP, was unsuitable for periplasmic fluorescence applications when targeted via the SecYEG translocon. However, Lee and Bernstein described an MBP signal sequence (MBP*1), which contains three amino acid mutations, optimized for efficient co-translational translocation across the Escherichia coli (E. coli) inner membrane(28). We exploited this optimized signal sequence to construct our periplasmic (peri)-sfGFP, peri-mGFP, peri-mCherry and cytoplasmic (cyt)-sfGFP and cyt-mCherry without a signal sequence, to differentiate between the localization patterns of the cytoplasm and periplasm of bacteria (Fig. 2a).
Figure 2.
(a) The periplasmic mGFP, mCherry and sfGFP bacterial expression constructs contain an optimized MBP signal sequence and the cytoplasmic sfGFP and mCherry constructs do not. (b) The epifluorescent micrographs (63X) of E. coli expressing peri-mCherry (750ms), cyt-mCherry (12ms), cyt-sfGFP (100ms) or peri-sfGFP (500ms) indicates peri-sfGFP localized to the periplasm and is much brighter than peri-mCherry. (c) GFP in peri-mGFP is not fluorescent (2s exposure) in either the periplasm or the cytoplasm. However, it is made and accumulates in the cytoplasm similar to reports by others (27). Scale bars are 10μm. (d) Each of our constructs is present in the expected E. coli fractions, except for peri-mGFP, which is only detected by immunoblot in the cytoplasm. We used anti-GroEL and anti-β-lactamase to label the cytoplasmic and periplasmic fractions, respectively. (e) Fluorimeter measurements of the periplasmic and cytoplasmic fractions of peri-sfGFP expressing E. coli. The majority of fluorescence activity was in the periplasmic fraction of peri-sfGFP as denoted by the much higher peak at approximately 510nm.
When expressed in E. coli, cyt-sfGFP and cyt-mCherry correctly localized to the cytoplasm of transformed E. coli. The positive control peri-mCherry fluoresced in a characteristic periplasmic ring pattern (Fig. 2b), whereas the negative control peri-mGFP did not fluoresce (Fig. 2c). Significantly, our peri-sfGFP was brightly fluorescent in the periplasm (Fig. 2b). The use of the more efficient signal sequence in combination with sfGFP, rather than mGFP, is sufficient for SecYEG pathway-targeted formation of fluorescent green fluorophore in the periplasm, which is visible even at modest camera exposures of 500ms. In contrast, peri-mGFP fluorescence cannot be detected even at camera exposures up to 2 seconds. Anti-GFP immunofluorescence of peri-mGFP confirmed the protein is, indeed, expressed in bacteria (Fig. 2c). Thus, while an improved signal sequence can enable correct targeting and folding of sfGFP, it is not sufficient to rescue folding of mGFP (Fig. 2c). Notably, Fisher and DeLisa (27) similarly observed failure of an EGFP variant containing the cycle 3 mutations to target to the periplasm or fluoresce.
We confirmed the localization of each construct by biochemical fractionation of the transformed E. coli followed by immunoblotting of SDS-PAGE separated fractions. Fractions were probed with anti-GroEL and anti-β-lactamase to label the cytoplasmic and periplasmic fractions, respectively. While cyt-sfGFP is detectable using anti-GFP in the cytoplasmic fraction as indicated by the presence of GroEL and the peri-sfGFP is labeled in the periplasmic fraction as marked by the β-lactamase (Fig. 2d). To further verify active fluorescence in the periplasm of peri-sfGFP expressing E. coli, we measured the fractions using a fluorimeter and determined the majority of fluorescence activity was in the periplasmic fraction of peri-sfGFP (Fig. 2e). Surprisingly, non-fluorescent peri-mGFP is only detected in the cytoplasmic fraction, but not in the periplasmic fraction (Fig. 2d). Tian et al. reported basic amino acids at the beginning of the mature domain of secretory proteins could impair co-translational translocation of the protein (29).
Our findings provide potential insights into the folding mechanisms differentiating EGFP and sfGFP in oxidizing environments. First, EGFP must be forming interchain disulfides before the β barrel forms. Therefore, sfGFP must form its β barrel or at least a protective or steric part of the barrel much faster than EGFP. Interestingly, two of the sfGFP mutations occur before either of the cysteines (S30R and Y39N). The crystal structure of sfGFP revealed these two mutations alter the conformations of the first three β-strands and provide the greatest improvement to sfGFP folding robustness (8). We hypothesize the folding of the first three b-strands are critical for GFP folding as it emerges from a translocation channel. It remains unclear whether sfGFP could correctly fold and fluoresce in an in vitro oxidizing environment (15).
Our characterization of sfGFP provides the first green, actively fluorescent, SecYEG translocated FP for the bacterial periplasm. In addition, sfGFP can be mutated to create cyan and yellow variants to dramatically expand the palette of FPs available for periplasmic and ER studies(8). The ability of sfGFP to circumvent disulfide bond formation in the ER establishes sfGFP and its chromatic variants as the essential GFP standard for studies of secretory proteins in cells.
Methods
Plasmid Constructions
Mammalian Primers and Constructs
ER-mGFP was constructed by fusing the bovine prolactin signal sequence and the amino acid following the signal cleavage site into our vector based on the Clontech N1-GFP backbone. The construct was modified by PCR to append a KDEL sequence at the -COOH terminus for localization of mGFP to the ER. ER-mGFP has been previously described(20) ER-sfGFP was constructed by swapping out mGFP KDEL with sfGFP KDEL (made with primers identical to those used for mGFP KDEL) with Age/Not1. All constructs were confirmed by sequencing.
Bacterial Primers and Constructs
peri-pEcoli-Cterm 6xHN vector
Forward:GATCCCATGGGTATGAAAATAAAAACAG GTGCACGCATCCTCGC Reverse:GATCGAATTCGGTCATCAAGATCTCGGC
peri-mGFP, -sfGFP and -mCherry
Forward:GATCGAATTCAGCGTGAGCAAGGGCGAG Reverse:GATCCTGCAGCCTTGTACAGCTCGTC
mCherry PstI site removal by site directed mutagenesis
Forward:GACCCAGGACTCCTCCCTCCAGGACGGC GAGTTC Reverse:GAACTCGCCGTCCTGGAGGGAGGAGTC CTGGGTC
cyt-mGFP and -sfGFP
Forward:GATCCCATGGGTATGGTGAGCAAGGGCG AGGAG Reverse:GATCCTGCAGCCTTGTACAGCTCGTC
cyt-mCherry
Forward:GATCGAATTCAGCGTGAGCAAGGGCGAG Reverse:GATCCCATGGGTATGGTGAGCAAGGGC GAGGAG
For periplasmic localization, the maltose binding protein (MBP*1) signal sequence was cloned by PCR from the previously described vector, Pgk(-2)(29) (a generous gift from Dr. Harris Bernstein, National Institutes of Health). The MBP signal sequence fragment was inserted into Clontech pEcoli-Cterm 6xHN vector via NcoI/EcoRI restriction sites to create our peri-pEcoli-Cterm 6xHN vector. The mGFP and sfGFP fragments were isolated by PCR and inserted into peri-pEcoli-Cterm 6xHN vector with EcoRI/PstI restriction sites. After the removing an internal PstI site by standard site directed mutagenesis, the mCherry fragment was cloned into our peri-pEcoli-Cterm 6xHN vector using the same strategy. To create cytoplasmic localized FPs, mGFP and sfGFP were cloned into pEcoli-Cterm 6xHN with NcoI/PstI restriction sites.
Mammalian Tissue Culture
U2OS cells were grown in RPMI lacking phenol red with 5mM L-glutamine, 10% heat inactivated fetal bovine serum at 37°C in 5% CO2. Cells were plated evenly in Lab-Tek chambered cover glass slides (Thermo) for live imaging.
Bacterial Immunofluorescence
BL21-RP (a gift from Jeff Chao, Albert Einstein College of Medicine) bacterial cell cultures were transformed using a standard heat shock protocol. Antibiotic selection was maintained on plates and in cultures using 100μg/ml ampicillin and 20μg/ml chloramphenicol. Cultures of the transformed bacteria were grown at 37°C with shaking overnight. Overnight cultures were diluted twenty-fold in SOC media for 2 hrs at 37°C and induced with 1mM IPTG for 1 hr. For immunofluorescence, cultures were spun down in a microfuge at max speed for 1 min, washed once with PBS and fixed in 3.7% formaldehyde for 1–2 hrs on ice. They were washed once with PBS and then pelleted and resuspended in GTE. We permeabilized the membrane with 1μg/ml of lysozyme in GTE for 5 min at room temperature (RT). Cells were pelleted as above, resuspended in fresh GTE, smeared onto pre-prepared glass cover slips and let dry. The coverslips of cells were blocked with 1% BSA for 30 min in a humid chamber. Anti-GFP (a gift from Ramanujan S. Hegde) was diluted 1:1000 in 1% BSA and incubated at RT for 1hr in a humid chamber after which the coverslips were washed 10 times with PBS. Alexa 555-conjugated anti-rabbit IgG secondary antibody (Molecular Probes) was diluted 1:2000 in 1% BSA and incubated at RT for 1hr in a humid chamber. The coverslips were again washed 10 times with PBS and mounted onto slides.
Fluorescence Microscopy
U2OS cells were imaged in phenol red-free RPMI supplemented with 10mM Hepes, 5mM glutamine and 10% fetal bovine serum. Live and fixed cells were imaged on a widefield microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) with either 20X objective or a 63X/1.4 NA oil objective and a 450–490nm excitation/500–550 emission bandpass filter using a Retiga 2000R camera. Composite figures were prepared using ImageJ (NIH), Photoshop CS4 and Illustrator CS4 software (Adobe).
Bacterial Fractionations & Fluorimetry
Transformed BL21-RP overnight cultures were prepared as before and used to inoculate to 50ml cultures with added drugs and 1mM IPTG for 4–5 hrs to a mid-exponential phase with a starting OD600 of ~0.1. Cells were pelleted at 4000g for 10 min at 4°C, resuspended in 7.5ml TES and incubated at RT for 10 min. Cells were pelleted as before and resuspended in 2ml ice cold 5mM MgSO4 and incubated for 20 min to generate spheroplasts. We pelleted the cells, collected the supernatant as the periplasmic fraction and resuspended the spheroplasts in 2ml ice cold 5mM MgSO4 and sonicated 5 × 20s 5mm amplitude bursts. The lysates were then centrifuged at 10,000g for 5min at 4°C, the supernatant was collected as the cytoplasmic fraction and a final concentration of 10mM Tris pH 8.0 was added to all fractions for storage at −20°C(25).
To quantitate active fluorescence, we measured periplasmic and cytoplasmic fractions in 5mM MgSO4, on a Fluorolog spectrofluorometer (Horiba Jobin Yvon). We subtracted the background signal with a blank measurement of 5mM MgSO4 and plotted the calibrated and background subtracted fluorescence signal.
Immunoblots
Total mammalian cell lysates for immunoblotting were prepared in SDS-PAGE buffer with 100mM DTT (reducing conditions) or no DTT (non-reducing conditions) using cells in 24 well plates at 80–90% confluence. For the reducing and non-reducing immunoblot, cells were first treated with 20mM NEM in PBS for 15 min at RT. Bacterial cell lysates were diluted with reducing SDS-PAGE buffer containing 100mM DTT. Approximations of equal loading of the bacterial fractions were determined by densitometric measurements of bands on pilot immunoblots. Proteins were separated using either 5% or 12% Tris-tricine gels, transferred to nitrocellulose, probed with the indicated antibodies and developed using enhanced chemiluminescent reagents from Pierce and exposed to X-ray film. Antibodies used included anti-GFP (a gift from Ramanujan S. Hegde), anti-GroEL, anti-β-lactamase (Abcam), and horseradish peroxidase-labeled anti-rabbit and anti-mouse (Jackson Immunoresearch Laboratories).
Supplementary Material
Supplemental Figure 1. (a) Fluorimetric measurements of the periplasmic and cytoplasmic fractions of peri-mCherry expressing E. coli. The majority of fluorescence activity was in the periplasmic fraction of peri-mCherry as denoted by the much higher peak at approximately 610nm. (b) Anti-GFP immunoblot of the fractionated samples used for the fluorimeter measurements in Fig. 2e. Note the significantly higher level of protein in the periplasmic fraction. The nature of the doublet band is unclear. The size difference is distinct from 2.6 kDa removed by cleavage of the signal peptide (data not shown). It may represent alternative signal cleavage at an amino acid adjacent to the primary cleavage site.
Acknowledgments
We thank Dr. Ramanujan S. Hegde (National Institutes of Health, NIH) for the anti-GFP antibody, Dr. Jeff Chao (Albert Einstein College of Medicine) for the competent BL21 RP bacteria, Dr. Harris Bernstein (National Institutes of Health) for the MBP*1 signal sequence and Dr. Louis Hodgson for use of and assistance with the fluorimeter. This work was supported by grants from the National Institute of General Medical Sciences (NIGMS) (R01GM086530–01) (E.L.S), a National Research Service Award Ruth L. Kirschstein Award from the NIGMS (F31GM089090) (D.E.A.), and an NIH Training Program in Cellular and Molecular Biology and Genetics Grant T32 GM007491 (D.E.A. and L.M.C). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH.
Abbreviations
- FP
fluorescent protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- mGFP
monomeric GFP
- sfGFP
superfolder GFP
- NEM
N-ethylmaleimide
- FRET
fluorescence resonance energy transfer
- U2OS
human osteosarcoma epithelial cells
- SRP
signal recognition particle
- MBP
maltose binding protein
Footnotes
Author Contributions
E.L.S., D.E.A. and L.M.C. conceived and designed the study. D.E.A. and L.M.C. performed the experiments. D.E.A. and E.L.S. analyzed the data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Contributor Information
Deborah E. Aronson, Email: Deb.Aronson@phd.einstein.yu.edu.
Lindsey M. Costantini, Email: Lindsey.Costantini@phd.einstein.yu.edu.
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 3.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273(5280):1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol. 2004;22(3):289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 6.Jain RK, Joyce PB, Molinete M, Halban PA, Gorr SU. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem J. 2001;360(Pt 3):645–649. doi: 10.1042/0264-6021:3600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182(14):4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24(1):79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 9.Duplay P, Hofnung M. Two regions of mature periplasmic maltose-binding protein of Escherichia coli involved in secretion. J Bacteriol. 1988;170(10):4445–4450. doi: 10.1128/jb.170.10.4445-4450.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snapp EL. Fluorescent proteins: a cell biologist’s user guide. Trends Cell Biol. 2009;19(11):649–655. doi: 10.1016/j.tcb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296(5569):913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 12.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163(2):257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molinete M, Lilla V, Jain R, Joyce PB, Gorr SU, Ravazzola M, Halban PA. Trafficking of non-regulated secretory proteins in insulin secreting (INS-1) cells. Diabetologia. 2000;43(9):1157–1164. doi: 10.1007/s001250051507. [DOI] [PubMed] [Google Scholar]
- 14.Paladino S, Sarnataro D, Pillich R, Tivodar S, Nitsch L, Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J Cell Biol. 2004;167(4):699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36(22):6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 16.Kaether C, Gerdes HH. Visualization of protein transport along the secretory pathway using green fluorescent protein. FEBS Lett. 1995;369(2–3):267–271. doi: 10.1016/0014-5793(95)00765-2. [DOI] [PubMed] [Google Scholar]
- 17.Dayel MJ, Hom EF, Verkman AS. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J. 1999;76(5):2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89(6):963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- 19.Jokitalo E, Cabrera-Poch N, Warren G, Shima DT. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J Cell Biol. 2001;154(2):317–330. doi: 10.1083/jcb.200104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snapp EL, Hegde RS. Curr Protoc Cell Biol. Unit 17. Chapter 17. 2006. Rational design and evaluation of FRET experiments to measure protein proximities in cells; p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138(6):1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55(4):1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 23.Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112(4):491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 24.Miot M, Betton JM. Protein quality control in the bacterial periplasm. Microb Cell Fact. 2004;3(1):4. doi: 10.1186/1475-2859-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol. 2001;39(1):47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 26.Cava F, de Pedro MA, Blas-Galindo E, Waldo GS, Westblade LF, Berenguer J. Expression and use of superfolder green fluorescent protein at high temperatures in vivo: a tool to study extreme thermophile biology. Environ Microbiol. 2008;10(3):605–613. doi: 10.1111/j.1462-2920.2007.01482.x. [DOI] [PubMed] [Google Scholar]
- 27.Fisher AC, DeLisa MP. Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS ONE. 2008;3(6):e2351. doi: 10.1371/journal.pone.0002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HC, Bernstein HD. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci U S A. 2001;98(6):3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian P, Bernstein HD. Identification of a post-targeting step required for efficient cotranslational translocation of proteins across the Escherichia coli inner membrane. J Biol Chem. 2009;284(17):11396–11404. doi: 10.1074/jbc.M900375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. (a) Fluorimetric measurements of the periplasmic and cytoplasmic fractions of peri-mCherry expressing E. coli. The majority of fluorescence activity was in the periplasmic fraction of peri-mCherry as denoted by the much higher peak at approximately 610nm. (b) Anti-GFP immunoblot of the fractionated samples used for the fluorimeter measurements in Fig. 2e. Note the significantly higher level of protein in the periplasmic fraction. The nature of the doublet band is unclear. The size difference is distinct from 2.6 kDa removed by cleavage of the signal peptide (data not shown). It may represent alternative signal cleavage at an amino acid adjacent to the primary cleavage site.