Abstract
Background
The study of speciation in the marine realm is challenging because of the apparent absence of physical barriers to dispersal, which are one of the main drivers of genetic diversity. Although phylogeographic studies using mitochondrial DNA (mtDNA) information often reveal significant genetic heterogeneity within marine species, the evolutionary significance of such diversity is difficult to interpret with these markers. In the northwestern (NW) Pacific, several studies have emphasised the potential importance of sea-level regression during the most recent glaciations as a driver of genetic diversity in marine species. These studies have failed, however, to determine whether the period of isolation was long enough for divergence to attain speciation. Among these marine species, the cosmopolitan estuarine-dependent fish Mugil cephalus represents an interesting case study. Several divergent allopatric mtDNA lineages have been described in this species worldwide, and three occur in sympatry in the NW Pacific.
Results
Ten nuclear microsatellites were surveyed to estimate the level of genetic isolation of these lineages and determine the role of sea-level fluctuation in the evolution of NW Pacific M. cephalus. Three cryptic species of M. cephalus were identified within this region (NWP1, 2 and 3) using an assignment test on the microsatellite data. Each species corresponds with one of the three mtDNA lineages in the COI phylogenetic tree. NWP3 is the most divergent species, with a distribution range that suggests tropical affinities, while NWP1, with a northward distribution from Taiwan to Russia, is a temperate species. NWP2 is distributed along the warm Kuroshio Current. The divergence of NWP1 from NWP2 dates back to the Pleistocene epoch and probably corresponds to the separation of the Japan and China Seas when sea levels dropped. Despite their subsequent range expansion since this period of glaciation, no gene flow was observed among these three lineages, indicating that speciation has been achieved.
Conclusions
This study successfully identified three cryptic species in M. cephalus inhabiting the NW Pacific, using a combination of microsatellites and mitochondrial genetic markers. The current genetic architecture of the M. cephalus species complex in the NW Pacific is the result of a complex interaction of contemporary processes and historical events. Sea level and temperature fluctuations during Plio-Pleistocene epochs probably played a major role in creating the marine species diversity of the NW Pacific that is found today.
Background
In the marine environment, fluctuations in sea level, water temperatures and sea ice caused by glacial cycles are believed to have had a major influence on species distributions and the population connectivity of marine species [1-3]. For instance, lowered sea levels during glacial maxima led to the emergence of land bridges, which fragmented marine ecosystems and isolated populations of aquatic species [4-6]. At the same time, temperature fluctuations in the Pleistocene epoch influenced population dynamics by promoting bottlenecks and the loss of genetic heterogeneity. In fact, much of the intraspecific genetic diversity that is exhibited by contemporary species is generally assumed to have derived from such events in the Pleistocene (for a review, see [7]).
The northwestern (NW) Pacific is characterised by marginal seas, which were particularly impacted by Plio-Pleistocene glacial cycles. The lowering of sea level caused the recurrent closure of the Japan Sea, the semi-closure of the South China Sea and the partial or full exposure of the East China and Yellow Seas [8]. The closure of the Japan Sea caused fluctuations in sea temperatures, by halting the influx of the warm Tsushima Current, a branch of the Kuroshio Current (Figure 1) that supplies a large amount of heat to northern areas [9]. As a consequence, seascape dynamics of the region are believed to have profoundly influenced both intraspecific genetic diversity and species diversity of marine organisms, as exemplified by the exceptional diversity found in SE Asia [10-12]. This hypothesis has been confirmed by recent phylogeographic investigations in the NW Pacific area (Table 1). The significance of the lowering of the sea level and glaciations for species diversity has generally, however, not been assessed. The overlapping distribution of multiple divergent lineages is a striking phylogeographic pattern that is common to most of the organisms that have been investigated in this region, and it raises the question of whether some of these lineages are in fact cryptic species. For example, some authors [13-15] considered the various divergent lineages to be different species of Pleistocene origin, while others [16,17] have interpreted these lineages as intraspecific polymorphisms, consistent with the existence of isolated populations during glacial maxima and secondary contact at the end of the Pleistocene. Finally, the comparison of phylogeographic information among different marine species inhabiting the same area can provide for conflicting interpretations of the role of decreasing sea level and Pleistocene glaciations, and highlights the difficulty of distinguishing species in the early stages of differentiation from populations experiencing secondary contact after an extended isolation.
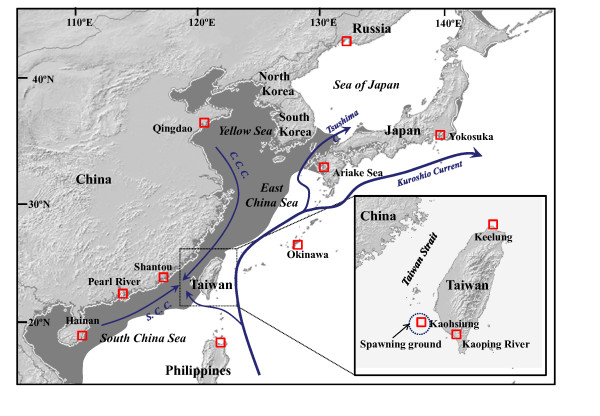
Figure 1.
Sampling localitions of Mugil cephalus in the northwestern Pacific. The shaded zone in dark grey is the area of the continental shelves that was exposed during periods of low sea levels. Blue arrows correspond to currents present in the area, C. C. C.: China Coastal Current, S. C. C.: South China Current.
Table 1.
List of phylogeographic studies that investigated the genetic diversity of NW Pacific marine species and summary of the relevant information reported in each study.
| Species | Organism | Genetic info. | N | Structure/Refugium | NL | % div. | cryptic sp. | References |
|---|---|---|---|---|---|---|---|---|
| Mugil cephalus | Fish | control region | 126 | East China Sea vs South China Sea | 2 | 48% | yes | [14] |
| Mugil cephalus | Fish | control region | 140 | East China Sea vs South China Sea | 2 | 55% | no | [21] |
| Mugil cephalus | Fish | cyto b | 98 | Japan Sea vs East China Sea vs South China Sea | 3 | 5.1-6.6% | no | [17] |
| Mugil cephalus | Fish | COI | 448 | Japan Sea vs South China Sea vs SW Pacific origin | 3 | 2.5-4.8% | yes | This study |
| Mugil cephalus | Fish | 16 S, 12 S | 9 | East China Sea vs South China Sea | 2 | 1.87%, 1.61% | yes | [71] |
| Mugil cephalus | Fish | microsatellites | 713 | Japan Sea vs South China Sea vs SW Pacific origin | 3 | 11.5-30.5% | yes | This study |
| Mugil cephalus | Fish | AFLP | 118 | Bohai Sea vs Yellow Sea vs East China Sea vs South China Sea | 4 | - | no | [23] |
| Chelon haematocheilus | Fish | control region | 272 | Japan Sea vs East China Sea vs South China Sea | 3 | 1.55-2.41% | no | [16] |
| Chelon haematocheilus | Fish | 16 S, 12 S | 8 | East China Sea vs South China Sea | 2 | 2.81%, 4.11% | yes | [71] |
| Odontamblyopus sp. | Fish | ND5 | 85 | East China Sea vs South China Sea | 3 | 2.37-8.96% | yes | [15] |
| Lateolabrax sp. | Fish | control region | 256 | Japan Sea vs East China Sea | 2 | 22.60% | yes | [72] |
| Lateolabrax sp. | Fish | cyto b | 256 | Japan Sea vs East China Sea | 2 | 7.80% | yes | [72] |
| Eriocheir sensu stricto | Crustacean | COII, cyto b | 154 | Japan Sea vs East China Sea vs South China Sea | 3 | 3.2-4.05% | yes | [13] |
| Eriocheir sensu stricto | Crustacean | COI, cyto b | 446 | Japan Sea vs East China Sea vs South China Sea vs Ryukyu | 4 | - | no | [54] |
| Penaeus japonicus | Crustacean | control region | 95 | Japan Sea vs East China Sea vs South China Sea | 3 | FCT = 0.04 | no | [73] |
| Cyclina sinensis | Mollusk | RAPD | 50 | Yellow Sea vs South China Sea | 2 | 10.90% | no | [74] |
| Cyclina sinensis | Mollusk | AFLP | 160 | Yellow Sea vs China Sea | 2 | - | no | [75] |
| Tegillarca granosa | Mollusk | COI | 38 | East China Sea vs South China Sea | 2 | 15% | no | [76] |
| Tegillarca granosa | Mollusk | RAPD | 96 | East China Sea vs South China Sea | 2 | 32.6 -45.1% | no | [77] |
| Helice sp. | Mollusk | COI, 16 S, 12 S, ITS2 | 123 | East China Sea vs South China Sea vs Ryukyu & Taiwan Islands | 3 | 0.16-1.13% | yes | [78] |
N: sample size, NL: number of lineage, % div.: percentage of divergence.
The flathead mullet, Mugil cephalus Linnaeus, 1758, is a euryhaline teleost distributed worldwide in coastal waters, lagoons, bays and estuaries between latitudes 42° N and 42° S [18]. It is subjected to intense and unregulated inshore fisheries in the NW Pacific because of the economic value of its roe. There have been drastic declines in the landings of M. cephalus since 1980 (6 865 metric tons in 1980 versus 159 metric tons in 2008; Taiwan Fisheries Agency), which has been suggested to be the result of overfishing and the impact of global warming [19,20]. Thus, a sound management program is urgently needed, which requires precise knowledge about the taxonomy and population structure of this over-exploited species.
Previous studies have shown that up to three highly divergent mitochondrial lineages exist in sympatry in NW Pacific M. cephalus populations [14,17,21]. This high inter-lineage divergence raises questions about the taxonomic status of M. cephalus in the NW Pacific, but studies have not reached consensus on this issue. The inter-lineage divergence of 48% observed at the mtDNA control region probably indicates a species complex, because such divergence coupled with the existence of different reproductive behaviours (resident versus migratory) demonstrates that these lineages are independent [14]. By contrast, an inter-lineage divergence of 5.3 to 6.7% was estimated using cytochrome b sequences, which would argue for the existence of a single species because the genetic divergence between mugilid species exceeds this by a factor of two [17]. However, because the mtDNA genome used in both studies is maternally inherited, it is not possible to determine whether the presence of divergent mitochondrial lineages in the same sample is a result of secondary contact after an extended period of isolation and/or the presence of two sibling species.
To our knowledge, only two studies on M. cephalus in this region have used biparentally inherited markers [22,23]. The first study used an allozyme locus, GPI-A (glucose-6-phosphate isomerase), which may be able to distinguish migratory from resident M. cephalus populations [22]. More recently, Liu et al. [23] found a high level of genetic structure in M. cephalus using AFLP, and four populations were identified among a set of 6 samples collected along the Chinese coast, with the southern samples (Hainan Island) being the most divergent. This marked genetic structuring in NW Pacific M. cephalus contrasts with findings from the Gulf of Mexico and the northwest Atlantic, which did not reveal any genetic heterogeneity over a similar geographic scale, and found only one mitochondrial lineage [24,25].
Therefore, given all of the above uncertainties, this study investigated the level of both historical and contemporary gene flow among M. cephalus samples collected in the NW Pacific, using a set of ten microsatellites and one mtDNA (COI) locus. The objective was to estimate the level of independence of the different M. cephalus mtDNA lineages, and reveal the existence of cryptic species. This provided an opportunity to address both the evolutionary forces and reproductive boundary uncertainties among and within the mitochondrial lineages. Finally, we aimed to better understand the importance of Pliocene and Pleistocene geologic and climatic events in the diversification of marine species in the NW Pacific.
Methods
Sampling and DNA extraction
A total of 713 sub-adult to adult M. cephalus were collected from 12 locations in the NW Pacific (Table 2, Figure 1). Additional temporal sampling (2005-2008) was conducted at three localities around Taiwan: the coast of Keelung (KL), which is located midway along the M. cephalus spawning migration route from the Eastern China Sea; the offshore waters of Kaohsiung (KS), where spawning occurs; and the Kaoping Estuary (KP), a tropical system located in southwestern Taiwan. Sampling locations, dates of collection, sample sizes for microsatellites analyses, mtDNA lineage (highlighted either by sequencing or multiplex COI haplotype-specific PCR), mean fork length (mm) and gonadosomatic index (GSI) data for the M. cephalus from Taiwan are shown in Table 2. Muscle tissues were preserved in 95% ethanol prior to DNA extraction. Genomic DNA was extracted from muscle tissue using a DNA Purification Kit (Bioman, Taipei, Taiwan), preserved in TE buffer and then quantified and diluted to 1 ng/μl for PCR.
Table 2.
Summary of the collected data and sample sizes of Mugil cephalus used in microsatellite (MS) and mitochondrial (mtDNA) COI gene analyses.
| Country | code | Locations/Habitat | Sampling date | Long. - Lat. | N indiv. | % mtDNA Lineage | Fork length | GSI | GSI mean ± se/ | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| microsat | COI/RS | 1 | 2 | 3 | mean ± se (mm) | mean ± se | mtDNA 1 | mtDNA 2 | mtDNA 3 | |||||
| Taiwan | 05 KL | Keelung/ | Dec. 2005 | 25°11'N 121°47'E | 50 | 50/50 | 92 | 8 | 0 | 464.80 ± 37.89 | 15.67 ± 5.97 | 15.91 ± 5.82 | 12.86 ± 7.94 | - |
| 06 KL | Coastal waters | Dec. 2006 | 48 | 11/48 | 98 | 2 | 0 | 478.77 ± 32.81 | 15.16 ± 7.80 | 15.32 ± 7.81 | 7.49 | - | ||
| 07 KL | Dec. 2007 | 47 | 47/47 | 77 | 19 | 4 | 484.6 ± 68.37 | 10.60 ± 6.45 | 12.31 ± 5.55 | 5.87 ± 6.58 | 1.03 ± 0.27 | |||
| 05 KS | Kaohsiung/ | Dec. 2005 | 22°37'N 119°30'E | 44 | 36/44 | 100 | 0 | 0 | 475.73 ± 34.55 | 20.61 ± 0.68 | 20.61 ± 0.68 | - | - | |
| 06 KS | Offshore waters | Dec. 2006 | 50 | 20/50 | 100 | 0 | 0 | 489.25 ± 25.37 | 19.75 ± 4.23 | 19.75 ± 4.23 | - | - | ||
| 07 KS | Dec. 2007 | 57 | 49/57 | 100 | 0 | 0 | 526.01 ± 38.07 | 18.03 ± 1.44 | 18.03 ± 1.44 | - | - | |||
| 05 KP | Kaoping River/ | Dec. 2005 | 22°28'N 120°25'E | 45 | 36/45 | 0 | 49 | 51 | 289.34 ± 19.58 | 0.81 ± 1.97 | - | 1.01 ± 1.52 | 0.73 ± 2.37 | |
| 07 KP | Estuary | Dec. 2007 | 49 | 39/49 | 14 | 63 | 23 | 501.07 ± 55.12 | 8.30 ± 7.50 | 7.31 ± 5.94 | 6.73 ± 6.19 | 13.37 ± 7.65 | ||
| 08 KP | Jan. 2008 | 38 | 37/38 | 13 | 71 | 16 | 481.69 ± 39.00 | 0.01 ± 0.01 | 0.02 ± 0.03 | 0 | 0.01 ± 0.01 | |||
| Japan | OK | Okinawa | Nov. 2005 | 26°24'N 127°54'E | 48 | 13/48 | 27 | 73 | 0 | - | - | - | - | - |
| YK | Yokosuka | Jan.-Feb. 2005 | 36°16'N 139°41'E | 43 | 8/43 | 35 | 65 | 0 | ||||||
| AS | Ariake Sea | Nov. 2007 | 33°00'N 130°26'E | 21 | 19/21 | 100 | 0 | 0 | - | - | - | - | - | |
| China | HN | Hainan | Apr. 2010 | 18°65'N 110°37'E | 32 | 32/32 | 0 | 41 | 59 | - | - | - | - | - |
| PR | Pearl River | Feb. 2005 | 22°45'N 113°37'E | 30 | 16/30 | 0 | 90 | 10 | ||||||
| ST | Shantou | Jan. 2008 | 23°20'N 116°44'E | 48 | 7/48 | 0 | 91 | 9 | - | - | - | - | - | |
| QD | Qingdao | Feb. 2006 | 36°02'N 120°21'E | 32 | 10/31 | 100 | 0 | 0 | - | - | - | - | - | |
| Philippines | PH | Philippines | Feb. 2007 | 18°21'N 121°37'E | 14 | 14/14 | 0 | 100 | 0 | - | - | - | - | - |
| Russia | RU | Russia | Jun. 2007 | 42°53'N 132°44'E | 4 | 4/4 | 100 | 0 | 0 | - | - | - | - | - |
| Jul.-Aug. 2009 | 13 | 0/13 | 100 | 0 | 0 | - | - | - | - | - | ||||
| Total | 713 | 448/712 | ||||||||||||
RS: rapid PCR screening of different lineages. The ratio of different lineages in each sample, their fork length and Gonadosomatic index (GSI) are also shown.
Mitochondrial DNA analysis
Partial mtDNA COI gene sequences (627 bp) were amplified by polymerase chain reaction (PCR) using the universal primers FishF1 and FishR1 [26]. To identify M. cephalus cytochrome b lineages [17], cytochrome b gene was also sequenced for 20 individuals belonging to three COI lineages using the cytochrome b primers MCglu-1.F: GGCTTGAAAAACCACCGTTG and MCcytbR: AGTACTGTGGCAGAGCTTGG. PCR was performed in a Biometra TGradient Thermocycler with a 15-μL reaction volume containing 0.2 μm dNTPs, 1.5 μL of 10× PCR buffer (Bioman, Taipei, Taiwan), 0.5 μm each of forward and reverse primer, 0.2 U Taq DNA polymerase (Bioman, Taipei, Taiwan), and 1.0 μL of template DNA. MtDNA amplification was carried out using the following PCR conditions: 35 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 15 s and extension at 72°C for 30 s after heating at 94°C for 5 min. The PCR products were electrophoresed on a 1.0% agarose gel (Bioman, Taipei, Taiwan) and stained with ethidium bromide for band characterisation via ultraviolet trans-illumination. All sequencing reactions were performed according to the manufacturer's protocol (Applied Biosystems, Foster City, CA, USA) using the same forward primer as used for PCR.
All COI sequences were automatically aligned using MAFFT version 6 [27] and manually corrected. The characterisation of the genetic variability of the COI gene sequences and the number of nucleotide substitutions, including transition/transversions, were conducted using ARLEQUIN version 3.01 [28]. Phylogenetic trees were reconstructed using the Bayesian, neighbour joining (NJ), maximum likelihood (ML) and maximum parsimony (MP) methods. The Bayesian method was applied using the program MrBayes 3.0 [29], and NJ, ML and MP were applied using the program PAUP 4.0 [30]. The transversion model with invariable sites HKY + I + G (I = 0.097, G = 0.244, Ts/Tv ratio = 5.4465) was selected for construction of the phylogenetic tree using Mugil curema as an outgroup (GenBank accession numbers: EU715464, [31]) and the Akaike information criterion (AIC) in MODELTEST 3.7 [32]. Nodes with bootstrap values ≥ 70% were considered well supported [33].
The fixation index (ΦST) for all pair-wise comparisons among different populations was calculated to investigate genetic diversity among M. cephalus populations, and a permutation test (10 000 permutations) was performed using ARLEQUIN version 3.01 [28]. Population genetic diversity was measured within each of the populations based on the number of distinct haplotypes, gene diversity (h) and mean nucleotide diversity (π), using ARLEQUIN version 3.01 [28].
The divergence time for the M. cephalus COI gene was estimated using Bayesian Evolutionary Analysis Sampling Trees (BEAST) ver. 1.5.2 [34] with 20 million steps in a Monte Carlo Markov Chain (MCMC) simulation (2 million step burn-in time). COI sequences of M. cephalus samples from disjunct locations in the western Atlantic (South Carolina: GenBank accession numbers: HQ149710 and Florida: GenBank accession numbers: HQ149711) and eastern Pacific (Peru: GenBank accession numbers: HQ149714 and Mexico: GenBank accession numbers: HQ149715) were used for the calibration, and assuming that the divergence of these populations corresponds to the closure of the Isthmus of Panama, 2.8 MY ago [35]. A strict molecular clock was assumed for this final run. The effective sample sizes (ESS) of parameters sampled from the MCMC were > 500 (acceptable ESS is >200). The results were viewed using TRACER 1.4 [36]. Historical demographic/spatial expansions of M. cephalus were explored using two different approaches. Tajima's D [37] and Fu's FS [38] tests were used to test population equilibrium. Deviations from the sudden population expansion model were tested using the Harpending's raggedness index RI of mismatch distributions [39].
Rapid screening of the different mtDNA lineages
A multiplex COI haplotype-specific PCR (MHS-PCR) was designed following the recommendations of previous studies [40,41] to develop a rapid screening method capable of detecting different mitochondrial lineages of M. cephalus. Specific mutations of the different M. cephalus mtDNA lineages were identified using 448 COI sequences of M. cephalus produced in this study. NWP1,2F (5' GCTTTTCCCCGAATAAAT 3') was the forward primer for both lineage 1 and lineage 2, while NWP3F (5'TACTGCCCTAAGCCTACTC 3') was the forward primer for lineage 3. NWP1,3R (5' CGATCTGTTAGGAGTATGG 3') was the reverse primer for lineage 1 and lineage 3, while NWP2R (5' CTCATACGAAAAGGGGTGTT 3') was the reverse primer for lineage 2. PCR was performed using a Biometra TGradient Thermocycler with a 15-μL reaction volume containing 0.2 μm of each dNTP, 1.5 μL 10× PCR buffer (Bioman, Taipei, Taiwan), 0.5 μm forward and reverse primers, 0.2 U Taq DNA polymerase (Bioman, Taipei, Taiwan), and 1.0 μL of template DNA. MtDNA amplification was carried out using the following PCR program: 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 15 s and extension at 72°C for 30 s after heating at 94°C for 5 min. The PCR products were electrophoresed in a 1.0% agarose gel (Bioman, Taipei, Taiwan) and stained with ethidium bromide. Lineage 1 had a PCR product of 362 bp, while lineage 2 and lineage 3 had PCR products of 283 bp and 549 bp, respectively (Additional file 1, Figure S1). The rest of the samples that were not sequenced were all rapidly screened to determine their lineages.
Microsatellite analysis
Ten microsatellite loci were used to screen the genetic diversity of the M. cephalus samples [42]. Reverse primers for each locus were labelled with fluorescent dyes (6-FAM, HEX and TAMRA), and multiplex PCR was performed in a 15-μl reaction volume containing 0.1 ng DNA, 1.25 pmole each of the three reverse primers labelled with different fluorescent dyes, 1.25 pmole of each forward primer, 5 mM dNTP, 1.5 mM MgCl2 and 0.5 U of Taq polymerase (Bioman, Taipei, Taiwan). Amplification was conducted as follows: initial denaturation at 95°C for 4 min, followed by 35 cycles at 94°C, 54-58°C and 72°C for 30 sec each. Locus polymorphisms were screened using an ABI PRISM 377 auto DNA sequencer (Applied Biosystems, Foster City, California, USA). Lengths of microsatellite alleles were determined using a TAMRA-labelled 100 bp standard (Perkin-Elmer, Waltham, Massachusetts, USA).
The calculation of the number of alleles, estimated (HE) and observed heterozygosities (HO) for each locality and for all samples, the genetic differentiation index (θ an estimator of f analogues of Wright's fixation indices [43]) among samples, locus-by-locus AMOVA and the exact test for deviations from Hardy-Weinberg equilibrium at every locus for each locality were performed using ARLEQUIN version 3.01 [28] and GENETIX software [44]. The significance level for multiple simultaneous comparisons was adjusted using the sequential Bonferroni technique [45]. The allelic richness of the minimum population size by a rarefaction method and Fis was estimated with FSTAT 2.9.3 [46]. The significance of the differences in allelic richness was tested by the Wilcoxon signed-rank test for paired observations (see [47] for power analysis). The program GENEPOP version 3.1 [48] was used to test for linkage disequilibrium among the ten loci analysed in this study (10 000 permutations, 1 000 dememorisation steps).
The presence of intraspecific genetic structure was tested using the model-based clustering method [49], as implemented in STRUCTURE VER. 2.1. For each value of K, which is the number of genetically distinct populations, the Markov chain Monte Carlo scheme was run with a burn-in period of 10 000 steps and a chain length of 100 000 replicates following the non-admixture model. Twenty runs were performed to evaluate the reliability of the results, with the number of populations being determined from posterior probabilities of K calculated as K = {1 ~ 6}. The K values could be incorrectly estimated if the migration rates between populations are not equal, so the values of ΔK were also calculated for each value of K [50]. Individuals were regarded as correctly assigned to a population when their q-value (i.e. the posterior probability) was at least 80% after subtracting the posterior probability assignment of another population [47].
Results
Mitochondrial DNA
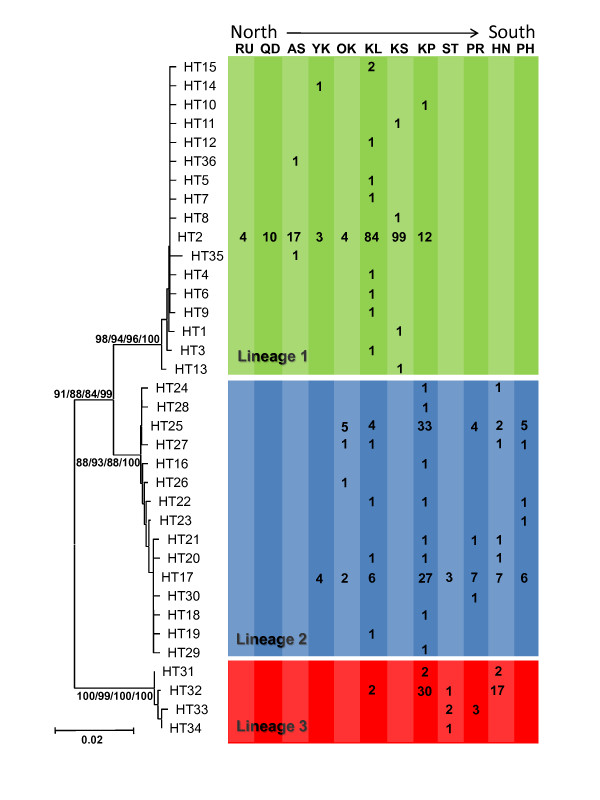
A total of 627 nucleotides of the COI gene were obtained from 448 individuals. In these sequences, 54 positions were variable (Additional file 2, Table S1), defining 36 haplotypes (Genbank accession numbers GU260664-GU260697, HQ149082-HQ149083). Phylogenetic trees reconstructed using either COI (Figure 2) or cytochrome b (Additional file 3, Figure S2) sequences [17] all supported the existence of three highly supported monophyletic lineages in NW Pacific M. cephalus. Seventeen COI haplotypes were found in lineage 1, including 15 nucleotide transitions and three nucleotide transversions. Of these, HT2 was the most frequent haplotype, being shared by the samples from Taiwan, Japan, Qingdao and Russia, and it was connected to the other 16 haplotypes (HT1-HT15, HT35-HT36) by one to two steps in a "star-like" haplotype network (Figure 3a). The other 16 COI haplotypes were minor and mainly found in Keelung and Kaohsiung. Fifteen COI haplotypes were found in lineage 2, including 11 nucleotide transitions and two nucleotide transversions. Two major haplotypes (HT17 and HT25) were shared by samples from Keelung, the Kaoping River, Okinawa, Yokosuka, the Pearl River, Shantou and the Philippines. These two main haplotypes had only two nucleotide differences and were mainly found at the Kaoping River site. Lineage 3 consisted of only four COI haplotypes, with the main haplotype (HT32) occurring at the Kaoping River and Hainan sites and in a few samples from Shantou and Keelung. The fixation index (ΦST) showed significant differentiation among the three lineages (ΦST = 0.973, P < 0.001). The pair-wise ΦST values were 0.960 (P < 0.001) between lineage 1 and lineage 2; 0.956 (P < 0.001) between lineage 2 and lineage 3 and 0.994 (P < 0.001) between lineage 1 and lineage 3.
Figure 2.
Phylogenetic trees of Mugil cephalus in the northwestern Pacific. Phylogenetic trees reconstructed from (A) 36 COI sequences haplotypes (HT1-HT36) of Mugil cephalus in the northwestern Pacific and its geographic distribution (for location names follow Table 1). The values above the branches are bootstrap values for the NJ, ML and MP analyses and the posterior probabilities for the Bayesian analysis.
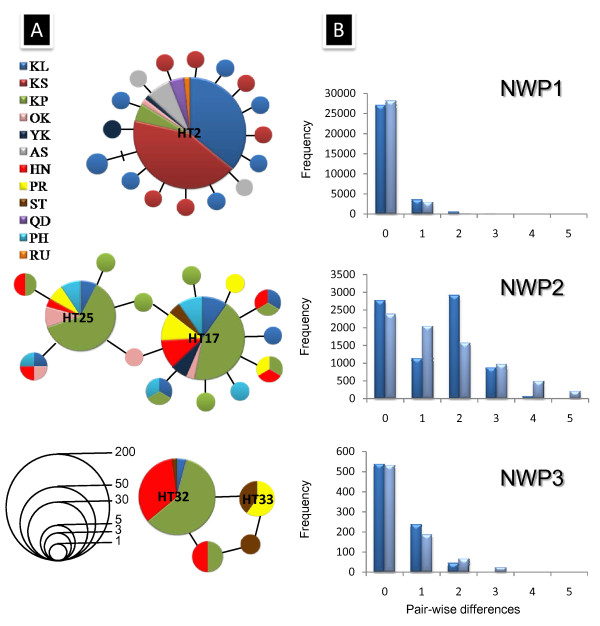
Figure 3.
Haplotype networks and mismatch distribution for three lineages of Mugil cephalus COI sequences. (A) Haplotype networks for three lineages of Mugil cephalus COI sequences. The lengths of the connecting lines are in relation to the number of mutations between haplotypes. Each circle represents a haplotype, with the diameter of the circle proportional to the number of sequences of that haplotype. The names of the major haplotypes for each cryptic species correspond to those listed in Table S1. (B) Mismatch distributions from the mtDNA COI sequences of M. cephalus from the ten sampling locations. Blue bar: observed distributions; Light blue bar: expected distributions from the sudden expansion model.
Fifteen variable sites were found within the amplified 627-bp COI fragment (2.5% of sites) between lineage 1 and lineage 2, with approximately 6.7% of the inferred changes being transversions. While 24 and 30 variable sites were found between lineages 2 and 3 (3.8%) and between lineages 1 and 3 (4.8%), respectively, with approximately 21% and 13% of the inferred changes being transversions. The calibrated mean mutation rate of the COI gene in M. cephalus was 1.6% mutations/site/MY. Therefore, the calibrated divergence time using BEAST was 1.607 MY (HPD 95% confidence interval were 2.383-0.892 MY, ESS = 1475) between lineages 1 and 2 and 4.200 MY (HPD 95% confidence interval were 5.708-2.801 MY, ESS = 876) between lineage 3 and the ancestor of lineages 1 and 2. BEAST also suggested that lineage 3 is much older than the others, even compared to M. cephalus populations present in the eastern Pacific and western Atlantic, and does not share the most recent common ancestor of lineages 1 and 2.
The mismatch distributions of lineage 1 and 3 were unimodal, with almost no pair-wise differences between them (Figure 3b). Lineage 3 exhibited a more uneven mismatch distribution compared to lineage 1. By contrast, the mismatch distribution of lineage 2 was bimodal (Figure 3b), with peaks at zero and two substitutions. RIs were not significant for any lineage. On the other hand, both Tajima's D and Fu's FS were negative and highly significant for lineage 1. For lineage 2, Tajima's D values were all negative, but only significant for the control region, while Fu's FS were all negative and significant for all genes. Finally, both Tajima's D and Fu's FS were negative, but not significant for the lineage 3 for any gene considered (Table 3).
Table 3.
Summary of the genetic diversity of Mugil cephalus.
| Species | Marker | n | Na/H | h/HO +/- se | π +/- se | RI | Tajima's D | Fu's Fs |
|---|---|---|---|---|---|---|---|---|
| NWP1 | CR1 | 79 | 79 | 1.0000 +/- 0.0020 | 0.0095 +/- 0.0050 | 0.008 | -2.793*** | -24.76*** |
| Cytob2 | 35 | 6 | 0.2689 +/- 0.0982 | 0.0003 +/- 0.0003 | 0.285 | -2.007** | -5.866*** | |
| COI | 250 | 17 | 0.1316 +/- 0.0295 | 0.0002 +/- 0.0004 | 0.581 | -2.439*** | -3.4 × 1038*** | |
| Microsatellite | 390 | 17.2 | 0.6097 +/- 0.2751 | - | - | - | - | |
| NWP2 | CR1 | 47 | 47 | 1.0000 +/- 0.0040 | 0.0364 +/- 0.0180 | 0.003 | -1.962* | -23.263*** |
| Cytob2 | 43 | 24 | 0.9513 +/- 0.0176 | 0.0030 +/- 0.0017 | 0.028 | -1.68 | -16.53*** | |
| COI | 138 | 15 | 0.6525 +/- 0.0264 | 0.0021 +/- 0.0015 | 0.149 | -1.169 | -7.212** | |
| Microsatellite | 255 | 13.2 | 0.5745 +/- 0.2702 | - | - | - | - | |
| NWP3 | CR1 | 0 | - | - | - | - | - | - |
| Cytob2 | 17 | 11 | 0.9044 +/- 0.0572 | 0.0052 +/- 0.0030 | 0.08 | -0.783 | -1.875 | |
| COI | 60 | 4 | 0.2989 +/0.0737 | 0.0006 +/- 0.0006 | 0.245 | -0.361 | -1.617 | |
| Microsatellite | 68 | 5.8 | 0.3310 +/- 0.2991 | - | - | - | - | |
Number of individuals (n), number of haplotype (H) or alleles (Na), haplotype diversity (h ± standard deviation) or observed Heterozygosity (HO ± standard deviation), nucleotide diversity (π ± standard deviation) for each group of samples. Tajima's D and Fu's Fs, the corresponding P value, and the raggedness index (RI) of these lineages were also indicated.
1 data from [14], 2 data from [17]. *P < 0.05, ** P < 0.01, ***P < 0.001
Microsatellites
A total of 180 alleles with an average of 18.0 alleles per locus were observed across the ten microsatellite loci (Additional file 4, Table S2). All loci were polymorphic, except for locus Mce-8, which was monomorphic in some of the samples. Twenty-one of 180 sample-locus combinations deviated significantly from Hardy-Weinberg proportions after Bonferroni correction. However, the deviations of these loci were not significant in most locations (including Kaoping and Hainan) when individuals were sorted according to one of the three mtDNA lineages, suggesting a Wahlund effect, which is the reduction of heterozygosity in a sample due to the mixture of genetically differentiated populations. No linkage disequilibrium was detected within the M. cephalus populations (P > 0.05).
AMOVA indicated that the genetic differentiation of M. cephalus among 18 spatial and temporal samples was highly significant (θ = 0.060, P < 0.001). A locus-by-locus AMOVA indicated that all loci could detect population differentiation among the samples. However, no temporal variation was observed in allelic frequencies among the KS, KL and KP locations where the temporal samples were obtained. During three to four years of sampling, no genetic heterogeneity was observed among the temporal samples collected at the same location, except in KP, and the genetic differentiation between locations remained stable (Table 4).
Table 4.
Temporal pair-wise θ test, using microsatellite loci between different sampling locations and years in Taiwan.
| KL | KS | KP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Year | (n) | 2005 | 2006 | 2007 | 2005 | 2006 | 2007 | 2005 | 2007 | 2008 |
| KL | 2005 | 50 | - | 0.000 | -0.003 | 0.003 | 0.004 | 0.001 | 0.131*** | 0.068*** | 0.061*** |
| 2006 | 48 | - | 0.004 | -0.001 | 0.002 | 0.000 | 0.150*** | 0.087*** | 0.079*** | ||
| 2007 | 47 | - | 0.003 | 0.007* | 0.007** | 0.116*** | 0.055*** | 0.047*** | |||
| KS | 2005 | 44 | - | 0.001 | 0.000 | 0.157*** | 0.090*** | 0.080*** | |||
| 2006 | 50 | - | 0.001 | 0.154*** | 0.088*** | 0.085*** | |||||
| 2007 | 57 | - | 0.165*** | 0.099*** | 0.091*** | ||||||
| KP | 2005 | 45 | - | 0.021* | 0.041** | ||||||
| 2007 | 49 | - | 0.003 | ||||||||
| 2008 | 38 | - | |||||||||
KL:Keelung; KS:Kaohsiung; KP:Kaoping River.
* < 0.05, ** < 0.01, *** < 0.001 in bold significant values after bonferroni correction [45]
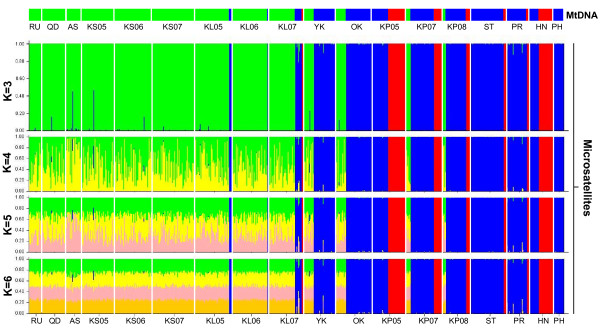
The results from the STRUCTURE assignment test supported three clusters based both on the log probability of the data [L(K)] and the statistic ΔK, as described in [49,50], respectively (Additional file 5, Table S3). With K = 3, almost all individuals were clearly assigned to one of the three clusters (Figure 4). The posterior probabilities corresponding to the assignment of individuals to cluster 3 (red bar, hereafter denoted as NWP3) were all unambiguous, while the assignments to one of the two other clusters (green bar for NWP1 and blue bar for NWP2) included several ambiguous individuals (7 over 645 individuals belonging either to NWP1 and NWP2). The individuals from Kaohsiung, Qingdao, the Ariake Sea and Russia were mostly assigned to NWP1; the individuals from the Philippines were all assigned to NWP2, while at least two clusters coexist in Keelung and the Kaoping River of Taiwan (NWP1, NWP2 and NWP3), Hainan, the Pearl River and Shantou of China (NWP2 and NWP3) and Okinawa and Yokosuka of Japan (NWP1 and NWP2) (Figure 5a). According to their occurrence areas, NWP1 is mainly observed in the north of our sampling area (Figure 5b, North China Seas and Japan), whereas the NWP3 is mostly found at southern stations (Figure 5d, South China Sea). NWP2 has the widest range; it is present from the north to the south of the sampling area (Figure 5c).
Figure 4.
Correspondence between mtDNA lineages and nuclear Structure clusters. First row is the distribution of the 3 mtDNA lineages (green: lineage 1, blue: lineage 2, red: lineage 3) in 18 samples of Mugil cephalus in NW pacific. Second to forth row are the results of the assignment test using STRUCTURE [49] for M. cephalus microsatellite data. Each cluster (K) is designated by a different colour with vertical bars representing individuals and the proportion of a bar assigned to a single colour representing the posterior probability that an individual is assigned to that cluster. Assignment results are shown with K = 3, 4, 5 and 6.
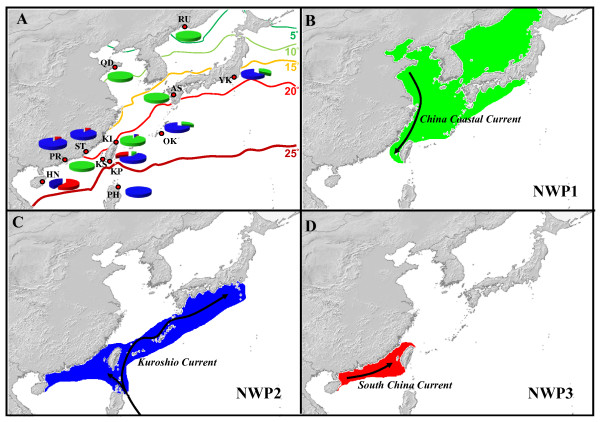
Figure 5.
The distribution of three cryptic species of Mugil cephalus in the northwestern Pacific. (A) the proportions of the three species in various locations; coloured lines indicate sea surface temperature in the NW Pacific in December, which is the period of reproductive migration of Mugil cephalus; (B) the distribution range of NWP1 and location of the China Coastal Current; (C) the distribution range of NWP2 and location of the Kuroshio Current; (D) the distribution range of NWP3 and location of the South China Sea Current.
Genetic variability in mtDNA and microsatellites
There was congruence between the two molecular markers when the results of the assignment test were compared to the COI sequence data (Figure 4). The results indicated that 385 of 390 individuals assigned to NWP1 harboured mtDNA representing lineage 1, 253 of 255 individuals assigned to NWP2 harboured mtDNA of lineage 2, and all 68 individuals assigned to NWP3 harboured lineage 3 mtDNA. This resulted in a successful assignment rate of 99.0%. Although there were seven ambiguous individuals in NWP1 and NWP2 below our assignment criteria, the mean q-values of assignment test for these two clusters were still over 0.99 (Additional file 6, Table S4).
In each location sampled, individuals belonging to the same mtDNA lineages were grouped together to further investigate the genetic structure of the three Operational Taxonomic Units (OTUs). Genetic variability analysis showed that only two of the 30 combinations, loci Mce-3 and Mce-8 of NWP3, deviated significantly from Hardy-Weinberg proportions after Bonferroni correction (Additional file 7, Table S5). The allelic richness per locus based on the minimum sample size (68 individuals of NWP3) was significantly lower at NWP3 than NWP2 (P < 0.001) and NWP1 (P < 0.001) and slightly lower at NWP2 than NWP1 (P < 0.05) for the ten microsatellite loci (HO was lower at NWP3 than the other two clusters). The genotypes at Mce-14 of NWP2 and Mce-10 of NWP3 were all homozygous.
Population genetic differentiation among these three clusters was highly significant (θ = 0.179, P < 0.001). Pair-wise θ comparison indicated that NWP3 was the most differentiated (θ NWP3-NWP1 = 0.296, P < 0.001; θ NWP3-NWP2 = 0.305, P < 0.001), while level of differentiation was lower but still highly significant between NWP1 and NWP2 (θ = 0.115, P < 0.001). In addition, no genetic heterogeneity was detected among samples belonging to any one of the three OTUs, with the exception of the Ariake Sea, which had some genetic heterogeneity among the samples of NWP1 (Table 5).
Table 5.
Pair-wise θ test, using microsatellite loci between different sampling locations under the same or different mtDNA lineages.
| Lineage 1 (L1) | Lineage 2 (L2) | Lineage 3 (L3) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locations | (n) | QD | AS | KL | KS | KP | YK | OK | KL | KP | YK | OK | ST | PR | HN | PH | KL | KP | ST | PR | HN | |
| RU | 17 | -0.002 | 0.008 | 0.003 | 0.001 | -0.003 | -0.001 | -0.002 | 0.096*** | 0.121*** | 0.113*** | 0.099*** | 0.122*** | 0.102*** | 0.130*** | 0.107*** | 0.268*** | 0.390*** | 0.311*** | 0.258*** | 0.279*** | |
| QD | 32 | 0.011* | 0.004 | 0.002 | -0.002 | -0.011 | 0.007 | 0.103*** | 0.116*** | 0.117*** | 0.101*** | 0.117*** | 0.102*** | 0.133*** | 0.109*** | 0.271*** | 0.372*** | 0.310*** | 0.261*** | 0.268*** | ||
| AS | 21 | 0.011** | 0.016*** | 0.008 | 0.000 | -0.003 | 0.086*** | 0.102*** | 0.098*** | 0.087*** | 0.100*** | 0.087*** | 0.112*** | 0.090*** | 0.247*** | 0.367*** | 0.290*** | 0.239*** | 0.255*** | |||
| L1 | KL | 129 | 0.001 | -0.006 | -0.003 | -0.001 | 0.107*** | 0.120*** | 0.121*** | 0.103*** | 0.119*** | 0.100*** | 0.133*** | 0.114*** | 0.261*** | 0.322*** | 0.296*** | 0.246*** | 0.248*** | |||
| KS | 151 | -0.002 | -0.003 | 0.001 | 0.112*** | 0.124*** | 0.124*** | 0.107*** | 0.122*** | 0.106*** | 0.138*** | 0.119*** | 0.273*** | 0.330*** | 0.309*** | 0.261*** | 0.257*** | |||||
| KP | 12 | -0.010 | 0.002 | 0.108*** | 0.121*** | 0.125*** | 0.103*** | 0.124*** | 0.101*** | 0.139*** | 0.111*** | 0.270*** | 0.399*** | 0.317*** | 0.258*** | 0.275*** | ||||||
| YK | 14 | 0.002 | 0.108*** | 0.115*** | 0.117*** | 0.100*** | 0.114*** | 0.100*** | 0.133*** | 0.108*** | 0.269*** | 0.395*** | 0.315*** | 0.258*** | 0.274*** | |||||||
| OK | 14 | 0.098*** | 0.125*** | 0.122*** | 0.100*** | 0.123*** | 0.105*** | 0.137*** | 0.109*** | 0.265*** | 0.395*** | 0.313*** | 0.253*** | 0.276*** | ||||||||
| KL | 14 | -0.001 | -0.002 | -0.004 | 0.004 | 0.005 | 0.005 | -0.013 | 0.274*** | 0.403*** | 0.324*** | 0.268*** | 0.290*** | |||||||||
| KP | 80 | 0.000 | 0.000 | -0.002 | 0.001 | 0.009 | 0.002 | 0.273*** | 0.342*** | 0.313*** | 0.268*** | 0.261*** | ||||||||||
| YK | 29 | 0.000 | 0.000 | 0.002 | -0.002 | 0.002 | 0.286*** | 0.381*** | 0.329*** | 0.282*** | 0.284*** | |||||||||||
| L2 | OK | 34 | -0.002 | -0.004 | 0.000 | -0.004 | 0.264*** | 0.365*** | 0.312*** | 0.263*** | 0.267*** | |||||||||||
| ST | 44 | -0.005 | -0.002 | 0.001 | 0.280*** | 0.355*** | 0.316*** | 0.269*** | 0.264*** | |||||||||||||
| PR | 27 | -0.002 | -0.002 | 0.255*** | 0.355*** | 0.299*** | 0.246*** | 0.253*** | ||||||||||||||
| HN | 13 | -0.008 | 0.289*** | 0.390*** | 0.327*** | 0.267*** | 0.282*** | |||||||||||||||
| PH | 14 | 0.251*** | 0.379*** | 0.303*** | 0.242*** | 0.267*** | ||||||||||||||||
| KL | 2 | 0.026 | -0.049 | -0.095 | 0.006 | |||||||||||||||||
| KP | 40 | -0.026 | -0.026 | 0.019* | ||||||||||||||||||
| L3 | ST | 4 | -0.118 | -0.003 | ||||||||||||||||||
| PR | 3 | -0.017 | ||||||||||||||||||||
| HN | 19 | |||||||||||||||||||||
See Table 1 for the complete name of locations.
* < 0.05, ** < 0.01, *** < 0.001 in bold significant values after bonferroni correction [45]
Discussion
Historical demography and phylogeography
Since the Plio-Pleistocene, successive periods of sea-level regression have directly impacted the species connectivity of the marginal seas of the NW Pacific. This has clearly been demonstrated by previous phylogeographic studies (Table 1), which have identified Pleistocene refugia in all marginal seas. Our results argue for the existence of three mtDNA lineages of M. cephalus in the NW Pacific but only two, NWP1 and NWP2, are of Pleistocene origin. The NWP3 lineage diverged from the NWP1 and NWP2 common ancestor during the Miocene or the Pliocene epoch, and we assume that the origin of NWP3 does not relate to geological events in the NW Pacific because this lineage is also observed in the southwestern (SW) Pacific (in Fiji and New Caledonia Islands; Durand, pers. com.). While it appears likely that the refugium of the NWP2 lineage was located in the South China Sea, the location of the NWP1 refugium is more difficult to assess. The closure of the Tsushima Straits (between Korea and Japan, Figure 1), which isolated the Japan Sea, may have been the powerful vicariant event that was at the origin of the NWP1 lineage. Reconstruction of the history of the Tsushima Current [9] demonstrated that during the interval between 3.5-1.7 MY, the current only flowed into the Sea of Japan periodically and the volume and salinity of water was lower than at present. After 1.7 MY, the Tsushima Current flowed throughout almost all interglacial periods and, more importantly, with a volume and water salinity similar to the present day. This date matches the calibrated dates of divergence estimated between NWP1 and NWP2, suggesting that the NWP1 ancestor was not able to enter in the Japan Sea before 1.7 MY. During each glacial episode, ancestors of the NWP1 lineage that were trapped in the Japan Sea probably experienced several demographic crashes due to decreases in surface water temperatures [51-53]. This hypothesis is consistent with the observed genetic diversity and Tajima's D and Fu's FS indices, which indicate bottleneck events in NWP1 (Table 3). Climatic oscillations during the Pleistocene epoch were probably less marked in more southerly latitudes, where the influence of the Kuroshio Current was still exerted, which would explain why NWP2 presents a higher genetic diversity than NWP1. Similar trends can be observed in the genetic diversity pattern of other NW Pacific species, such as Chelon haematocheilus and Odontamblyopus lacepedii sensu lato, suggesting that this scenario may have affected all species inhabiting the area [15,16].
Lastly, the East China Sea, which was exposed during the Pleistocene glaciations, seems to be a post-glacial contact zone for M. cephalus. While the Yellow Sea was recolonised by NWP1 from a northern route of dispersion through the Tsushima Strait, the Ryukyu Islands and East Japan were recolonised by both NWP1 and NWP2 from southern routes that followed the Kuroshio Current (but see below). If the East China Sea experienced a post-glacial colonisation wave from the Japan Sea, it appears that in species such as Chelon haematocheilus and Eriocheir sensu stricto, older colonisation waves were also possible, which allowed the emergence of a specific East China Sea lineage in these species [16,54].
Cryptic M. cephalus species in the NW Pacific
It has previously been hypothesised that M. cephalus is a species complex rather than a unique panmictic unit. To date, however, this has not been proven, due to inadequate sampling schemes with samples separated by thousands of km, or the use of only maternally inherited markers [14,17,25,31,55-57]. The present study demonstrates that M. cephalus is indeed composed of at least three genetically divergent species in the NW Pacific. While COI inter-lineage divergence in M. cephalus exceeds intra-lineage divergence (0.4 to 0.2%) by a ratio greater than 10, which is an indicator of cryptic species according to the 10× rule [58], the nuclear bi-parentally inherited markers provide the most definitive evidence of cryptic species. The assignment tests strongly supported the existence of three groups where 99% of M. cephalus were significantly assigned (posterior probabilities <10% and >90%). Furthermore, the complete congruence between nuclear groups and mitochondrial lineages suggests that lineage sorting of ancestral polymorphisms has been completed. Lastly, further to the argument that the M. cephalus mitochondrial lineages are true species, the gene flow estimated with microsatellite loci appeared to be more limited among sympatric individuals belonging to different mtDNA lineages than between geographically remote individuals sharing the same mtDNA lineage. These results reveal the marked genetic isolation of the three M. cephalus species in the NW Pacific.
If, as suggested earlier, the genetic differentiation of these cryptic species is related to past physical barriers to gene flow, the absence of genetic introgression suggests active reproductive isolation where they coexist sympatrically. Previous studies have reported various different reproductive behaviours in M. cephalus in the NW Pacific. While some individuals migrate over large geographic scales, others appear to be year-round residents of estuaries [59-64]. Migration was observed over the winter solstice (middle of December-early January), individuals of of 3-4 years of age (FL = 43-55 cm) migrate with the cold North China Coastal Current from the Eastern China Sea to south of the Taiwan Strait to spawn offshore of Kaohsiung, Taiwan [59-61]. The newly hatched larvae drift with the coastal currents and then recruit into the estuaries of the western coast of Taiwan at one to two months post-hatching [59,62-64]. The presence of M. cephalus juveniles in Taiwanese estuaries earlier in the year (end of autumn) was interpreted, therefore, as evidence of the existence of a resident M. cephalus population [22].
In this study, all individuals sampled from the supposed spawning ground of migrant M. cephalus (offshore waters of Kaohsiung) in the spawning season (end of December), over three successive years, belonged to NWP1. The high GSI of these individuals suggested that they were spawning or would spawn soon. By contrast, M. cephalus collected in estuarine or coastal waters in the same period belonged to NWP1, NWP2 and NWP3, but all exhibited low GSI (Table 2). This suggests that genetic isolation among M. cephalus species is maintained by spatial and temporal differences in spawning migration patterns.
Population structure and contemporary gene flow
No nuclear genetic heterogeneity was recorded within the cryptic M. cephalus species among the locations sampled within the NW Pacific. This findings contrast with those of Liu et al. [23], who documented up to four regional groups of M. cephalus along the Chinese coast. However, because we analysed samples from the same locations (Hainan, Shantou and Qingdao, which is a station close to Lianyungang), it is evident that the population structure revealed using AFLP [23] relied mostly on the abundant difference of the three cryptic species. Whereas the Hainan, Shantou and Qingdao samples were characterised by high abundances of NWP3, NWP2 and NWP1, respectively, Liu et al. [23] described them as belonging to 3 populations: IV, II and III, respectively. If there was no experimental artefact, according to Liu et al. [23], a northern population of NWP2 is expected to be isolated in the Bohai Sea (population I), as this population is genetically close to population II.
Finally, if all cryptic M. cephalus species revealed in this study are able to disperse and migrate at sea, low genetic structuring is not surprising because large oceanographic current systems in the NW Pacific are expected to greatly facilitate larval dispersal over considerable distances. However, dynamic oceanographic systems can also profoundly restrict connectivity among groups, genetic heterogeneity is generally increased in marine zones with predictable oceanographic fronts [65-67].
The distributional range of the three NW Pacific M. cephalus species appears mainly to be shaped by three major oceanographic current systems, namely the South China, North China Coastal and Kuroshio Currents. The range of the NWP2 species appears to match the circulation pattern of the Kuroshio Current, which brings warm tropical water northeastward from the Luzon Straits, past both the west and east coasts of Taiwan and, finally, to Japan, where a branch of the current (the Tsushima Current) enters the Japanese Sea [68,69]. As the world's second-largest ocean current, the Kuroshio has a major effect on other current systems in the area. It, however, mainly remains offshore, whereas the other currents are located on the Taiwanese and North China shelves, and are more coastal. Thus, the South China Current, which flows northward from the South China Sea to the Taiwan Strait, brings warm water which mixes with cold water from the North China Coastal Current flowing southward from the Yellow Sea and East China Sea. The NWP3 distribution range seems to follow the warm water of the South China Current, while the NWP1 species appears to be restricted to the cold waters of the North China Coastal Current, which flows southward (with the northeastern monsoon) during the winter. Such close relationships between oceanographic current systems and fish species distributions presumably reflect temperature preferences, because differences between the Kuroshio Current and the China Coastal Current can range from 6 to 11°C ([70], Figure 5a). While the Kuroshio Current remains warm throughout the year (23-26°C), the China Coastal Current exhibits a wide variation in temperature, from 12°C in March to 20°C in September. In this context, NWP1 may provide a particularly good indicator species for global warming. NWP1 individuals are assumed to spawn at the front (19-21°C) between the cold southward flowing North China Coastal Current and a warm branch of the northward flowing Kuroshio Current, with the front shifting according to the strength of the North China Coastal Current [20,59,60].
Conclusions
This study successfully identified three cryptic M. cephalus species in the NW Pacific Ocean, using both microsatellites and mitochondrial genetic markers. The genetic architecture and current distribution ranges of these species suggest a complex interaction of contemporary and evolutionary factors. Allopatric and adaptive speciation probably occurred during the Pliocene and Pleistocene epochs, when the Sea of Japan was periodically isolated and influenced by cold northern oceanographic currents. After the last glacial maxima, species range expansions were probably facilitated by changes in the oceanographic current structure and the species' temperature preferences. Currently, these species overlap in their distribution ranges, and it is likely that the genetic integrity of each is maintained by temporal and spatial isolation during the spawning period. This study emphasizes the importance of historical separations of marginal seas, in conjunction with fluctuating temperatures, in creating aquatic biodiversity in the NW Pacific. Isolation and demographic fluctuations are expected to constitute a powerful evolutionary force that increases speciation processes in the marine environment. These regional results also shed new light on the potential genetic diversity of M. cephalus worldwide.
Authors' contributions
KNS carried out the molecular genetic studies, performed the statistical analysis and drafted the manuscript. BWJ and CCH participated in the collection of samples and biological data. WNT participated in the design of the study and obtained funding. JDD conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors have read and approved the final version of the manuscript.
Supplementary Material
Table S1. Variable positions in the 627 bp mitochondrial COI gene segment of Mugil cephalus from 12 locations in the northwestern Pacific. Dots represent identical nucleotides relative to haplotype 1. Frequency of each haplotypes for each lineages (NWP1, NWP2 and NWP3) are also shown. Different color means the locations of the lineage specific nucleotides.
Table S2. Genetic variability at ten microsatellite loci of Mugil cephalus collected in the northwestern Pacific for spatial and temporal genetic structure test. Table-wide significance levels were applied using the sequential Bonferroni technique [45].
Table S3. Log probability and ΔK [50] for each number of clusters in the Bayesian assignment test as implemented in STRUCTURE [49].
Table S4. The mean q-values and standard deviations (sd) for assignment test of 3 Mugil cephalus cryptic species (NWP1, NWP2 and NWP3) as implemented in STRUCTURE [49].
Table S5. Genetic variability at ten microsatellite loci of Mugil cephalus among 3 cryptic species. Table-wide significance levels were applied using the sequential Bonferroni technique [45].
Figure S1. Ethidium bromide stained 2% agarose gel showing the multiplex COI haplotype-specific PCR (MHS-PCR) for the rapid screening of three Mugil cephalus cryptic species in the NW Pacific. 1-13: unidentified individuals belonging either to lineage NWP1 (2), NWP2 (3, 8, 12, 13) or NWP3 (1, 4-7, 9-11). M: 100-bp DNA ladder.
Figure S2. Phylogenetic relationships within Mugil cephalus recovered from 1140 bp of the cytochrome b sequences according to the neighbour-joining tree using Kimura 2 parameter distance. Leaves of the tree correspond to haplotypes of M. cephalus observed by Ke et al. [17] (H1-H43) and this study (H44-H55, in bold). The values above the branches are bootstrap support (500 replicates). Bootstrap supports higher than 50% are displayed.
Contributor Information
Kang-Ning Shen, Email: shenkangning@yahoo.com.tw.
Brian Wade Jamandre, Email: bwjamandre@gmail.com.
Chih-Chieh Hsu, Email: d98b45005@ntu.edu.tw.
Wann-Nian Tzeng, Email: r96b45029@ntu.edu.tw.
Jean-Dominique Durand, Email: Jean-Dominique.Durand@ird.fr.
Acknowledgements
This study could not have been carried out without the generosity in collecting specimens of those listed below: J.X. Liu, X.P. Nie, X.C. Liu, H.P. Chen and X. Qi from China; S. Chow and H. Imai from Japan; M.P. Garcia from the Philippines; Y. Kartavtsev from Russia; K.T. Shao and C.W. Chang from Taiwan. This study was financially supported by the National Science Council of the Republic of China (NSC 98-2313-B-002-016-MY3). The authors would like to thank Z.C. Lin for microsatellite genotyping and DNA sequencing, O. Sadio for statistical advice and A.K. Whitfield, F. Lecomte, N. Leander and D. McKenzie for their comments on the manuscript.
References
- Sala-Bozano M, Ketmaier V, Mariani S. Contrasting signals from multiple markers illuminate population connectivity in a marine species. Mol Ecol. 2009;18:4811–4826. doi: 10.1111/j.1365-294X.2009.04404.x. [DOI] [PubMed] [Google Scholar]
- Kelly RP, Palumbi SR. Genetic structure among 50 species of the Northeastern Pacific intertidal community. PLoS One. 2010;5:e8594. doi: 10.1371/journal.pone.0008594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko PB, Hoffman JM, Emme SA, McGovern TM, Keever CC, Cox LN. The 'Expansion-Contraction' model of pleistocene biogeography: rocky shores suffer a sea change? Mol Ecol. 2010;19:146–169. doi: 10.1111/j.1365-294X.2009.04417.x. [DOI] [PubMed] [Google Scholar]
- Wilson AB. Genetic signature of recent glaciation on populations of a near-shore marine fish species (Syngnathus leptorhyncus) Mol Ecol. 2006;15:1857–1871. doi: 10.1111/j.1365-294X.2006.02911.x. [DOI] [PubMed] [Google Scholar]
- Haig JA, Connolly RM, Hughes JM. Little shrimp left on the shelf: the roles that sea-level change, ocean currents and continental shelf width play in the genetic connectivity of a seagrass-associated species. J Biogeog. 2010;37:1570–1583. [Google Scholar]
- Kaiser TS, Neumann D, Heckel DG, Berendonk TU. Strong genetic differentiation and postglacial origin of populations in the marine midge Clunio marinus (Chironomidae, Diptera) Mol Ecol. 2010;19:2845–2857. doi: 10.1111/j.1365-294X.2010.04706.x. [DOI] [PubMed] [Google Scholar]
- Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Väinölä R, Viard F, Wares J. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology. 2008;89(Suppl 11):S108–S122. doi: 10.1890/08-0257.1. [DOI] [PubMed] [Google Scholar]
- Wang P. Response of Western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol. 1999;156:5–39. doi: 10.1016/S0025-3227(98)00172-8. [DOI] [Google Scholar]
- Kitamura A, Kimoto K. History of the inflow of the warm Tsushima Current into the Sea of Japan between 3.5 and 0.8 Ma. Paleogeogr Palaeocl Palaeoecol. 2006;236:355–366. doi: 10.1016/j.palaeo.2005.11.015. [DOI] [Google Scholar]
- McManus JW. Marine speciation, tectonics and sea-level changes in southeast Asia. Proceedings of the Fifth International Coral Reef Congress: 27 May-1 June 1985; Tahiti. pp. 133–138.
- Benzie JAH. In: Biogeography and Geological Evolution. Hall R, Holloway JD, editor. Leiden, The Netherlands: Backhuys Publishers; 1998. Genetic structure of marine organisms and SE Asian biogeography; pp. 197–209. [Google Scholar]
- Woodruff DS. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today's pattern and the future of the remaining refugal-phase biodiversity. Biodiversity Conserv. 2010;19:919–941. doi: 10.1007/s10531-010-9783-3. [DOI] [Google Scholar]
- Wang M, Zhang X, Yang T, Han Z, Yanagimoto T, Gao T. Genetic diversity in the mtDNA control region and population structure in the Sardinella zunasi Bleeker. Afr J Biotechnol. 2008;7:4384–4392. [Google Scholar]
- Jamandre BW, Durand JD, Tzeng WN. Phylogeography of the flathead mullet Mugil cephalus in the north-west Pacific as inferred from the mtDNA control region. J Fish Biol. 2009;75:393–407. doi: 10.1111/j.1095-8649.2009.02332.x. [DOI] [PubMed] [Google Scholar]
- Tang W, Ishimatsu A, Fu C, Yin W, Li G, Chen H, Wu Q, Li B. Cryptic species and historical biogeography of eel gobies (Gobioidei: Odontamblyopus) along the northwestern Pacific coast. Zool Sci. 2010;27:8–13. doi: 10.2108/zsj.27.8. [DOI] [PubMed] [Google Scholar]
- Liu JX, Gao TX, Wu SF, Zhang YP. Pleistocene isolation in the northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845) Mol Ecol. 2007;16:275–288. doi: 10.1111/j.1365-294X.2006.03140.x. [DOI] [PubMed] [Google Scholar]
- Ke HM, Lin WW, Kao HW. Genetic diversity and differentiation of grey mullet (Mugil cephalus) in the coastal waters of Taiwan. Zool Sci. 2009;26:421–428. doi: 10.2108/zsj.26.421. [DOI] [PubMed] [Google Scholar]
- Thomson JM. A bibliography of systematic references to the flathead mullets (Mugilidae) Tech Pap Div Fish Oceanogr, CSIRO. 1964;16:1–127. [Google Scholar]
- Huang CS, Lin JC, Huang JJ. Investigation and surveillance of grey mullet resource. FRI Newsletter. 2005;9:1–5. [Google Scholar]
- Hung CM, Shaw D. The impact of upstream catch and global warming on the grey mullet fishery in Taiwan: a non-cooperative game analysis. Mar Res Econ. 2006;21:285–300. [Google Scholar]
- Liu JY, Brown CL, Yang TB. Population genetic structure and historical demography of grey mullet, Mugil cephalus, along the coast of China, inferred by analysis of the mitochondrial control region. Biochem Syst Ecol. 2009;37:556–566. doi: 10.1016/j.bse.2009.09.002. [DOI] [Google Scholar]
- Huang CS, Weng CF, Lee SC. Distingishing two types of gray mullet, Mugil cephalus L. (Mugiliformes: Mugilidae), by using glucose-6-phosphate isomerase (GPI) allozymes with special reference to enzyme activities. J Comp Physiol B. 2001;171:387–394. doi: 10.1007/s003600100187. [DOI] [PubMed] [Google Scholar]
- Liu JY, Lun ZR, Zhang JB, Yang TB. Population genetic structure of striped mullet, Mugil cephalus, along the coast of China, inferred by AFLP fingerprinting. Biochem Syst Ecol. 2009;37:266–274. doi: 10.1016/j.bse.2009.04.010. [DOI] [Google Scholar]
- Campton DE, Mahmoudi B. Allozyme variation and population structure of striped mullet (Mugil cephalus) in Florida. Copeia. 1991;91:485–492. doi: 10.2307/1446596. [DOI] [Google Scholar]
- Rocha-Olivares A, Garber NM, Stuck KC. High genetic diversity, large inter-oceanic divergence and historical demography of the striped mullet. J Fish Biol. 2000;57:1134–1149. doi: 10.1111/j.1095-8649.2000.tb00476.x. [DOI] [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia's fish species. Phil Trans Roy Soc Lond B Biol Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misazawa K, Kuma KI, Takashi M. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods). version 4.0.b10. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Heras S, Roldán MI, Castro MG. Molecular phylogeny of Mugilidae fishes revised. Rev Fish Biol Fish. 2009;19:217–231. doi: 10.1007/s11160-008-9100-3. [DOI] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in Phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Drummond AJ, Rambaut A. "BEAST: Bayesian evolutionary analysis by sampling trees". BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios HA . The great American schism: divergence of marine organisms after the rise of the central American isthmus. Annu Rev Ecol Evol Syst. 2008;36:63–91. doi: 10.1146/annurev.ecolsys.38.091206.095815. [DOI] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007. http://beast.bio.ed.ac.uk/Tracer
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending RC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- Rocha-Olivares A, Chavez-Gonzalez JP. Molecular identification of dolphinfish species (genus Coryphaena) using multiplex haplotype-specific PCR of mitochondrial DNA. Ichthyol Res. 2008;55:389–393. doi: 10.1007/s10228-008-0040-7. [DOI] [Google Scholar]
- Durand JD, Diatta AM, Diop K, Trape S. Multiplex 16 S rRNA haplotype-specific PCR, a rapid and convenient method for fish species identification: an application to West African Clupeiform larvae. Mol Ecol Res. 2010;10:568–572. doi: 10.1111/j.1755-0998.2009.02776.x. [DOI] [PubMed] [Google Scholar]
- Molecular Ecology Resources Primer Development Consortium et al. Permanent Genetic Resources added to Molecular Ecology Resources Database 1 April 2010-31 May 2010. Mol Ecol Res. 2010;10:1098–1105. doi: 10.1111/j.1755-0998.2010.02898.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX: logiciel sous Windows TM pour la génétique des populations. 4.02. Laboratoire Genome Populations, Interactions CNRS, Université de Montpellier II; 1996. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.2307/2409177. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices Version 2.9.3. Lausanne University, Lausanne, Switzerland; 2001. [Google Scholar]
- Luikart GL, Cornuet JM. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol. 1998;12:228–237. doi: 10.1046/j.1523-1739.1998.96388.x. [DOI] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact test and ecumenism. J Hered. 1995;86:248–250. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Takano O, Takata H, Omote H. Late Pliocene early Pleistocene paleoceanographic evolution of the Japan Sea. Paleogeogr Palaeocl Palaeoecol. 2001;172:81–98. doi: 10.1016/S0031-0182(01)00272-3. [DOI] [Google Scholar]
- Yamamoto H. Submarine geology and post-opening tectonic movements in the southern region of the Japan Sea. Mar Geol. 1993;112:133–150. doi: 10.1016/0025-3227(93)90165-R. [DOI] [Google Scholar]
- Tada R. Paleoceanographic evolution of the Japan Sea. Paleogeogr Palaeocl Palaeoecol. 1994;108:487–508. doi: 10.1016/0031-0182(94)90248-8. [DOI] [Google Scholar]
- Xu JW, Chan TY, Tsang LM, Chu KH. Phylogeography of the mitten crab Eriocheir sensu stricto in East Asia: Pleistocene isolation, population expansion and secondary contact. Mol Phylogenet Evol. 2009;52:45–56. doi: 10.1016/j.ympev.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Crosetti D, Avise JC, Placidi F, Rossi AR, Sola L. Geographic variability in the grey mullet Mugil cephalus: preliminary results of mtDNA and chromosome analyses. Aquaculture. 1993;111:95–101. doi: 10.1016/0044-8486(93)90028-W. [DOI] [Google Scholar]
- Crosetti D, Nelson WS, Avise JC. Pronounced genetic structure of mitochondrial DNA among populations of the circumglobally distributed grey mullet (Mugil cephalus Linaeus) J Fish Biol. 1994;44:47–58. doi: 10.1111/j.1095-8649.1994.tb01584.x. [DOI] [Google Scholar]
- Rossi AR, Capula M, Crosetti D, Sola L, Campton DE. Allozyme variation in global populations of striped mullet, Mugil cephalus (Pisces: Mugilidae) Mar Biol. 1998;131:203–212. doi: 10.1007/s002270050312. [DOI] [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung IH. On the fishery biology of gray mullet, Mugil cephalus L., in Taiwan. Rep Inst Fish Biol Ministry Econ Affairs Natl Taiwan Univ. 1981;3:38–102. [Google Scholar]
- Chen WY, Su WC. In: Study on the resource of flathead mullet in Taiwan, 1983-1985. Su WC, editor. Kaohsiung: Kaohsiung Branch TFRI; 1986. Reproductive biology of the flathead mullets, Mugil cephalus L. of Taiwan; pp. 73–80. [Google Scholar]
- Huang CS, Su WC. Studies on the fluctuations of fishing conditions for flathead mullet (Mugil cephalus Linnaeus) from the western coast of Taiwan. J Fish Soc Taiwan. 1989;16:47–83. [Google Scholar]
- Lee CY, Kuo CL. Kinds, local names and seasonal appearance of the fingerlings of grey mullet, Mugil cephalus L., in coastal waters of west Taiwan. China Fish Monthly. 1990;448:13–18. [Google Scholar]
- Chang CW, Tzeng WN, Lee YC. Recruitment and hatching dates of gray mullet (Mugil cephalus L.) juveniles in the Tanshui estuary of northwest Taiwan. Zool Stud. 2000;39:99–106. [Google Scholar]
- Chang CW, Tzeng WN. Species composition and seasonal occurrence of mullets (Pisces, Mugilidae) in the Tanshui estuary northwest Taiwan. J Fish Soc Taiwan. 2000;27:253–262. [Google Scholar]
- Buonaccorsi VP, Kimbrell CA, Lynn EA, Vetter RD. Population structure of copper rockfish (Sebastes caurinus) reflects postglacial colonization and contemporary patterns of larval dispersal. Can J Fish Aquat Sci. 2002;59:1374–1384. doi: 10.1139/f02-101. [DOI] [Google Scholar]
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelc RA, Warner RR, Gaines SD. Geographical patterns of genetic structure in marine species with contrasting life history. J Biogeogr. 2009;36:1881–1890. doi: 10.1111/j.1365-2699.2009.02138.x. [DOI] [Google Scholar]
- Hu J, Kawamura H, Hong H, Qi Y. A review on the currents in the South China Sea: seasonal circulation, South China Sea Warm Current and Kuroshio intrusion. J Oceanogr. 2000;56:607–624. doi: 10.1023/A:1011117531252. [DOI] [Google Scholar]
- Teagle DAH, Bickle MJ, Alt JC. Recharge flux to ocean-ridge black smoker systems: a geochemical estimate from ODP Hole 504B. Earth Planet Sci Let. 2003;210:81–89. doi: 10.1016/S0012-821X(03)00126-2. [DOI] [Google Scholar]
- Teague WJ, Jacobs GA, Ko DS, Tang TY, Chang KI, Suk MS. Connectivity of the Taiwan, Cheju, and Korea straits. Cont Shelf Res. 2003;23:63–77. doi: 10.1016/S0278-4343(02)00150-4. [DOI] [Google Scholar]
- Liu JY, Brown CL, Yang TB. Phylogenetic relationships of mullets (Mugilidae) in China Seas based on partial sequences of two mitochondrial genes. Biochem Syst Ecol. 2010;38:647–655. doi: 10.1016/j.bse.2010.01.007. [DOI] [Google Scholar]
- Liu JX, Gao TX, Yokogawa K, Zhang YP. Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Mol Phylogenet Evol. 2006;39:799–811. doi: 10.1016/j.ympev.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Tzeng TD, Yeh SY, Hui CF. Population genetic structure of the kuruma prawn (Penaeus japonicus) in East Asia inferred from mitochondrial DNA sequences. ICES J Mar Sci. 2004;61:913–920. doi: 10.1016/j.icesjms.2004.06.015. [DOI] [Google Scholar]
- Pan BP, Song LS, Bu WJ, Sun JS. Studies on genetic diversity and differentiation between two allopatric populations of Cyclina sinensis. Acta Hydrobiol Sin. 2005;29:372–387. [Google Scholar]
- Zhao YM, Li Q, Kong LF, Bao ZM, Wang RC. Genetic diversity and divergence among clam Cyclina sinensis populations assessed using amplified fragment length polymorphism. Fish Sci. 2007;73:1338–1343. [Google Scholar]
- Zheng WJ, Zhu SH, Shen XQ, Liu BQ, Pan ZC, Ye YF. Genetic Differentiation of Tegillarca granosa Based on Mitochondrial COI Gene Sequences. Zool Res. 2009;30:17–23. doi: 10.3724/SP.J.1141.2009.01017. [DOI] [Google Scholar]
- Li CH, Li TW, Song LS, Su XR. RAPD analysis on intra-species differentiation of Tegillarca granosa populations to the south and north of Fujian province. Zool Res. 2003;24:362–366. [Google Scholar]
- Yin W, Fu C, Guo L, He Q, Li J, Jin B, Wu Q, Li B. Species delimitation and historical biogeography in the genus Helice (Brachyura: Varunidae) in the Northwestern Pacific. Zool Sci. 2009;26:467–475. doi: 10.2108/zsj.26.467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Variable positions in the 627 bp mitochondrial COI gene segment of Mugil cephalus from 12 locations in the northwestern Pacific. Dots represent identical nucleotides relative to haplotype 1. Frequency of each haplotypes for each lineages (NWP1, NWP2 and NWP3) are also shown. Different color means the locations of the lineage specific nucleotides.
Table S2. Genetic variability at ten microsatellite loci of Mugil cephalus collected in the northwestern Pacific for spatial and temporal genetic structure test. Table-wide significance levels were applied using the sequential Bonferroni technique [45].
Table S3. Log probability and ΔK [50] for each number of clusters in the Bayesian assignment test as implemented in STRUCTURE [49].
Table S4. The mean q-values and standard deviations (sd) for assignment test of 3 Mugil cephalus cryptic species (NWP1, NWP2 and NWP3) as implemented in STRUCTURE [49].
Table S5. Genetic variability at ten microsatellite loci of Mugil cephalus among 3 cryptic species. Table-wide significance levels were applied using the sequential Bonferroni technique [45].
Figure S1. Ethidium bromide stained 2% agarose gel showing the multiplex COI haplotype-specific PCR (MHS-PCR) for the rapid screening of three Mugil cephalus cryptic species in the NW Pacific. 1-13: unidentified individuals belonging either to lineage NWP1 (2), NWP2 (3, 8, 12, 13) or NWP3 (1, 4-7, 9-11). M: 100-bp DNA ladder.
Figure S2. Phylogenetic relationships within Mugil cephalus recovered from 1140 bp of the cytochrome b sequences according to the neighbour-joining tree using Kimura 2 parameter distance. Leaves of the tree correspond to haplotypes of M. cephalus observed by Ke et al. [17] (H1-H43) and this study (H44-H55, in bold). The values above the branches are bootstrap support (500 replicates). Bootstrap supports higher than 50% are displayed.