Abstract
Among the polycyclic aromatic hydrocarbon class of chemical carcinogens, dibenzo[a,l]pyrene (DB[a,l]P) is the most potent tumorigen that has been identified to date. Structurally, it is bulky with six aromatic rings and it contains the non-planar fjord-region. The conformational properties of DB[a,l]P-derived DNA adducts responsible for its extraordinary carcinogenicity are hence of great interest. We have carried out molecular modeling and MD simulations for the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts derived from the reactions of the DB[a,l]P diol epoxides with adenine in double-stranded DNA. The structures are based on the classically intercalated NMR solution structures of the analogous fjord-region benzo[c]phenanthrene B[c]Ph-derived -N6-dA adducts. One objective was to gain insight on the impact of the more bulky DB[a,l]P ring system on the structural characteristics of the intercalative adduct conformations. A further objective was to elucidate the effect of the flexible twist associated with the sterically hindered aromatic ring in the fjord-region on the intercalated conformations, for comparison with the intercalated but planar, bay-region benzo[a]pyrene B[a]P-derived -N6-dA adducts. For the DB[a,l]P-N6-dA adducts, our results show that the 14R (+)-adduct is more favorably intercalated on the 5′-side of the modified adenine than the stereoisomeric 14S (–)-adduct, intercalated on its 3'-side. The 14R (+)-adduct manifests better van der Waals stacking interactions with flanking base pairs, less perturbed Watson-Crick hydrogen bonding, less local groove enlargement, less unwinding, and a lower solvent exposure than the 14S (–)-adduct. These structural findings are consistent with observed thermodynamic melting data, UV absorption properties, and fluorescence quenching studies. By contrast, the NMR solution structures for the analogous but less bulky B[c]Ph-derived adducts reveal no such stereoisomeric effect, while the planar bay-region B[a]P-derived-N6-dA adducts do. Differences in nucleotide excision repair susceptibilities of the fjord and bay region adducts stem from distinctions in their intercalative conformations, produced by the intrinsic topological variations in their polycyclic aromatic ring systems.
Keywords: dibenzo[a,l]pyrene adenine adducts; MD simulations; structures
Introduction
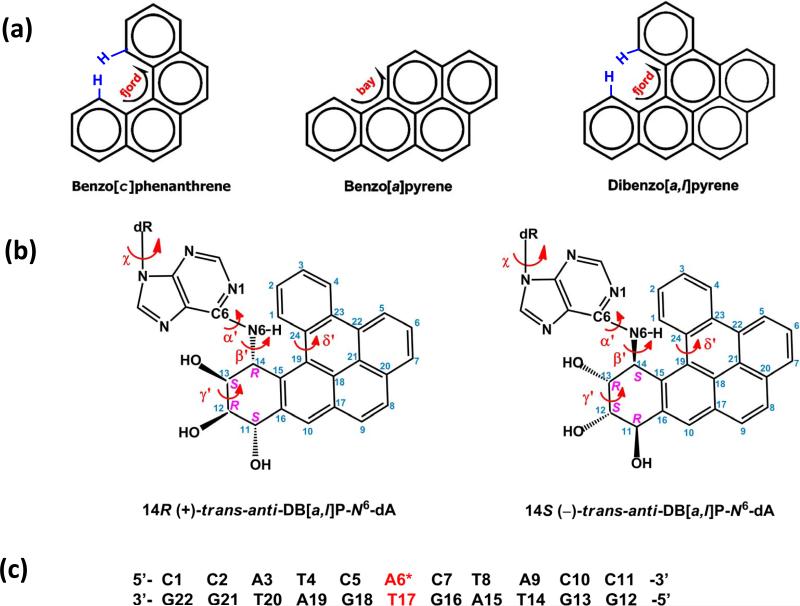
The successful repair of adducts derived from reactions of diol epoxide metabolites of B[a]P and structurally related polycyclic aromatic hydrocarbons (PAH) with DNA in cellular environments (1-4) is an important factor that contributes to the genotoxic characteristics of these DNA lesions. The structural features of such PAH-DNA adducts and their impact on the efficiencies of recognition and excision repair (NER) by the mammalian nucleotide excision apparatus are of great interest for assessing the carcinogenic activities of stereoisomeric PAH diol epoxide (DE) -DNA adducts (5, 6). A remarkable example is the difference in NER efficiencies in human cell extracts of N6-dA adducts derived from B[a]P and the structurally similar dibenzo[a,l]pyrene (DB[a,l]P) that differ from one another by a single aromatic ring (Figure 1a). The B[a]P-N6-dA adducts are good substrates of NER, while the stereochemically identical DB[a,l]P-N6-dA (Figure 1b) adducts have been reported to be resistant to NER (7). In this work, we examine the structural features of the DB[a,l]P-N6-dA adducts in double-stranded DNA that are likely to contribute to their unusual resistance to NER.
Figure 1.
(a) Chemical structures of B[c]Ph, B[a]P and DB[a,l]P with designation of bay region for B[a]P, and fjord regions for B[c]Ph and DB[a,l]P. (b) Chemical structures of the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts. The lesion-DNA linkage site torsion angles α′ and β′, glycosidic torsion angle, χ, fjord region twist torsion angle δ′ and benzylic ring pucker torsion angle γ′ are defined as follows: α′, N1-C6-NC14(DB[a,l]P); β′, C6-N-C14(DB[a,l]P)-C13(DB[a,l]P); χ, O4′-C1′-N9-C4; δ′, C15-C19-C24-C1; γ′, C14-C13-C12-C11. (c) The base sequence context in which the lesion is embedded. A6* designates the lesion modified adenine.
DB[a,l]P is an environmental contaminant that is present in cigarette smoke, smoky coal particulates and soil or sediment samples (8), and is the most potent PAH tumorigen yet identified (8-10). Remarkably, it is about 100-times more carcinogenic than the much more widely studied planar bay region B[a]P (Figure 1a) (8). As a member of the fjord region PAHs, DB[a,l]P is non-planar, which avoids a steric clash between the hydrogen atoms at C1 and C14 in DB[a,l]P, and the analogous protons of B[c]Ph across the sterically crowded fjord region (Figure 1a). The metabolic activation of DB[a,l]P via the diol epoxide pathway yields predominantly the (–)-anti-DB[a,l]P enantiomer (11, 12), with lesser amounts of the (+)-syn metabolite (11, 12), and minor amounts of the (+)-anti and (–)-syn metabolites (13). The diol epoxides derived from DB[a,l]P are highly tumorigenic (11, 14, 15), and cause mutations targeted to adenine and guanine (8, 16). These diol epoxides react with the exocyclic amino groups of adenine and guanine to form covalent DNA adducts. The proportions of adenine and guanine adducts depend on the diol epoxide absolute configurations and other variables (1, 8, 17-21); however there is a frequent predominance of adenine adducts. In addition, DB[a,l]PDE-derived adducts have been observed in lung tissues of rodents (17, 22). It is likely that these very bulky adducts block replicative polymerases and would be transited by error-prone bypass polymerases of the Y-family (23). Work from the Guengerich laboratory has shown with primer extension studies utilizing lesions of increasing size that adducts larger than ethyl groups are primarily blocking (24). However, primer extension studies with DNA polymerases and oligonucleotides that have been site-specifically modified with the DB[a,l]P adducts do not appear to have been performed yet. The 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts (Figure 1b) are of special significance because, along with the stereochemically identical fjord adenine adducts derived from benzo[c]phenanthrene (B[c]Ph) (Figure 1a) and benzo[g]chrysene (B[g]C), these adducts are essentially resistant to nucleotide excision repair (NER) in human HeLa cell extracts (7, 25). Structural characterizations of N6-dA adducts derived from the bay region B[a]P (Figure 1a and Figure S1a, Supporting Information) (26-32), and the fjord region B[c]Ph (Figure 1a and Figure S1b, Supporting Information) (33-35) and B[g]C (36) by NMR methods and computational approaches show that the S stereoisomers are intercalated on the 3’-side, while the R stereoisomers are intercalated on the 5’-side of the modified adenine. All base pairs, including the somewhat distorted modified A:T pair, are of the Watson-Crick type. It is particularly noteworthy that, in the case of the fjord region adducts, the non-planar aromatic ring system is somewhat flexible and therefore can optimize stacking interactions with adjacent base pairs; therefore, the twist of the non-planar distal ring is twisted in opposite directions in the R and S stereoisomeric N6-dA adducts (33, 34).
From a topological perspective, the DB[a,l]P-N6-dA adducts are especially interesting because they are resistant to NER (7), although they are bulky with six aromatic rings. Furthermore, the NER-resistant stereoisomeric DB[a,l]P-N6-dA adducts have identical absolute configurations (Figure 1a) to those of the analogous B[a]P-N6-dA adducts that are good substrates of NER. While structural information is available for the B[a]P-N6-dA adducts (26-32), this is not the case for the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts. Here, we have used molecular modeling and molecular dynamics (MD) simulations to explore their structural properties. The fjord region 1R (+)- and 1S (–)-trans-anti-B[c]Ph-N6-dA adducts (33, 34, 37, 38) and the benzo[g]chrysene diol epoxide-derived N6-dA adducts (36), as well as the bay region 10R (+)- and 10S (–)-trans-anti-B[a]P-N6-dA adducts are known to adopt classical intercalation conformations in which the aromatic ring systems are inserted between neighboring base pairs without base displacement (26-32, 38, 39). Our objectives were to investigate how the additional aromatic ring in DB[a,l]P-N6-dA adducts impacts the intercalative conformation as compared to the stereochemically similar B[c]Ph- and B[a]P-N6-dA adducts. While the polycyclic aromatic ring systems are intercalated in all three cases, we show that there are distinctions within the classical intercalation family that provide insights into the structural features that account for the successful repair of the B[a]P-N6-dA adducts by the human NER system (29), while the fjord region adducts resist repair (7, 25).
Results
As detailed in the Methods section, we have employed molecular modeling and MD simulations to analyze classically intercalated conformations of the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts in double-stranded DNA (Figure 1c). However, as detailed below, the unique topology of the DB[a,l]P aromatic ring system is characterized by striking differences in the structural characteristics of stereoisomeric 14R and 14S adducts; moreover, these are different from those observed in the case of the stereochemically similar B[c]Ph-N6-dA and B[a]P-N6-dA adducts (Figure 1a).
Opposite directionality
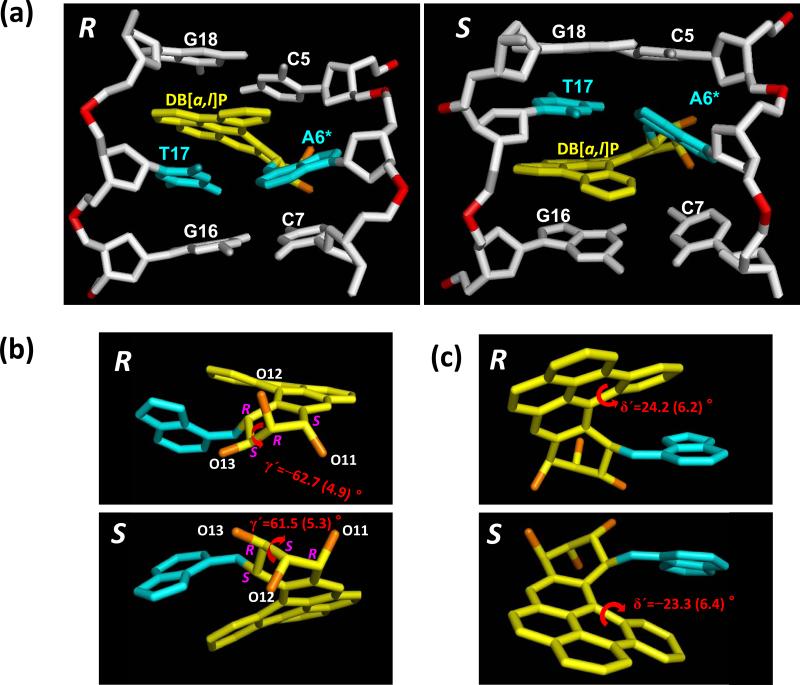
The classical intercalation conformations adopted by the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts share the overall features of this conformational family: 5’-side intercalation for the 14R (+)- and 3’-side intercalation for the 14S (–)-adducts with perturbed Watson-Crick pairing maintained; intercalation, from the major groove side, is achieved through stretching and unwinding of the double helix (Figure 2a and Movies S1 and S2, Supporting Information). Figure S1c, Supporting Information, shows the full modified duplex 11mers as well as the unmodified control duplex 11mer, which highlight the stretching and unwinding induced by the lesion. A manifestation of the opposite orientations of the two stereoisomers is the inversion of the torsion angles (similar absolute values with opposite signs) that characterize the structures and orientations of the DB[a,l]P moiety (Table S1, Supporting Information). The cyclohexene-type benzylic ring is half-chair in each stereoisomer, but they are near mirror images as reflected in their inverted torsion angles γ′, C14-C13-C12-C11 (Figure 1 and Supporting Information, Figure S2 and Table S1), and seen in Figure 2b. Of particular interest is the fjord region twist δ′, C15-C19-C24-C1 (Figures 1 and 2c); it is inverted in the two stereoisomers (Figure 2c, Table S1 and Figure S2, Supporting Information), reflecting the flexible adaptability of the DB[a,l]P ring system to allow optimal stacking for each stereoisomer with an adjacent base pair. Also of note are the inverted values of the DB[a,l]P-DNA linkage torsion angles, α′ and β′ (Table S1 and Figure S3, Supporting Information), which are in the ideal domains computed on the nucleoside level for R and S trans-anti-N6-dA adducts derived from B[a]P and B[c]Ph (40, 41). Additionally, the helicoidal parameters Buckle and Propeller at the damaged base pair are inverted (Figure 3 and Table S1, Supporting Information).
Figure 2.
(a) Views into the minor groove of the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adduct structures. These are the best representative structures for the 3.0 – 30.0 ns range of the MD simulations (See Methods). The central 3-mers are shown. (b) View into the minor groove side of the DB[a,l]P-modified adenine (c) View into the major groove side of the DB[a,l]P-modified adenine. The near mirror image nature of the stereoisomer pair is displayed; panel (b) emphasizes the fjord region twist and (c) emphasizes the benzylic ring pucker. For the DB[a,l]P moiety, the carbons are yellow and oxygen atoms are orange, the damaged bases are cyan. The DNA duplexes are white, except for the phosphorus atoms, which are red. Hydrogen atoms and pendant phosphate oxygen atoms are not shown for clarity. See also Movies S1 and S2, Supporting Information.
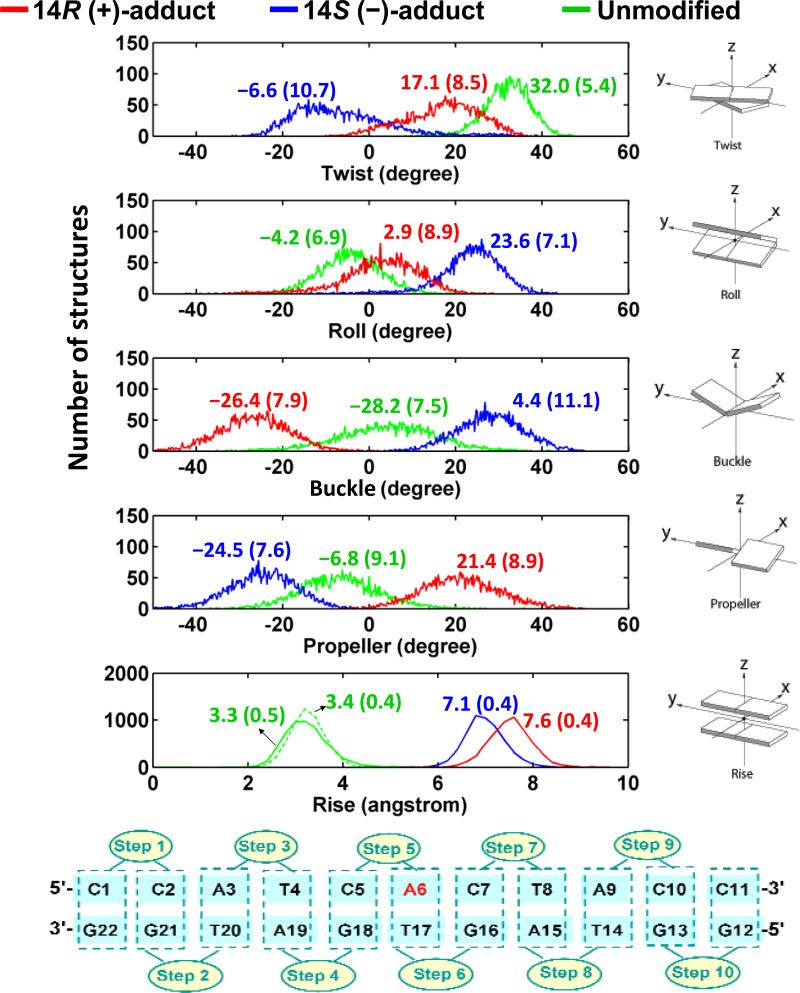
Figure 3.
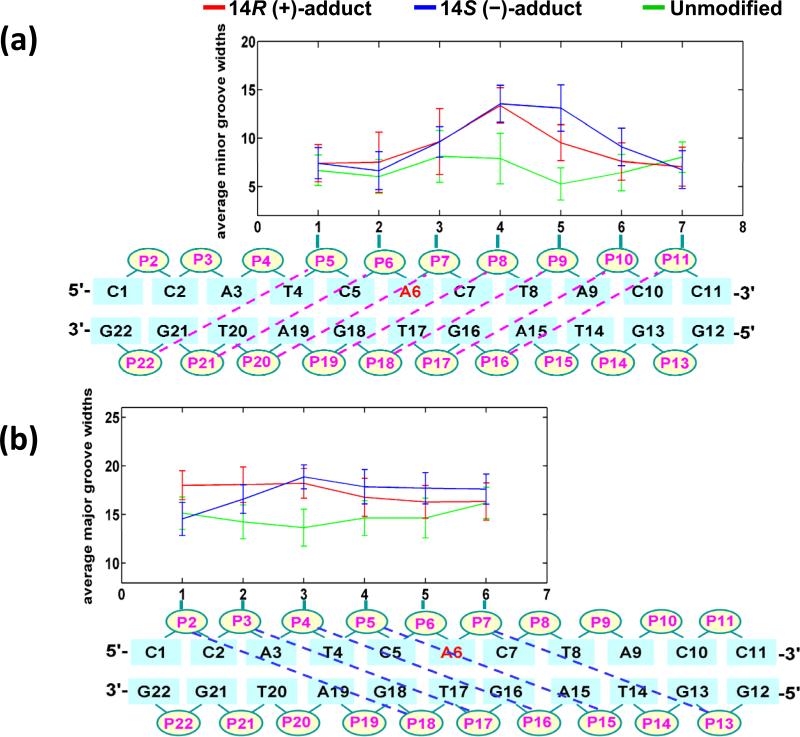
Population distributions of helicoidal parameters at the lesion site. Ensemble averages and standard deviations (in parentheses) are given. The Rise at the intercalation pocket is Step 5 for the R adduct and its unmodified control (solid line), and is Step 6 for the S adduct and its corresponding unmodified control step (dashed line). Helicoidal parameter definitions are shown on the right side: the cartoons are reprinted with permission from Lu et al, Nucleic Acids Res. 31 (17): 5108-5121, Figure 1, Copyright © 2003, Oxford University Press (69). The step numbering scheme is given at the bottom of the figure.
Helicoidal parameters
The stretching and unwinding induced by intercalation (42, 43) differs in the R and S stereoisomers. Helix stretching and unwinding are revealed on the molecular level by the helicoidal parameters Rise and Twist (Table S1, Supporting Information). We note that the Rise is greater and the untwisting is lower for the R than the S stereoisomer in their respective intercalation pockets. The ensemble average values of Rise are 7.6 ± 0.4 Å and 7.1 ± 0.4 Å (Figure 3 and Supporting Information Figure S4 and Table S1) for the 14R (+)- and 14S (–)-adducts respectively. For the unmodified duplex at the analogous steps, the values are those of normal B-DNA: 3.3 ± 0.5 Å and 3.4 ± 0.4 Å (44) (Figure 3 and Table S1, Supporting Information), respectively. For Twist, the ensemble average values are 17.1 ± 8.5° and –6.6 ± 10.7°, respectively, for the 14R (+)- and 14S (–)-stereoisomers. Average Twist is 36.0° for ideal B-DNA, whose structure is sequence-independent (44), and the value is 32.0 ± 5.4° for the analogous step in the unmodified duplex (Figure 3 and Table S1, Supporting Information). The helicoidal parameters Roll and Tilt are also more distorted in the 14S (–)- than the 14R (+)- stereoisomers, as shown in Figure S4 and Table S2, Supporting Information.
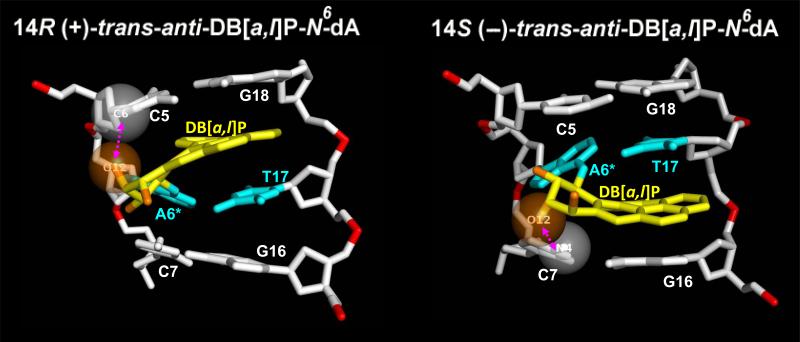
These differences between the R and S stereoisomer structures result from different steric effects due to the benzylic ring hydroxyl groups in 5’-side (R) vs. 3’-side (S) intercalation relative to the modified adenine (Figure 4 and Supporting Information, Movies S3 and S4). The key group is the axial carbon 12-OH (Figure 1). For the R stereoisomer, this group is positioned 5’-to the damaged A6* (Figure 2), within van der Waals contact of C5 and oriented along the helix axis. To avoid collision between C5 and the bulky, axial carbon 12-OH group, the DNA backbone has to stretch more than in the S case. For the S stereoisomer, however, the aromatic rings are on the 3’-side of A6*, and the adjacent C7:G16 base pair is rotated by two base pair steps (~72°, ~ 36° × 2) relative to the C5:G18 base pair. Additionally, the benzylic ring half-chair configuration is a near-mirror image of the R-stereoisomer and the axial carbon 12-OH is now oriented towards C7 in the base pair plane (Figure 2). Consequently, collision between the carbon 12-OH and C7 is avoided mainly by the large helix unwinding (Figure 5 and Table S2), without requiring as much stretch as in the R stereoisomer (Tables S1 and S2, Supporting Information, and Figure 4).
Figure 4.
Steric crowding governs the structural distortions in the lesion-containing duplexes. For the 14R (+)-adduct, steric crowding between the hydroxyl group O12 and 5’-flanking base C5 (atom C6) produces the enlarged Rise. For the 14S (–)-adduct, the crowding between the hydroxyl O12, pointing toward the 3’-side of the lesion, and the 3’-flanking base C7 (atom N4) causes the untwisting. The hydroxyl O12, C6 atom of the C5 base in the 14R (+)-adduct, and the N4 atom of the C7 base in the 14S (–)-adduct are shown in semi-transparent CPK. See also Movies S3 and S4, Supporting Information.
Figure 5.
Ensemble average groove widths with standard deviations for the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts and the unmodified duplex. Groove widths are the smallest phosphorus-phosphorus separations between the two strands across the minor and major groove. These widths are conventionally measured as the phosphorus atom center-to-center distances minus 5.8 Å to account for the van der Waals radii of the two phosphate groups (44, 66).
Groove widths
Interestingly, both major and minor grooves are enlarged relative to the unmodified control in both stereoisomers, as shown in Figure 5 and Supporting Information, Figure S5 and Table S3. Enlargement of the major groove results from the intercalation of both stereoisomers from the major groove side of the duplex where the adenine amino group resides. Minor groove enlargement results from protrusion of the bulky DB[a,l]P ring system into the duplex toward the minor groove. For each groove, there is greater enlargement for the S than for the R stereoisomer adduct, which correlates with its greater untwisting (45).
Stacking and solvent accessible surface area
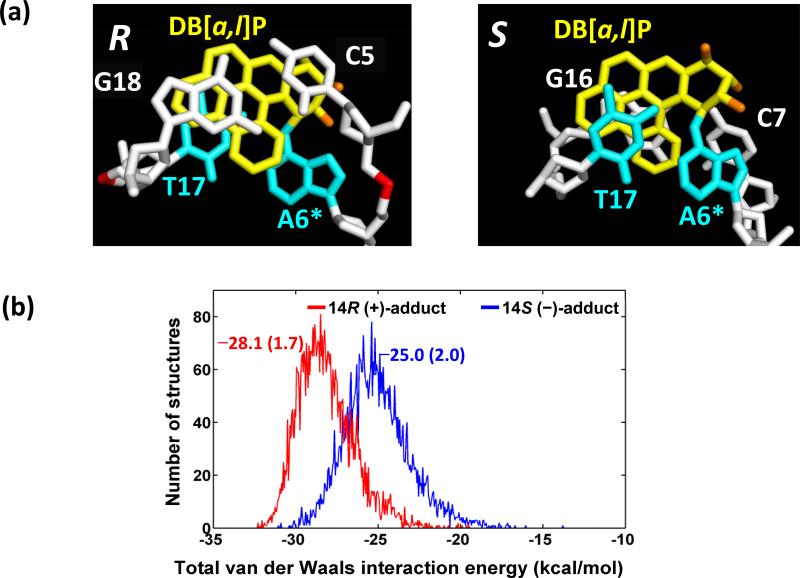
The 14S (–)-adduct has less van der Waals stacking interactions and greater solvent accessible surface area (SASA) than the 14R (+)-stereoisomer. A view looking down the helix axis of the central two base pair d(C5-A6*)•d(T17-G18) segment in the best representative structure of the 14R (+)-trans-anti-DB[a,l]P-N6-dA 11-mer duplex (Figure 6a) reveals the overlap geometry between the DB[a,l]P ring system and the C5:G18 base pair. A similar view for the 14S (–)-trans-anti-DB[a,l]P-N6-dA adduct at the analogous step d(A6*-C7)•d(G16-T17) is also shown (Figure 6a). We note that in the 14R (+)-stereoisomer, there are substantial stacking interactions between the DB[a,l]P aromatic rings and the C5 base, flanking the damaged A6* in the intercalation pocket on the 5’-side of the lesion. However, in the 14S (–)-case, the interactions between the DB[a,l]P aromatic rings and the flanking base C7 on the 3’-side are less pronounced (Figure 6a, and Figures S6 and S7, Supporting Information); this conclusion is supported by the differences in van der Waals interaction energies: the ensemble averages are –5.5 ± 1.1 kcal·mol-1 and –2.2 ± 0.7 kcal·mol-1, respectively. This difference is the dominant factor that contributes to the differences between the ensemble average total van der Waals interaction energies of the DB[a,l]P aromatic rings with the base pairs in the intercalation pocket, –28.1 ± 1.7 kcal/mol for the 14R (+)-adduct, and –25.0 ± 2.0 kcal/mol for the 14S (–)-adduct (Figure 6b, and Supporting Information Tables S1 and S4). The smaller stacking interactions in the S case are correlated with a greater extent of unwinding (Figure S8, Supporting Information) with a correlation coefficient of 0.72. Furthermore, stacking interaction energies between the DB[a,l]P aromatic rings and the damaged base pair A6:T17 (Figure 6a and Figure S6, Supporting Information) are essentially identical in the 14R (+) and 14S (–) stereoisomers, with ensemble average values of –12.1 (1.3) and –11.8 (1.3) kcal/mol, respectively (Table S4, Supporting Information). Therefore, the difference in SASA, discussed below, is not impacted.
Figure 6.
(a) Views looking down the helix axis of the intercalation pockets. Note the substantial overlap between C5 and the DB[a,l]P rings in the 14R (+)-isomer, while the analogous C7 in the 14S (–)-isomer is poorly stacked. The structures shown are the best representative structures from the 3.0 – 30.0 ns MD simulations. Color schemes are the same as Figure 2. (b), population distribution of total van der Waals interaction energy between the DB[a,l]P aromatic rings and the intercalation pocket. Ensemble average values and standard deviation (in parentheses) (kcal/mol) are given.
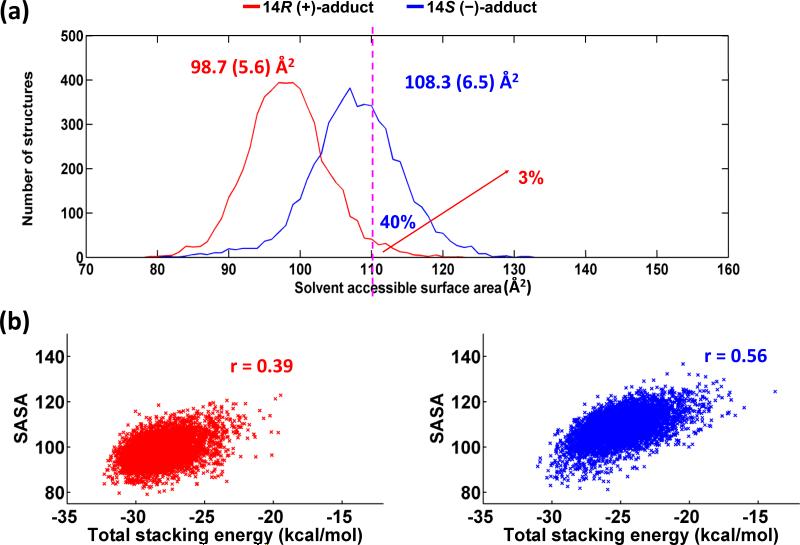
Greater interaction of the aromatic rings with the flanking base pairs leads to more shielding from solvent in the better-stacked structures. We observe that the aromatic ring system is less solvent-exposed in the 14R (+)-adduct than in the 14S (–)-adduct: the ensemble average SASA is 98.7 ± 5.6 Å2 for the 14R (+)-adduct, and 108.3 ± 6.5 Å2 for the 14S (–)-adduct. Figure 7a shows the population distributions of the SASA for the two stereoisomeric adducts. We note that the population distribution for the 14S (–)-stereoisomer is shifted towards greater solvent exposure. A manifestation of this shifted population distribution is highlighted by the observation that 40% of the 14S (–)-adduct population has a SASA of 110 Å2 or greater, while only 3% of the 14R (+)-adduct population has at least a 110 Å2 SASA (Figure 7a). However, both S and R stereoisomers contain small populations of structures with the aromatic rings largely protruding into the major groove, with little stacking and large SASA. Figure S7, Supporting Information, shows structures from the ensembles that have maximal and minimal stacking.
Figure 7.
(a) Population distributions of the solvent accessible surface areas for the DB[a,l]P aromatic rings in the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts. Ensemble average values and standard deviations (in parentheses) are given. The vertical magenta line shows that 40 percent of the population has a SASA greater than 110 Å2 for the 14S (–)-adduct, whereas only 3 percent of the population has a SASA greater than this value for the 14R (+)-adduct. (b) Scatter plots showing the correlation between total van der Waals interaction energies in the intercalation pocket and SASA. Correlation coefficients are shown.
Hydrogen bonding
Table S5, Supporting Information, shows hydrogen bond occupancies of all base pairs, in both stereoisomers and the unmodified duplex. We note that all hydrogen bonds are essentially maintained in both adducted duplexes and the unmodified control, although the C7:G16 base pair in the 14S (–)-adduct is perturbed (Table S5, Supporting Information). Despite the perturbations in Buckle and Propeller (Figure 3 and Table S1, Supporting Information), these distortions do not cause rupturing of the Watson-Crick base pairing.
Discussion
Classical intercalation conformations share common features but differ as ring system topologies vary
The classical intercalation conformation is characteristic of PAH-derived N6-dA adducts. This conformational family shares common features. The aromatic ring system is shielded from the solvent upon intercalation of the hydrophobic aromatic ring system between adjacent base pairs from the spacious major groove side of the DNA duplex; the aromatic rings thus achieve favorable stacking interactions with adjacent base pairs in the intercalation pocket. The double helix stretches and unwinds to accommodate the inserted aromatic ring system without breaking the Watson-Crick pairs at the lesion site. However, adduct stereochemistry and aromatic ring system topology influence other structural characteristics. Stereochemistry at the lesion site has a profound effect on the nature of the intercalation by the adducted aromatic ring system. In all cases, the R stereoisomer adduct is intercalated on the 5’-side and the S stereoisomers on the 3’-side of the damaged base. The fundamental origin of this opposite-orientation phenomenon has been elucidated for the classical intercalation and other conformational families (40, 41, 46, 47): steric hinderance between the benzylic ring of the adduct with the covalently-linked base impedes adoption of the stereochemically disfavored conformation. However, it has been previously noted for the case of the B[a]P-N6-dA adducts that there is conformational heterogeneity only in the S stereoisomer (31). This has been attributed to steric crowding between the benzylic ring and the adjacent 5’-side base in this 3’-intercalated adduct; this produces a syn-anti equilibrium, in which the glycosidic bond conformation can oscillate between syn and anti regions to relieve the crowding (27, 29, 39). Sequence context determines how rapidly the two conformational domains interchange, that is, the barrier height between syn and anti (29, 39). The R stereoisomer of the B[a]P-N6-dA adduct is intercalated on the 5’-side of the modified adenine residue, and does not exhibit any steric crowding between the hydroxyl groups and the base (C7) on its 3’-side since the right-handed twist of the DNA helix has now rotated C7 by ~72 degrees relative to C5.
We found a similar conformational heterogeneity only in the 3’-intercalated S stereoisomer for the DB[a,l]P-N6-dA adduct. In this case, benzylic ring-imposed crowding produces a less-well intercalated structure, as evidenced by a number of structural differences from the 5’-intercalated R stereoisomer. The DB[a,l]P rings are less well stacked within the intercalation pocket (Figure 6). This is also manifested in the greater solvent exposure of this adduct (Figure 7). Hydrogen bonding is also more perturbed (Table S5, Supporting Information). Additionally, the adduct causes a much greater untwisting, combined with greater disturbances in other helicoidal parameters, namely Roll and Tilt (Table S1, Supporting Information). Thus, to avoid steric crowding, the price paid by the lesion-containing duplex involves unwinding, diminished stacking and greater solvent exposure.
Our simulated results indicating intercalative conformations for both 14S (–)- and 14R (+)- stereoisomers, with greater solvent exposure, distortion, and conformational heterogeneity for the 14S (–)- adduct, are supported by and explain several experimental findings (48). The UV absorption spectrum of intercalated B[a]P-DNA adducts is characterized by a red-shifted absorption spectrum (49, 50). Both DB[a,l]P-N6-dA adducts exhibit red-shifted UV absorbance maxima that are indicative of intercalative conformations for both stereoisomeric adducts ((48) and A. Schwaid et al., manuscript in preparation). ). However, thermal melting (Tm) data demonstrate a notable difference in the thermodynamic stabilities of duplexes of the 14S (–)- and 14R (+)-trans-anti-DB[a,l]P-N6-dA adducts. In a particular 11-mer oligonucleotide duplex, it was found that the 14R stereoisomer stabilizes the DNA duplex by + 8 °C, while the S stereoisomer destabilizes the duplex by – 6 °C (The Tm value of the unmodified duplex was 43.8 ± 0.5 °C) (51). In addition, fluorescence quenching studies show a solvent exposed component of the conformational population only for the S stereoisomer, consistent with our finding of a much greater solvent-exposed population for this 14S stereoisomeric adduct (48).
Comparisons of the intercalative conformations of PAH N6-dA adducts reveal that the structural impact of steric crowding in the case of the S stereoisomers is dependent on PAH ring system topology. For the bay-region 10S (+)-trans-anti-B[a]P-N6-dA adduct, a syn-anti equilibrium in the glycosidic bond conformation relieves the crowding imposed by steric hindrance between the benzylic ring hydroxyl groups and the DNA backbone (27, 29). However, the DB[a,l]P S adduct does not require a syn-anti equilibrium, because the flexible twist of the distal fjord region aromatic ring permits relief of crowding without rotation to the syn domain. For the small, well-intercalated fjord region 1R (+)- and 1S (–)-trans-anti-B[c]Ph-N6-dA adducts, the benzylic ring hydroxyl groups can be easily accommodated and no conformational heterogeneity is observed in the NMR solution structures in either case (33, 34). Figure S1b, Supporting Information, presents these NMR solution structures in 11mer duplexes.
Variable NER susceptibilities result from differences in intercalative conformations that are governed by topology of the aromatic ring system
Our findings point to some intriguing considerations regarding the initial recognition of the XPC/hHR23B NER factor; according to the crystal structure of the Rad4/Rad23 yeast homolog bound to a DNA complex bearing a single UV light-induced cyclobutane pyrimidine lesion (52), a β-hairpin is inserted between the two strands at the lesion site, and the two opposite bases flip out of the duplex and bind to Rad4/Rad23. NER experiments with HeLa cell extracts indicate that the 14R (+) and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts, as well as their B[c]Ph-derived analogues (7, 25, 53) are resistant to NER in HeLa cell extracts; however, the B[a]P-derived adducts are susceptible to NER, and the 10S (+)-trans-stereoisomeric adduct is better repaired than the 10R (–)-trans-anti-B[a]P-N6-dA adduct with base sequence context playing an important role in their relative incision efficiencies (39, 53). Representative data are reviewed in (39, 53). Thermal melting data suggest only modest destabilization or stabilization compared to the unmodified duplexes in the case of the DB[a,l]P- (48, 51) and B[c]Ph-derived-N6-dA adducts (51, 54); however, the analogous B[a]P-derived N6-dA adducts are destabilizing by ~13 °C for the 10R stereoisomers and by ~20 °C for the 10S cases (39, 53). Our structural studies suggest that the stacking interactions within the intercalation pocket in both the 14R and 14S DB[a,l]P adducts studied here, as well as in the R and S stereoisomeric B[c]Ph adenine adducts (33, 34) do not allow sufficient local thermodynamic destabilization to permit hairpin insertion and base flipping that facilitate the NER recognition process; however, the rigid and planar 10R (–)- and 10S (+)-trans-anti-B[a]P-N6-dA adenine adducts, lacking the flexible twist of the fjord region aromatic ring, are not accommodated as well within the intercalation pocket (29-32, 39, 55), are more destabilizing and hence better facilitate the partner base flipping and β-hairpin insertion needed for lesion recognition; in the case of the 10S (+)-trans-anti-B[a]P-N6-dA adduct, the significant steric hinderance between the benzylic ring and the DNA backbone produces a syn-anti glycosidic torsion bond equilibrium (29, 31), which makes it even more NER susceptible than the 10R stereoisomeric adduct (29, 39). We conclude that significant differences in intercalative conformations can result from a combination of topological and stereochemical effects in different PAH-derived N6-dA adducts. In turn, the conformational distinctions can yield striking NER susceptibility differences that may play an important role in varying carcinogenic potencies. We hypothesize that steric occlusion to β-hairpin insertion per se does not preclude lesion recognition, which is facilitated by thermodynamic destabilization and may occur via induced fit and partner base-flipping in a cooperative manner (52).
Methods
Starting structures and force field
The initial models for the 14R (+)- and 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts were built from the high resolution NMR solution structures of the fjord region B[c]Ph analogs (Figure 1a) with the same stereochemistry at the linkage site, namely, 1R (+)-trans-anti-B[c]Ph-N6-dA (33) and 1S (–)-trans-anti-B[c]Ph-N6-dA (34), respectively. Figure S1b, Supporting Information, presents these NMR solution structures in 11mer duplexes. Molecular modeling was employed to add two rings to the B[c]Ph to create the DB[a,l]P initial model (Figure 1a). These models on the nucleotide level were subjected to geometry optimization utilizing the Gaussian 03 package from Gaussian, Inc (56). The optimized DB[a,l]P lesions were cut from the optimized nucleotide structures and modeled into the duplexes, with the lesion-DNA linkage site torsion angles α′, β′ and χ (Figure 1a) assuming the same values as in the initial B[c]Ph adduct NMR structures (R: α′ = 154.0°, β′ = 103.6°, χ = –128.4°; S: α′ = 130.6°, β′ = 110.7°, χ = –66.0°). The damaged adenine base and sugar remained the same as the initial B[c]Ph adduct NMR structures. Base sequences were modified to reflect those shown in Figure 1c.
MD simulations were carried out using SANDER in the AMBER 9 simulation package (57). The Cornell et al. force field (58) and the parm99.dat parameter set (59) modified by parmbsc0 (60) were employed for all simulations. Partial charges for the 14R (+)- and the 14S (–)-trans-anti-DB[a,l]P-N6-dA adducts were calculated on the nucleotide level with α′, β′ and χ ( values the same as in the initial models, using Hartree Fock quantum mechanical calculations with 6-31G* basis set without geometry optimization, employing the Gaussian 03 package from Gaussian, Inc (56). The charges were then fitted to each atomic center with the RESP algorithm (61, 62). The fitted charges, together with missing bond angle parameters that were added to the force field are shown in Table S6, Supporting Information.
MD computation protocols
Details of the MD protocols are given in Supplementary Materials. The stability of the MD simulation was evaluated for each model. For each sequence context, the root-mean-square deviation (RMSD) of each snapshot in the trajectory relative to its respective starting structure was plotted as a function of time and is shown in Figure S9, Supporting Information. The ensemble averages and standard deviations for the RMSDs are shown. In this calculation, two base pairs at each end were excluded. For all cases, MD achieved good stability, fluctuating around the mean after 3.0 ns, and we employed the structural ensembles from the 3.0 ns to 30.0 ns time frame for further analyses.
Structural analyses
The PTRAJ module of the AMBER 9 package (57) was employed for structural analyses. The CARNAL module of the AMBER 7 package (63) was utilized to compute hydrogen bond occupancies. Frames were selected at 5 ps intervals from the last 27.0 ns of simulation. DNA duplex helicoidal parameters and groove dimensions were analyzed using MD Toolchest (64, 65); for the groove dimensions, 5.8 Å was subtracted from the pairwise phosphorous-phosphorous distances to account for the van der Waals radius of the P atoms (66). The first and last base pairs were excluded in this analysis due to possible end effects. Stacking interactions were estimated by computing the van der Waals interaction energies between the DB[a,l]P aromatic rings and all adjacent base pairs, and pairwise between all adjacent base pairs of the duplex excluding two base pairs at each end, utilizing the ANAL module of the AMBER 9 package (57). The SASA for the DB[a,l]P aromatic rings was calculated with the Connolly algorithm (67) with the INSIGHTII 2005 program (Accelrys Software, Inc.), adopting default parameters: probe radius of 1.4 Å and atom radius scale of 1.0 Å. Visualization and model building were performed in the INSIGHTII 2005 program (Accelrys Software, Inc.). PyMOL (Delano Scientific, LLC.) (68) was employed to make molecular images and movies. Computations were carried out on our own cluster of Silicon Graphic Origin and Altix high-performance computers.
Supplementary Material
Acknowledgments
Funding Support
This research was supported by NIH Grants CA-28038 to S.B. and CA-099194 to N.E.G. Computational infrastructure and systems management was partially supported by CA-75449 to S.B. We gratefully acknowledge TeraGrid resources provided by the Texas Advanced Computing Center supported by the National Science Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations
- PAHs
polycyclic aromatic hydrocarbons
- DB[a,l]P
dibenzo[a,l]pyrene
- B[c]Ph
benzo[c]phenanthrene
- B[a]P
benzo[a]pyrene
- B[g]C
benzo[g]chrysene
- DE
diol epoxide
- anti-DB[a,l]PDE
r11,t12-dihydrodiol-t13,14-epoxide-11,12,13,14-tetrahydrodibenzo[a,l]pyrene
- MD
molecular dynamics
- NER
nucleotide excision repair
- RMSD
root mean square deviation
Footnotes
Supporting Information
MD computation protocols; methods for analyzing best representative structures and hydrogen bond quality indexes; Table S1 through S7, Figure S1 through S9, and Movies S1 through S4. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Dreij K, Seidel A, Jernstrom B. Chem. Res. Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 2.Lagerqvist A, Hakansson D, Prochazka G, Lundin C, Dreij K, Segerback D, Jernstrom B, Tornqvist M, Seidel A, Erixon K, Jenssen D. DNA Repair (Amst) 2008;7:1202–1212. doi: 10.1016/j.dnarep.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Spencer WA, Singh J, Orren DK. Chem Res Toxicol. 2009;22:81–89. doi: 10.1021/tx8001675. [DOI] [PubMed] [Google Scholar]
- 4.Zhong Q, Amin S, Lazarus P, Spratt TE. DNA Repair (Amst) 2010;9:1011–1016. doi: 10.1016/j.dnarep.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Cai Y, Patel DJ, Broyde S, Geacintov NE. J Nucleic Acids 2010. 2010 doi: 10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y, Geacintov NE, Broyde S. In: The Chemical Biology of DNA Damage. Geacintov NE, Broyde S, editors. Wiley-VCH; 2010. [Google Scholar]
- 7.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 8.Luch A. EXS. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 9.Cavalieri EL, Higginbotham S, RamaKrishna NV, Devanesan PD, Todorovic R, Rogan EG, Salmasi S. Carcinogenesis. 1991;12:1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- 10.Cavalieri EL, Higginbotham S, Rogan EG. Polycycl. Aromat. Comp. 1994;6:177–183. [Google Scholar]
- 11.Ralston SL, Lau HH, Seidel A, Luch A, Platt KL, Baird WM. Cancer Res. 1994;54:887–890. [PubMed] [Google Scholar]
- 12.Gill HS, Kole PL, Wiley JC, Li KM, Higginbotham S, Rogan EG, Cavalieri EL. Carcinogenesis. 1994;15:2455–2460. doi: 10.1093/carcin/15.11.2455. [DOI] [PubMed] [Google Scholar]
- 13.Luch A, Seidel A, Glatt H, Platt KL. Chem. Res. Toxicol. 1997;10:1161–1170. doi: 10.1021/tx970005i. [DOI] [PubMed] [Google Scholar]
- 14.Amin S, Desai D, Dai W, Harvey RG, Hecht SS. Carcinogenesis. 1995;16:2813–2817. doi: 10.1093/carcin/16.11.2813. [DOI] [PubMed] [Google Scholar]
- 15.Amin S, Krzeminski J, Rivenson A, Kurtzke C, Hecht SS, el-Bayoumy K. Carcinogenesis. 1995;16:1971–1974. doi: 10.1093/carcin/16.8.1971. [DOI] [PubMed] [Google Scholar]
- 16.Mahadevan B, Dashwood WM, Luch A, Pecaj A, Doehmer J, Seidel A, Pereira C, Baird WM. Environ. Mol. Mutagen. 2003;41:131–139. doi: 10.1002/em.10136. [DOI] [PubMed] [Google Scholar]
- 17.Arif JM, Smith WA, Gupta RC. Carcinogenesis. 1999;20:1147–1150. doi: 10.1093/carcin/20.6.1147. [DOI] [PubMed] [Google Scholar]
- 18.Todorovic R, Devanesan P, Rogan E, Cavalieri E. Chem. Res. Toxicol. 2005;18:984–990. doi: 10.1021/tx049681s. [DOI] [PubMed] [Google Scholar]
- 19.Spencer WA, Singh J, Orren DK. Chem. Res. Toxicol. 2009;22:81–89. doi: 10.1021/tx8001675. [DOI] [PubMed] [Google Scholar]
- 20.Ralston SL, Seidel A, Luch A, Platt KL, Baird WM. Carcinogenesis. 1995;16:2899–2907. doi: 10.1093/carcin/16.12.2899. [DOI] [PubMed] [Google Scholar]
- 21.Devanesan P, Ariese F, Jankowiak R, Small GJ, Rogan EG, Cavalieri EL. Chem. Res. Toxicol. 1999;12:796–801. doi: 10.1021/tx980203p. [DOI] [PubMed] [Google Scholar]
- 22.Mahadevan B, Luch A, Bravo CF, Atkin J, Steppan LB, Pereira C, Kerkvliet NI, Baird WM. Cancer Lett. 2005;227:25–32. doi: 10.1016/j.canlet.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 23.Friedberg EC, Lehmann AR, Fuchs RP. Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 24.Choi JY, Guengerich FP. J Biol Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 25.Kropachev K, Kolbanovskiy M, Rodriguez FA, Cai Y, Ding S, Zhang L, Amin S, Broyde S, Geacintov NE. American Chemical Society 238th National Meeting & Exposition Washington, D.C., USA Chem. Res. Toxicol..2009. p. 282. [Google Scholar]
- 26.Mao B, Gu Z, Gorin A, Chen J, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Biochemistry. 1999;38:10831–10842. doi: 10.1021/bi991212f. [DOI] [PubMed] [Google Scholar]
- 27.Schurter EJ, Yeh HJ, Sayer JM, Lakshman MK, Yagi H, Jerina DM, Gorenstein DG. Biochemistry. 1995;34:1364–1375. doi: 10.1021/bi00004a031. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz JL, Rice JS, Luxon BA, Sayer JM, Xie G, Yeh HJ, Liu X, Jerina DM, Gorenstein DG. Biochemistry. 1997;36:11069–11076. doi: 10.1021/bi971306u. [DOI] [PubMed] [Google Scholar]
- 29.Yan S, Shapiro R, Geacintov NE, Broyde S. J. Am. Chem. Soc. 2001;123:7054–7066. doi: 10.1021/ja0043035. [DOI] [PubMed] [Google Scholar]
- 30.Yeh HJ, Sayer JM, Liu X, Altieri AS, Byrd RA, Lakshman MK, Yagi H, Schurter EJ, Gorenstein DG, Jerina DM. Biochemistry. 1995;34:13570–13581. doi: 10.1021/bi00041a037. [DOI] [PubMed] [Google Scholar]
- 31.Zegar IS, Chary P, Jabil RJ, Tamura PJ, Johansen TN, Lloyd RS, Harris CM, Harris TM, Stone MP. Biochemistry. 1998;37:16516–16528. doi: 10.1021/bi9817616. [DOI] [PubMed] [Google Scholar]
- 32.Zegar IS, Kim SJ, Johansen TN, Horton PJ, Harris CM, Harris TM, Stone MP. Biochemistry. 1996;35:6212–6224. doi: 10.1021/bi9524732. [DOI] [PubMed] [Google Scholar]
- 33.Cosman M, Fiala R, Hingerty BE, Laryea A, Lee H, Harvey RG, Amin S, Geacintov NE, Broyde S, Patel D. Biochemistry. 1993;32:12488–12497. doi: 10.1021/bi00097a029. [DOI] [PubMed] [Google Scholar]
- 34.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 35.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suri AK, Mao B, Amin S, Geacintov NE, Patel DJ. J. Mol. Biol. 1999;292:289–307. doi: 10.1006/jmbi.1999.2974. [DOI] [PubMed] [Google Scholar]
- 37.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 38.Lukin M, de Los Santos C. Chem. Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 39.Yan S, Wu M, Buterin T, Naegeli H, Geacintov NE, Broyde S. Biochemistry. 2003;42:2339–2354. doi: 10.1021/bi0270081. [DOI] [PubMed] [Google Scholar]
- 40.Tan J, Geacintov NE, Broyde S. Chem. Res. Toxicol. 2000;13:811–822. doi: 10.1021/tx000094q. [DOI] [PubMed] [Google Scholar]
- 41.Wu M, Yan SF, Tan J, Patel DJ, Geacintov NE, Broyde S. Front Biosci. 2004;9:2807–2818. doi: 10.2741/1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee A, Lavery R, Bagchi B, Hynes JT. J. Am. Chem. Soc. 2008;130:9747–9755. doi: 10.1021/ja8001666. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Cooper VR, Thonhauser T, Lundqvist BI, Langreth DC. J. Phys. Chem. B. 2009;113:11166–11172. doi: 10.1021/jp905765c. [DOI] [PubMed] [Google Scholar]
- 44.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1983. p. 226. [Google Scholar]
- 45.Johnson RC, Stella S, Heiss JK. In: Protein-Nucleic Acid Interactions. Rice PA, Correll CC, editors. The Royal Society of Chemistry; Cambridge: 2008. p. 397. [Google Scholar]
- 46.Xie XM, Geacintov NE, Broyde S. Biochemistry. 1999;38:2956–2968. doi: 10.1021/bi9825605. [DOI] [PubMed] [Google Scholar]
- 47.Xie XM, Geacintov NE, Broyde S. Chem. Res. Toxicol. 1999;12:597–609. doi: 10.1021/tx990021a. [DOI] [PubMed] [Google Scholar]
- 48.Schwaid A, Liu Z, Y.J. T, Rodriguez FA, Kolbanovskiy A, Cai YQ, Ding S, Amin S, Broyde S, Geacintov NE. American Chemical Society 238th National Meeting & Exposition Washington, D.C., USA. Chem. Res. Toxicol..2009. pp. 281–282. [Google Scholar]
- 49.Huang W, Amin S, Geacintov NE. Chem. Res. Toxicol. 2002;15:118–126. doi: 10.1021/tx010135y. [DOI] [PubMed] [Google Scholar]
- 50.Huang X, Colgate KC, Kolbanovskiy A, Amin S, Geacintov NE. Chem. Res. Toxicol. 2002;15:438–444. doi: 10.1021/tx015588h. [DOI] [PubMed] [Google Scholar]
- 51.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Chem. Res. Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 52.Min JH, Pavletich NP. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 53.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 54.Laryea A, Cosman M, Lin JM, Liu T, Agarwal R, Smirnov S, Amin S, Harvey RG, Dipple A, Geacintov NE. Chem. Res. Toxicol. 1995;8:444–454. doi: 10.1021/tx00045a017. [DOI] [PubMed] [Google Scholar]
- 55.Volk DE, Rice JS, Luxon BA, Yeh HJ, Liang C, Xie G, Sayer JM, Jerina DM, Gorenstein DG. Biochemistry. 2000;39:14040–14053. doi: 10.1021/bi001669l. [DOI] [PubMed] [Google Scholar]
- 56.Frisch JM, Trucks WG, Schlegel BH, Scuseria EG, Robb AM, Cheeseman RJ, Zakrzewski GV, Montgomery AJ, Stratmann ER, Burant CJ, Dappprich S, Millam MJ, Daniels DA, Kudin NK, Strain CM, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson AG, Ayala YP, Cui Q, Morokuma K, Malick KD, Rabuck DA, Raghavachari K, Foresman BJ, Cioslowski J, Ortiz VJ, Baboul GA, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Comperts R, Martin LR, Fox JD, Keith T, Al-Laham AM, Peng YC, Nanayakkara A, Gonzalez C, Challacombe M, Gill WMP, Johnson B, Chen W, Wong WM, Andres LJ, Head-Gordon M, Replogle SE, Pople AJ. Gaussian 98. Gaussian, Inc.; Pittsburgh: 1998. [Google Scholar]
- 57.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco, CA: 2006. [Google Scholar]
- 58.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 59.Cheatham TE, Cieplak P, Kollman PA. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 60.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayly CI, Cieplak P, Cornell WD, Kollman PA. J. Phys. Chem.-Us. 1993;97:10269–10280. [Google Scholar]
- 62.Cieplak P, Cornell WD, Bayly C, Kollman PA. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 63.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER 7. University of California; San Francisco, CA: 2002. [Google Scholar]
- 64.Ravishanker G, Beveridge DL. MD Toolchest 2.0. Wesleyan University; Middletown, CT 06459: 1993. [Google Scholar]
- 65.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 66.Fratini AV, Kopka ML, Drew HR, Dickerson RE. J. Biol. Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 67.Connolly ML. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 68.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 69.Lu XJ, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.