Abstract
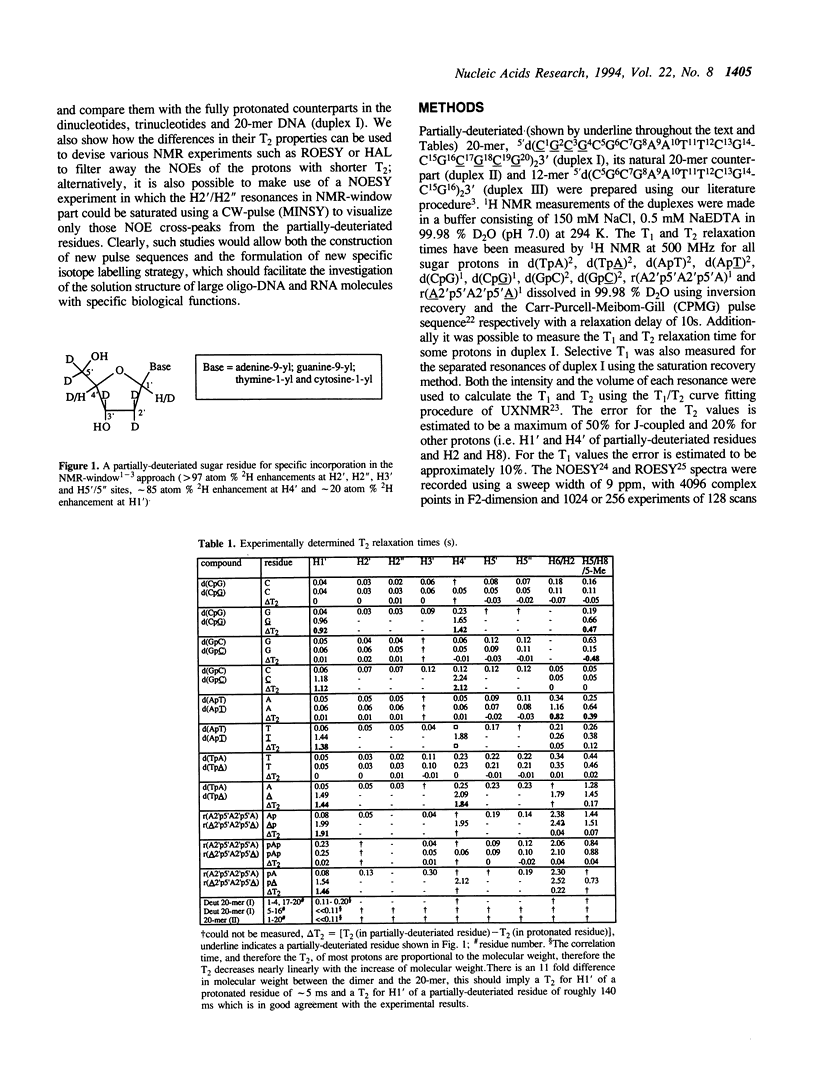
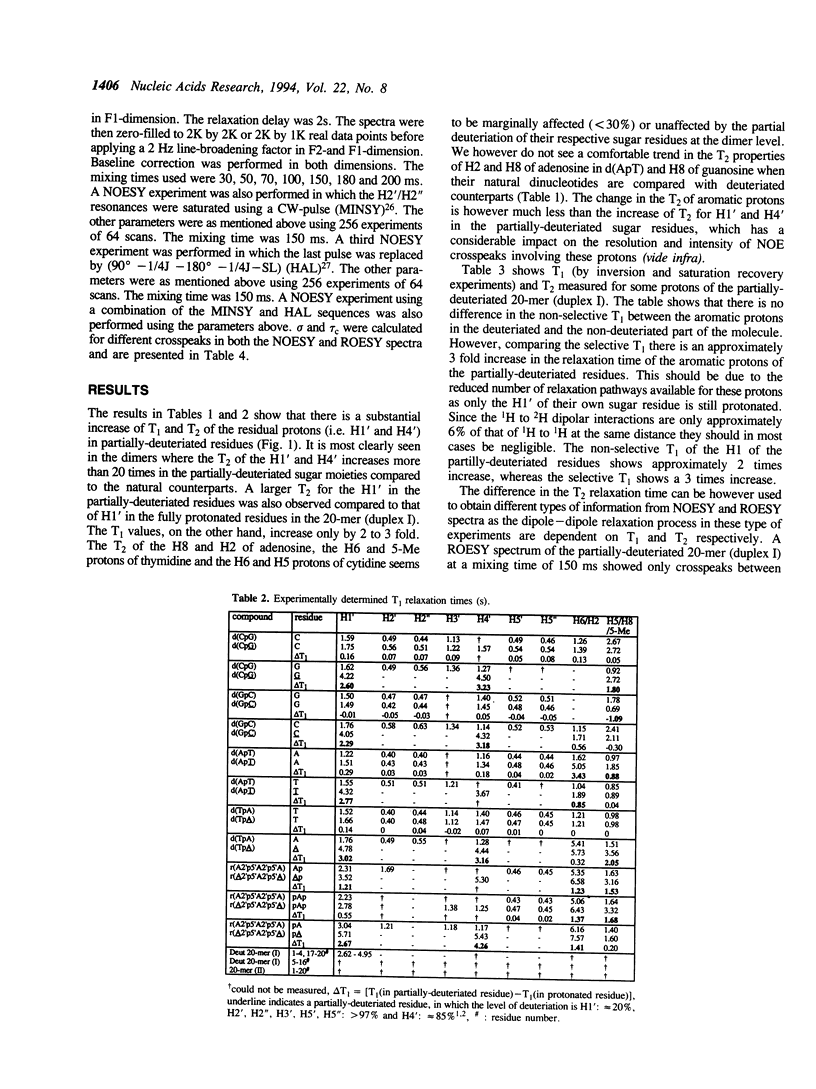
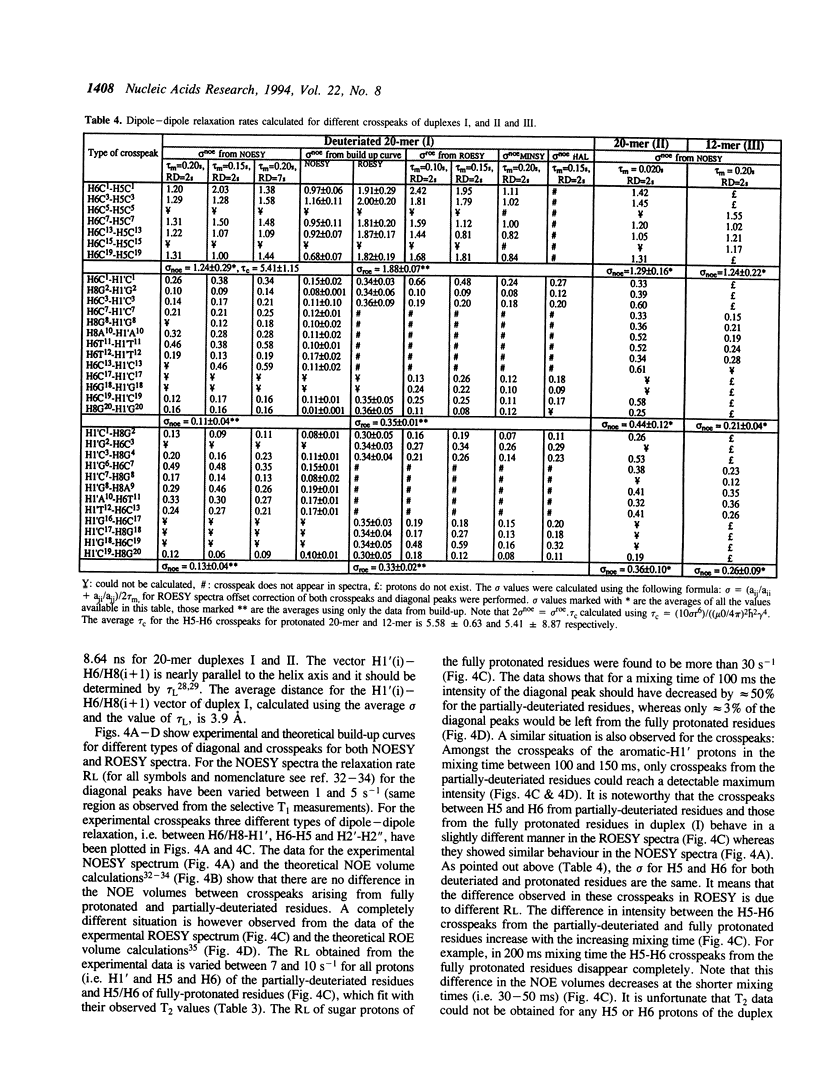
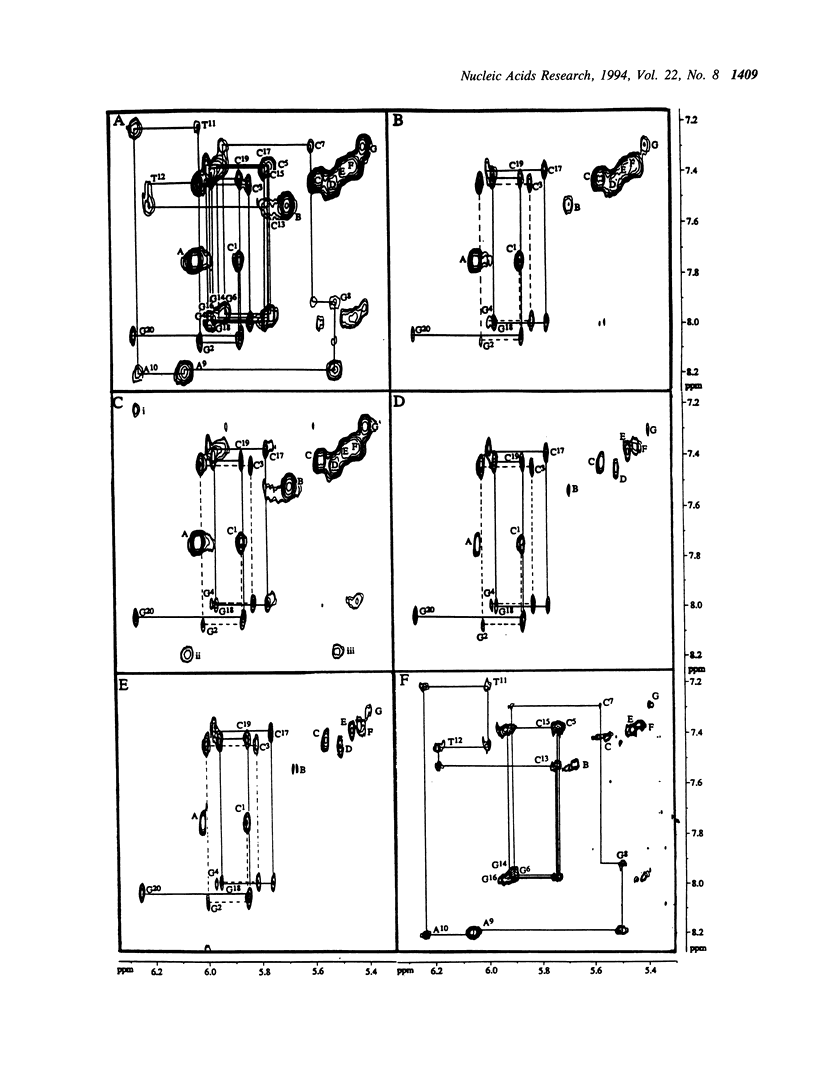
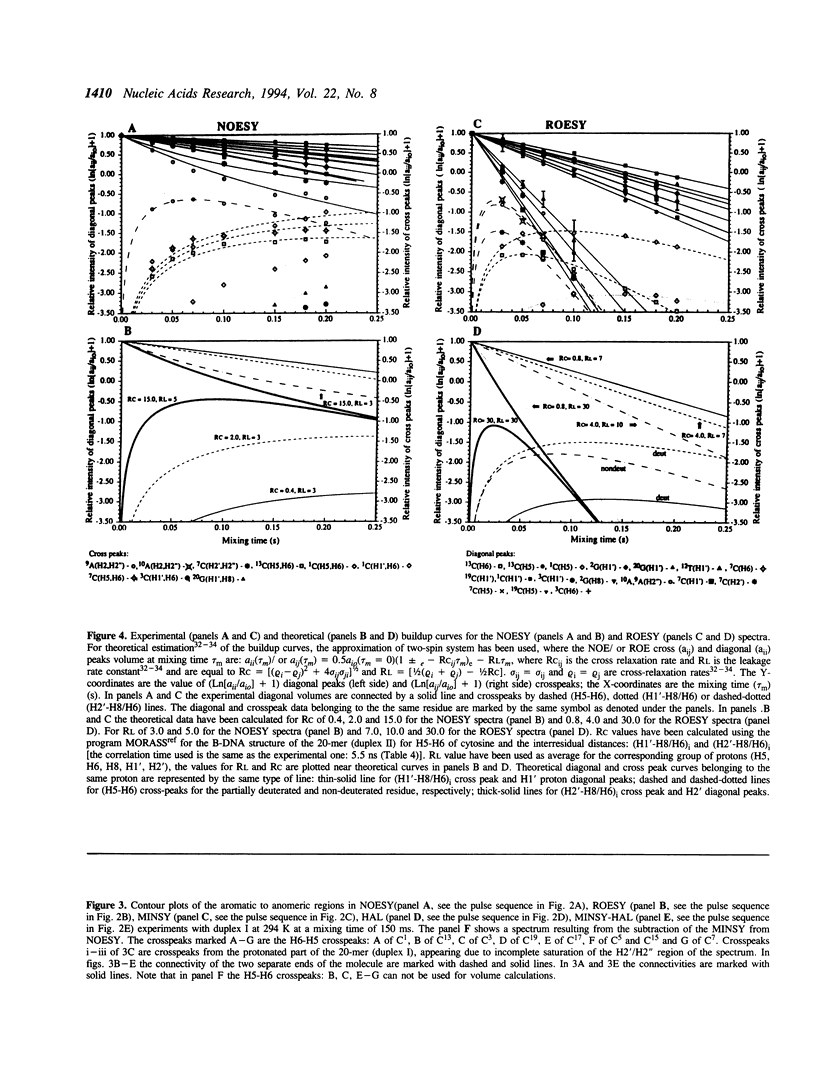
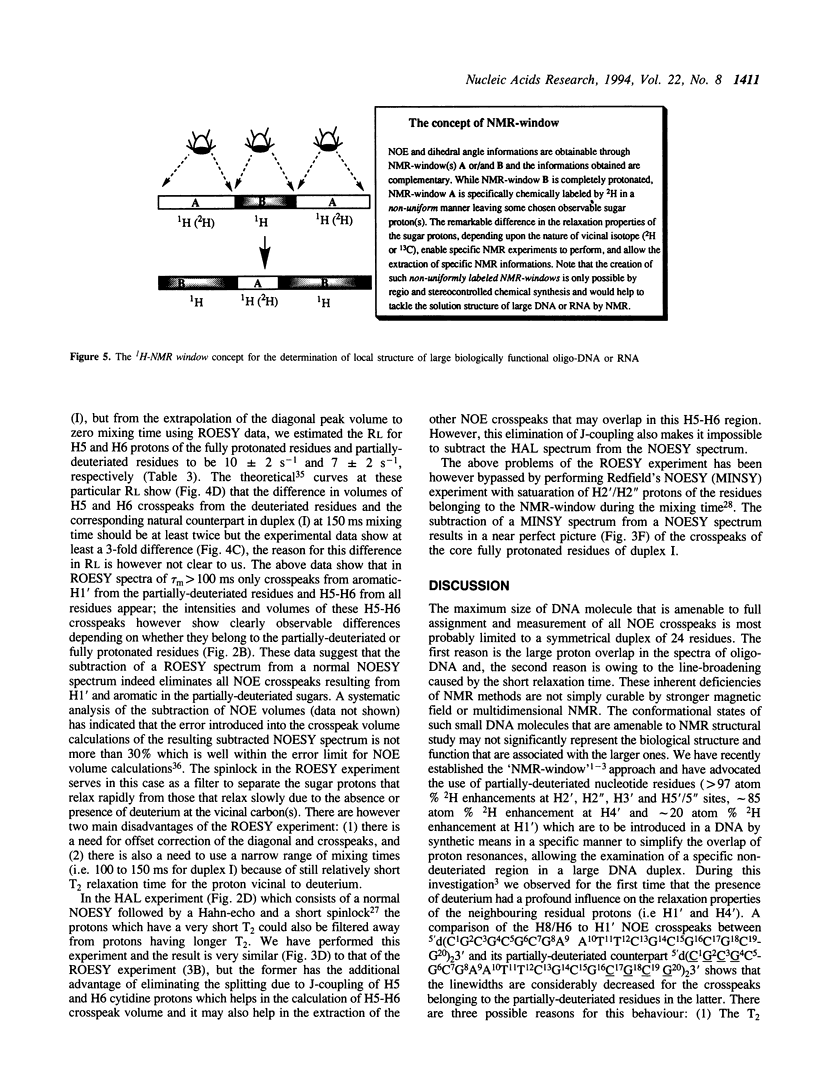
Selective incorporation of the stereospecifically deuteriated sugar moieties (> 97 atom % 2H enhancements at H2', H2'', H3' and H5'/5'' sites, approximately 85 atom % 2H enhancement at H4' and approximately 20 atom % 2H enhancement at H1') in DNA and RNA by the 'NMR-window' approach has been shown to solve the problem of the resonance overlap [refs. 1, 2 & 3]. Such specific deuterium labelling gives much improved resolution and sensitivity of the residual sugar proton (i.e. H1' or H4') vicinal to the deuteriated centers (ref. 3). The T2 relaxation time of the residual protons also increases considerably in the partially-deuteriated (shown by underline) sugar residues in dinucleotides [d(CpG), d(GpC), d(ApT), d(TpA)], trinucleotide r(A2'p5'A2'p5'A) and 20-mer DNA duplex 5'd(C1G2C3-G4C5G6C7G8A9A10T11T12C13G14C15G16C17G18C19G20)(2) 3'. The protons with shorter T2 can be filtered away using a number of different NMR experiments such as ROESY, MINSY or HAL. The NOE intensity of the cross-peaks in these experiments includes only straight pathway from H1' to aromatic proton (i-i and i-i + 1) without any spin-diffusion. The volumes of these NOE cross-peaks could be measured with high accuracy as their intensity is 3 to 4 times larger than the corresponding peaks in the fully protonated residues in the normal NOESY spectra. The structural informations thus obtainable from the residual protons in the partially-deuteriated part of the duplex and the fully protonated part in the 'NMR window' can indeed complement each other.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Batey R. T., Inada M., Kujawinski E., Puglisi J. D., Williamson J. R. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992 Sep 11;20(17):4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu X. H., Avison M. J., Shulman R. G. Nuclear magnetic resonance relaxation of glycogen H1 in solution. Biochemistry. 1993 Sep 14;32(36):9417–9422. doi: 10.1021/bi00087a021. [DOI] [PubMed] [Google Scholar]
- Kushlan D. M., LeMaster D. M. Resolution and sensitivity enhancement of heteronuclear correlation for methylene resonances via 2H enrichment and decoupling. J Biomol NMR. 1993 Nov;3(6):701–708. doi: 10.1007/BF00198372. [DOI] [PubMed] [Google Scholar]
- LeMaster D. M. Deuterium labelling in NMR structural analysis of larger proteins. Q Rev Biophys. 1990 May;23(2):133–174. doi: 10.1017/s0033583500005527. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E. P., Pardi A. Three-dimensional heteronuclear NMR studies of RNA. Nature. 1992 Jan 9;355(6356):184–186. doi: 10.1038/355184a0. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E. P., Sirr A., Legault P., Jucker F. M., Baer L. M., Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992 Sep 11;20(17):4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Nakamura H., Yamazaki T., Nagayama K., Yoshida M., Kanaya S., Ikehara M. 1H NMR studies of deuterated ribonuclease HI selectively labeled with protonated amino acids. J Biomol NMR. 1992 Mar;2(2):137–147. doi: 10.1007/BF01875525. [DOI] [PubMed] [Google Scholar]
- Pachter R., Arrowsmith C. H., Jardetzky O. The effect of selective deuteration on magnetization transfer in larger proteins. J Biomol NMR. 1992 Mar;2(2):183–194. doi: 10.1007/BF01875529. [DOI] [PubMed] [Google Scholar]
- Varani G., Tinoco I., Jr RNA structure and NMR spectroscopy. Q Rev Biophys. 1991 Nov;24(4):479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Hinck A. P., Loh S. N., LeMaster D. M., Markley J. L. Solution studies of staphylococcal nuclease H124L. 2. 1H, 13C, and 15N chemical shift assignments for the unligated enzyme and analysis of chemical shift changes that accompany formation of the nuclease-thymidine 3',5'-bisphosphate-calcium ternary complex. Biochemistry. 1992 Jan 28;31(3):921–936. doi: 10.1021/bi00118a039. [DOI] [PubMed] [Google Scholar]
- Wang J. F., LeMaster D. M., Markley J. L. Two-dimensional NMR studies of staphylococcal nuclease. 1. Sequence-specific assignments of hydrogen-1 signals and solution structure of the nuclease H124L-thymidine 3',5'-bisphosphate-Ca2+ ternary complex. Biochemistry. 1990 Jan 9;29(1):88–101. doi: 10.1021/bi00453a011. [DOI] [PubMed] [Google Scholar]
- Weisz K., Shafer R. H., Egan W., James T. L. The octamer motif in immunoglobulin genes: extraction of structural constraints from two-dimensional NMR studies. Biochemistry. 1992 Aug 25;31(33):7477–7487. doi: 10.1021/bi00148a007. [DOI] [PubMed] [Google Scholar]


