Abstract
The inner ear is composed of a cochlear duct and five vestibular organs in which mechanosensory hair cells play critical roles in receiving and relaying sound and balance signals to the brain. To identify novel genes associated with hair cell differentiation or function, we analyzed an archived gene expression dataset from embryonic mouse inner ear tissues. Since atonal homolog 1a (Atoh1) is a well known factor required for hair cell differentiation, we searched for genes expressed in a similar pattern with Atoh1 during inner ear development. The list from our analysis includes many genes previously reported to be involved in hair cell differentiation such as Myo6, Tecta, Myo7a, Cdh23, Atp6v1b1, and Gfi1. In addition, we identified many other genes that have not been associated with hair cell differentiation, including Tekt2, Spag6, Smpx, Lmod1, Myh7b, Kif9, Ttyh1, Scn11a and Cnga2. We examined expression patterns of some of the newly identified genes using real-time polymerase chain reaction and in situ hybridization. For example, Smpx and Tekt2, which are regulators for cytoskeletal dynamics, were shown specifically expressed in the hair cells, suggesting a possible role in hair cell differentiation or function. Here, by reanalyzing archived genetic profiling data, we identified a list of novel genes possibly involved in hair cell differentiation.
Keywords: Inner ear, Hair cell, Atoh1, Differentiation
Introduction
The mammalian inner ear is a highly intricate organ composed of a cochlear duct and vestibular organs, responsible for hearing and balance, respectively (Fig. 1A). The hair cells residing in each sensory organ of the inner ear play a primary role in sensory transduction by converting sound and balance information into electrical signals, which are then transmitted to their associated neurons to convey the signals to the brain. In humans, these hair cells can easily be damaged by various causes including noise, drugs, aging, or genetic mutations. The damage is irreversible in most cases, leading to permanent hearing loss [1]. Thus far, efforts to preserve or regenerate hair cells using stem cells or gene therapy have met with only limited success [2-4]. A better understanding of normal hair cell development may help overcome such limitations.
Fig. 1.
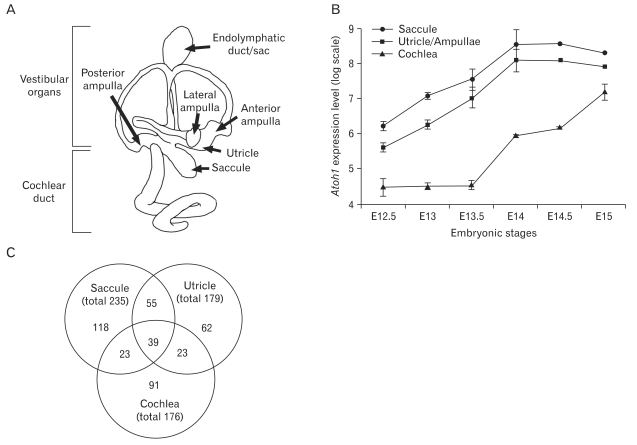
Atoh1 expression patterns during inner ear development and the numbers of genes expressed in a similar pattern with Atoh1. (A) Diagrammatic representation of the substructures of the mouse inner ear. (B) Expression levels of Atoh1 in the cochlea, saccule, and utricle/ampullae mixture from embryonic day 12.5 (E12.5) to E15 extracted from the dataset GSE7536 and plotted on a logarithmic scale. (C) The number of genes expressed in a similar pattern with Atoh1 in each organ analyzed from the dataset GSE7536 and the number of genes found in two or more organs.
Inner ear hair cells are formed via a series of cell fate decision and differentiation processes which are tightly regulated by a combination of extrinsic and intrinsic factors [5]. Among such factors, the basic helix-loop-helix transcription factor, Atoh1 (Atonal homolog, also known as Math1), has been shown to be a major determinant in hair cell differentiation. In Atoh1 knockouts, both cochlear and vestibular hair cells failed to differentiate [6], while delivering the Atoh1 gene induced formation of ectopic hair cells in tissue cultures as well as animal models [3, 7, 8]. These results indicate a crucial role of Atoh1 in hair cell differentiation, yet how Atoh1 functions to dictate hair cell differentiation remains poorly understood.
In this study, to identify novel genes that play a role in hair cell differentiation, we analyzed an archived gene expression dataset, generated by extensive microarray analysis using mouse inner ear tissues at several embryonic stages [9]. Since Atoh1 plays a key role in hair cell differentiation [5], we assumed that genes involved in hair cell differentiation would be regulated directly or indirectly by Atoh1 and their expression patterns would be closely associated with that of Atoh1 during inner ear development. For example, expression patterns of Gfi1, Pou4f3, and Myo7a, which are important for hair cell differentiation, have been shown to resemble that of Atoh1 during inner ear development [10, 11]. We identified several genes that showed similar expression patterns to that of Atoh1 during inner ear development, some of which were confirmed as being specifically expressed in developing hair cells.
Materials and Methods
Data analysis
We analyzed a gene expression dataset (NCBI Gene Expression Omnibus; GSE7536), generated by microarray analysis (mouse MOE430A_2 gene chip; Affymetrix, Santa Clara, CA, USA) using inner ear tissues from CBA/J mouse embryos, which were micro-dissected at half-day intervals from embryonic day 9 (E9) to E15 [9]. The experimental procedures and statistical analysis used for the GSE7536 dataset have been described in detail in the original paper [9]. In particular, we focused our analysis on expression profiles between E12.5 to E15, when Atoh1 expression begins and rapidly increases. In the dataset, the cochlea, saccule, and utricle/ampullae mixtures were divided and profiled separately [9]. In addition, non-ear tissues were also isolated from each stage and profiled [9]. We first applied Pearson's correlation to the dataset to eliminate genes exhibiting low degrees of correlation (-0.5<r<0.5) compared to the Atoh1 expression pattern. We then segregated the profiles into two groups by developmental stage according to Atoh1 expression levels (E12.5, E13, E13.5 vs. E14, E14.5, E15) for cochlear samples and (E12.5, E13 vs. E14, E14.5, E15) for utricle and saccule samples. Significant analysis of microarrays [12] was applied to identify genes differentially expressed between the two groups with at least two-fold changes and an estimated false discovery rate of ≤1%. We particularly focused on genes positively correlated with Atoh1 expression patterns. Information regarding gene ontology was extracted from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/).
Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR)
Timed pregnant C57BL/6 mice were euthanized at E13.5, E15.5, and E17.5 at which time the embryos were collected in ice-cold phosphate buffered saline (PBS). Embryonic inner ear tissues from each stage were micro-dissected under a stereomicroscope (SZX10, Olympus, Tokyo, Japan) and the vestibular and cochlear tissues were collected separately. Total RNAs were isolated from each tissue sample using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). RNA quality was assessed by agarose gel electrophoresis of an aliquot of total RNA. The cDNA was synthesized from each RNA sample using poly(dT)20 and a Superscript III RT kit according to the manufacturer's instructions (Invitrogen). Real-time PCR was performed using SYBR Green PCR kits and an ABI 7500 machine (Applied Biosystems, Carlsbad, CA, USA). Primers used for real-time PCR are listed in Table 1. Relative expression levels in each stage were determined by comparative methods.
Table 1.
Primers used in real-time polymerase chain reaction
In situ hybridization
In situ hybridization was performed as previously described [13]. Briefly, embryos were fixed overnight in 4% paraformaldehyde in PBS, dehydrated in 30% sucrose, and embedded in O.C.T. (Tissue-Tek, Sakura Finetechnical Co., Tokyo, Japan). Embryos were then sectioned at a thickness of 12-µm onto Superfrost slides (VWR Scientific, West Chester, PA, USA). The slides were dried at room temperature overnight. Before in situ hybridization, slides were rehydrated, post-fixed, and permeabilized using 10 µg/ml proteinase K for 2-5 minutes. Hybridization was performed in Seal-a-Meal bags (Kapak, Minneapolis, MN, USA). Each bag contained four slides and 5-7 ml of hybridization solution with a DIG-labeled RNA probe. RNA probes for Tekt2 were generated from a fragment of mouse Tekt2 cDNA containing -46 to +531 nucleotides (GenBank accession number: NM_011902.2), and for Smpx, from a 664 base pair (bp) mouse Smpx cDNA fragment containing the entire open reading frame as well as 112 bp 5' and 294 bp 3' untranslated regions (GenBank accession number: NM_025357.2).
Results
Search for genes concordantly expressed with Atoh1 in the developing inner ear
To identify genes involved in differentiation of mechanosensory hair cells, we took advantage of the vast amount of gene expression profiles available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). We chose dataset GSE7536, in which mouse inner ear tissues from E9 to E15 had been micro-dissected at half-day intervals and subjected to microarray analysis [9]. This dataset provides comprehensive gene expression profiles for substructures of the inner ear tissues including the cochlea, saccule, and utricle/ampullae mixture ("utricle" hereafter) during critical periods of hair cell differentiation.
Hair cells in the vestibular organs such as saccule and utricle differentiate earlier than those in the cochlea. Consistently, Atoh1 expression in the vestibular organs is obvious around E12.5, but not in the cochlea until E14.5 [14, 15]. Our analysis of the GSE7536 dataset confirmed such expression patterns of Atoh1 during inner ear development (Fig. 1B). While Atoh1 expression levels in the vestibular organs (utricle and saccule) gradually increased from E12.5 and plateaued at E14, cochlear Atoh1 expression remained low until E13.5 and began to increase at E14 (Fig. 1B). In order to identify genes exhibiting expression patterns similar to Atoh1 during these developmental stages, we divided the expression profiles into two groups by developmental stage according to Atoh1 expression levels. For cochlear samples, expression profiles of E12.5, E13, and E13.5 were combined and designated as 'low Atoh1 stages' and those of E14, E14.5, and E15 were combined and designated as 'high Atoh1 stages.' For saccule and utricle samples, expression profiles of E12.5 and E13 were combined as 'low Atoh1 stages' and those of E14, E14.5, and E15 were combined as 'high Atoh1 stages.' The mean expression levels of Atoh1 were at least three fold higher in the 'high Atoh1 stages' compared to the 'low Atoh1 stages,' indicating that expression levels of Atoh1 significantly differ between the two groups.
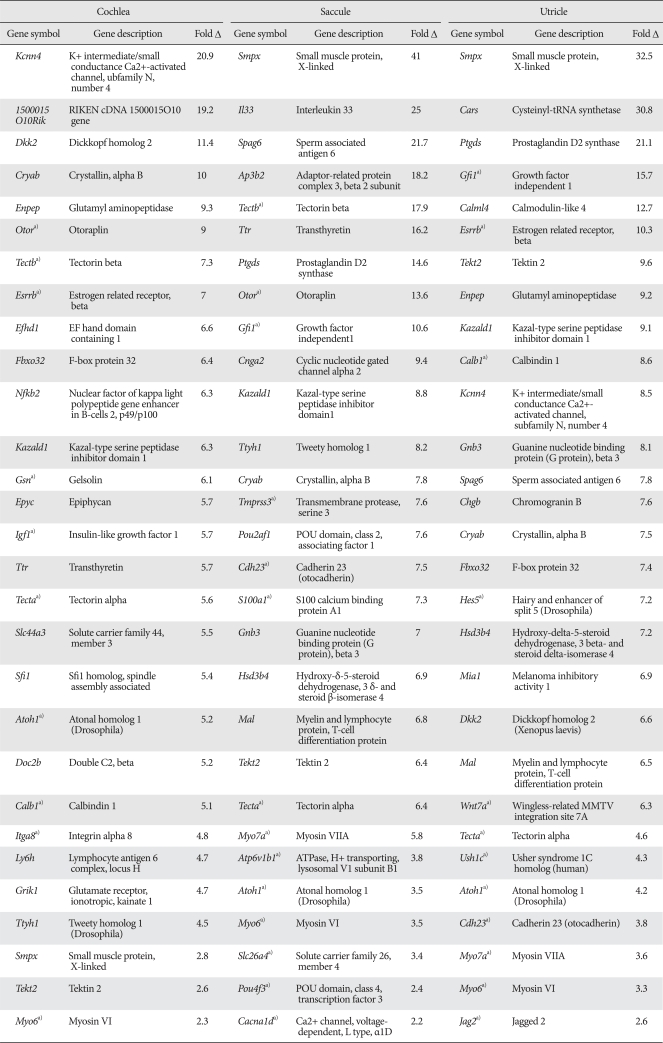
By applying Pearson's correlation and comparing fold-change differences, we searched for genes whose expression patterns positively correlated with that of Atoh1 and showed at least a two-fold difference between 'low Atoh1 stages' and 'high Atoh1 stages' from each organ sample. A total of 176 cochlea, 179 utricle, and 235 saccule genes were identified from each organ, and 140 genes were found in at least in two organs and 39 genes were found in all three organs (Fig. 1C). Some examples exhibiting high-fold changes from each organ are listed in Table 2. The list contains several genes shown to be involved in inner ear development, hair cell differentiation or hereditary deafness such as Myo6, Tecta, Tmprss3, Atp6v1b1, Slc26A4, Pou4f3, Gfi1, Tecta, Tectb, Hes5 and Jagged 2 [16-20].
Table 2.
Examples of the genes expressed concordantly with Atoh1 during inner ear development
a)Genes previously associated with inner ear development or deafness.
Genes not previously associated with hair cell differentiation or hearing loss
The fact that many genes previously associated with hair cell differentiation or hearing loss are included in our list (Table 2) demonstrates the validity of our methodology and also suggests that novel genes previously unrecognized, yet playing important roles in hair cell differentiation, may be included in the list.
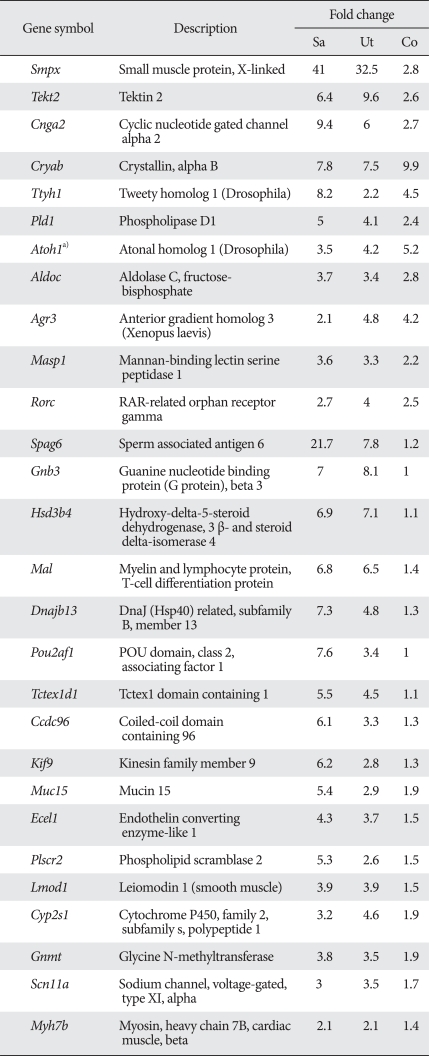
To increase our chance of identifying novel inner ear genes, we removed the genes previously associated with inner ear development such as Atoh1, Myo6, Myo7a, Wnt7a, and Gfi1, and also excluded genes that are abundantly expressed in non-inner ear tissues. Thus, the resulting list contains genes that are expressed in a similar pattern with Atoh1 but have not previously been associated with inner ear development or function (Table 3). These genes include Tekt2 and Spag6, which are implicated in ciliary or flagellar motility [21-23], Smpx, Lmod1, Myh7b, and Kif9, implicated in muscle contraction or cytoskeleton dynamics [24-28], transmembrane ion transporters Ttyh1, Scn11a, and Cnga2 [29-33], and transcription factors such as Rorc and Pou2af1 [29-33].
Table 3.
Examples of genes concordantly expressed with Atoh1 in ear tissues but not in non-ear tissues
Some seemingly interesting genes that were changed <two-fold in the cochlea were also included. a)Genes previously associated with inner ear development or deafness were excluded from the list, except for Atoh1.
Real-time PCR analyses for the selected genes
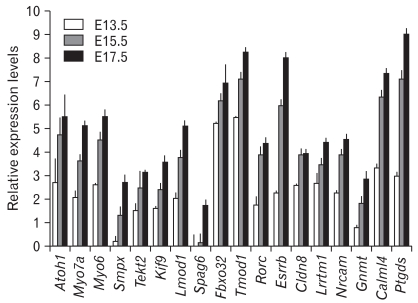
To confirm that the genes included in Tables 2 and 3 are indeed expressed in a similar pattern with Atoh1 during inner ear development, we dissected mouse inner ear tissues at E13.5, E15.5, and E17.5 and performed quantitative RT-PCR (Fig. 2). Since the gradual increase of Atoh1 expression levels was more obvious in the cochlea (Fig. 1), we used the cochlear tissues for our RT-PCR experiments. The examined genes included some known hair cell-specific genes as controls (Atoh1, Myo7a, Myo6), genes associated with cytoskeletal structures or regulation (Smpx, Tekt2, Kif9, Lmod1, Spag6, Fbxo32, Tmod1), and genes encoding nuclear proteins (Rorc, Esrrb), membrane proteins (Cldn8, Lrrtm1, Nrcam), and various other functional proteins (Gnmt, Calml4, Ptgds).
Fig. 2.
Real-time polymerase chain reaction analysis of the selected genes. Total RNAs isolated from mouse cochlear tissues at E13.5, E15.5, and E17.5 subjected to quantitative real-time polymerase chain reaction. Expression levels of the genes examined gradually increased similar to that of Atoh1.
Real-time PCR results confirmed that genes associated with hair cell differentiation such as Myo7a and Myo6 showed similar expression patterns with Atoh1 during inner ear development (Fig. 2). Other genes tested also gradually increased similar to Atoh1, although relative expression levels of each gene were different from each other to some extent. These results demonstrate that the microarray results from the GEO dataset, GSE7536, closely represent gene expression patterns in vivo (Fig. 2).
Expression of Smpx and Tekt2 in developing hair cells
We next examined whether the genes confirmed by real-time PCR are expressed specifically in the developing hair cells by in situ hybridization. As examples, we chose a few genes associated with cytoskeletal dynamics or architecture which would be directly related to differentiation or maturation of the hair cells.
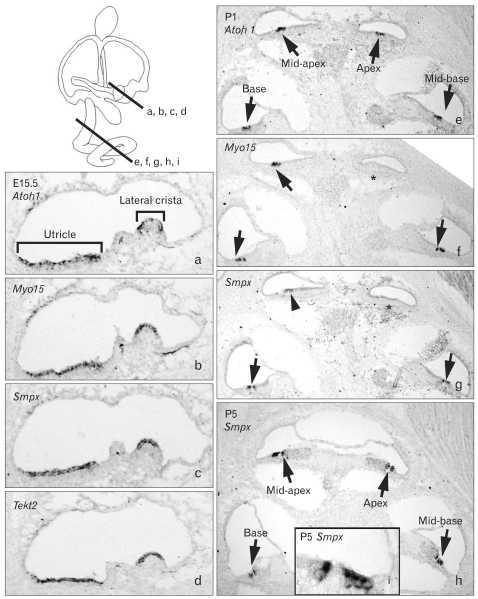
Small muscle protein, X-linked (Smpx) showed the highest fold changes in both saccule and utricle during inner ear development (Table 3). Smpx expression was observed in the hair cells of the vestibular organs including utricle, saccule, and all three cristae at E15.5 (Fig. 3a-c) and continued to be present at least up to P5 (data not shown). In the cochlea, Smpx expression was barely detected at E15.5 (data not shown), probably because differentiation of the cochlear hair cells occurs later than that of the vestibular hair cells [14, 15]. Thus, we compared the cochlear expression patterns of Smpx in later stages with Atho1 as well as Myo15, which is known to be essential for hair cell maturation [34]. At P1, Atoh1 expression was detected in all cochlear turns from the base to apex (Fig. 3e, arrow), but Myo15 expression was only detected up to the mid-apical turn (Fig. 3f, arrows), showing the wave of hair cell differentiation in a base to apex progression [14, 35]. At this stage (P1), Smpx expression was clearly found in the base and mid-base of the cochlear duct (Fig. 3g, arrows) and faintly in the mid-apex (Fig. 3g, arrowhead), indicating that the onset of Smpx expression is slightly later than that of Myo15. By P5, Smpx was strongly expressed in the hair cells of all cochlear turns (Fig. 3h, i).
Fig. 3.
Expression patterns of Smpx and Tekt2 in the developing inner ear. Expression patterns of Smpx and Tekt2 in the developing inner ear analyzed with in situ hybridization at E15.5 (a-d), P1 (e-g), and P5 (h, i). Lines in the inner ear diagram show the levels of sections. Atoh1 (a, e) and Myo15 (b, f), well known markers for hair cells, were used to locate the hair cells in the inner ear tissues. At E15.5, expressions of Smpx and Tekt2 were found in the developing hair cells of the vestibular organs and continued up to P5 (a-d; data not shown), but were barely detected in the cochlea at this stage (data not shown). At P1, Atoh1 expression was found in the hair cells of all cochlear turns (e, arrows) and Myo15 expression was found in the base, mid-base, and mid-apex of the cochlea (f, arrows), but not yet in the apex (f, asterisk). Smpx expression was found in hair cells of basal and mid-basal turns (g, arrows) and weakly in the mid-apical turn (g, arrowhead). At P5, Smpx expression was detected in hair cells of all cochlear turns (h, i, arrows).
Another gene found to be expressed in the hair cells is Tekt2, which encodes Tektin 2, a member of the Tektin family of proteins [21, 22]. Tektins are known as constitutive proteins of microtubules contributing to the stability and structural complexes of axonemal microtubules [22]. Tekt2 expression was observed in the hair cells of all vestibular organs from E15.5 to P5 (Fig. 3d) (data not shown), whereas its expression in the cochlea was observed faintly in the basal and mid-basal turns at E15.5 and barely in the neonatal cochlea (data not shown). These expression patterns of Tekt2 may be related to the transient appearance of the microtubule-based kinocilium in the cochlear hair cells, in contrast to the vestibular hair cells that retain kinocilium [36].
Discussion
To identify novel genes playing a role in hair cell differentiation, we re-analyzed the gene expression dataset GSE7536 [9] and searched for genes concordantly expressed with Atoh1 during hair cell differentiation. As expected, the list includes many genes previously associated with inner ear development, hair cell differentiation or hereditary deafness (Table 2). For example, Myo6, Tecta, and Tmprss3 have been associated with autosomal recessive or dominant deafness [16-18]. Also included were Atp6v1b1 and Slc26A4, which are involved in maintenance of pH and ion composition in the inner ear fluids [19, 20]. Several other genes, such as transcription factors Pou4f3 and Gfi1, extracellular matrix (tectorial membrane) components Tecta and Tectb, and components of the Notch signaling pathway Hes5 and Jagged 2 were also included.
Interestingly, some genes known to be important for shaping the stereocilia of the hair cells such as Myo7a, Cdh23, and Ush1c were identified as being concordantly expressed with Atoh1 only in the vestibular organs but not in the cochlea from the evaluated dataset. This suggests that since the dataset used in this study covers expression profiles only up to E15 [9], the genes included in the dataset do not accurately represent gene expression changes in later stages (i.e., later than E15) and may not include some important genes involved in maturation of the hair cells, especially in the cochlea. Thus, careful attention should be paid when comparing the genes identified from the vestibular organs and the cochlea.
From the list of genes concordantly expressed with Atoh1, we further excluded ones previously associated with inner ear as well as ones abundantly expressed in non-inner ear tissues to increase our chance to identify novel genes important for hair cell differentiation (Table 3). However, not all genes in the list were expressed in the hair cells but were in other parts of the inner ear such as cochleovestibular ganglion neurons, supporting cells, or mesenchyme-derived structures (data not shown). This is because hair cell differentiation dictated by Atoh1 occurs in harmony with development of the entire inner ear as a whole. Thus, specific expression domains in the developing inner ear should be carefully examined for each candidate.
As examples, we found that Smpx and Tekt2 were specifically expressed in the hair cells of developing inner ear. Smpx encodes an 85-amino acid protein, which is associated with actin and focal adhesion complexes and functions as a regulator of cytoskeletal dynamics [25, 27]. The onset of Smpx expression in the developing hair cells appeared to be slightly later than Myo15 (Fig. 3), suggesting a possible role of Smpx in maturation or maintenance, rather than early specification, of hair cells. Tekt2 encodes Tektin 2, also known as Tektin-t. It has been shown that Tekt2 is expressed in sperm flagellum and the loss of Tekt2 leads to a defect in the dynein arms of flagellar axonemal microtubules, resulting in immotile sperm and male infertility [21, 22, 37]. Interestingly, male infertility is often associated with hearing loss, especially in patients suffering from primary ciliary dyskinesia [38]. Since our observation of Tekt2 expression in the developing hair cells suggests a possible role of Tekt2 in hair cell differentiation, especially in kinocilium mainly composed of microtubules, it will be interesting to examine whether the loss of Tekt2 causes hearing loss in addition to male infertility.
Taking advantage of gene expression profiles that are readily available in the GEO database [9], we identified several genes not previously associated with, yet possibly involved in hair cell differentiation. Further investigation of the genes identified in this study will enhance our understanding of how the hair cells normally develop and function.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (KRF-2008-331-E00010 and 2009-0066470).
References
- 1.Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003;9:1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- 3.Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 4.Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 6.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 7.Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 8.Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sajan SA, Warchol ME, Lovett M. Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics. 2007;177:631–653. doi: 10.1534/genetics.107.078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- 11.Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yücel R, Frankel WN, Rechavi G, Möröy T, Friedman TB, Kelley MW, Avraham KB. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 15.Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 16.Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet. 1999;8:409–412. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- 17.Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed ZM, Morell RJ, Riazuddin S, Gropman A, Shaukat S, Ahmad MM, Mohiddin SA, Fananapazir L, Caruso RC, Husnain T, Khan SN, Riazuddin S, Griffith AJ, Friedman TB, Wilcox ER. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet. 2003;72:1315–1322. doi: 10.1086/375122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett LA, Morsli H, Wu DK, Green ED. Expression pattern of the mouse ortholog of the Pendred's syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc Natl Acad Sci U S A. 1999;96:9727–9732. doi: 10.1073/pnas.96.17.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover EH, Borthwick KJ, Bavalia C, Eady N, Fritz DM, Rungroj N, Giersch AB, Morton CC, Axon PR, Akil I, Al-Sabban EA, Baguley DM, Bianca S, Bakkaloglu A, Bircan Z, Chauveau D, Clermont MJ, Guala A, Hulton SA, Kroes H, Li Volti G, Mir S, Mocan H, Nayir A, Ozen S, Rodriguez Soriano J, Sanjad SA, Tasic V, Taylor CM, Topaloglu R, Smith AN, Karet FE. Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss. J Med Genet. 2002;39:796–803. doi: 10.1136/jmg.39.11.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iguchi N, Tanaka H, Fujii T, Tamura K, Kaneko Y, Nojima H, Nishimune Y. Molecular cloning of haploid germ cell-specific tektin cDNA and analysis of the protein in mouse testis. FEBS Lett. 1999;456:315–321. doi: 10.1016/s0014-5793(99)00967-9. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004;24:7958–7964. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Jones BH, Tang W, Moss SB, Wei Z, Ho C, Pollack M, Horowitz E, Bennett J, Baker ME, Strauss JF., 3rd Dissecting the axoneme interactome: the mammalian orthologue of Chlamydomonas PF6 interacts with sperm-associated antigen 6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Proteomics. 2005;4:914–923. doi: 10.1074/mcp.M400177-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 25.Kemp TJ, Sadusky TJ, Simon M, Brown R, Eastwood M, Sassoon DA, Coulton GR. Identification of a novel stretch-responsive skeletal muscle gene (Smpx) Genomics. 2001;72:260–271. doi: 10.1006/geno.2000.6461. [DOI] [PubMed] [Google Scholar]
- 26.Piddini E, Schmid JA, de Martin R, Dotti CG. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001;20:4076–4087. doi: 10.1093/emboj/20.15.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindeler A, Lavulo L, Harvey RP. Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp Cell Res. 2005;307:367–380. doi: 10.1016/j.yexcr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 28.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trudeau MC, Zagotta WN. Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J Biol Chem. 2003;278:18705–18708. doi: 10.1074/jbc.R300001200. [DOI] [PubMed] [Google Scholar]
- 30.Priest BT, Murphy BA, Lindia JA, Diaz C, Abbadie C, Ritter AM, Liberator P, Iyer LM, Kash SF, Kohler MG, Kaczorowski GJ, MacIntyre DE, Martin WJ. Contribution of the tetrodotoxin-resistant voltage-gated sodium channel NaV1.9 to sensory transmission and nociceptive behavior. Proc Natl Acad Sci U S A. 2005;102:9382–9387. doi: 10.1073/pnas.0501549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki M. The Drosophila tweety family: molecular candidates for large-conductance Ca2+-activated Cl- channels. Exp Physiol. 2006;91:141–147. doi: 10.1113/expphysiol.2005.031773. [DOI] [PubMed] [Google Scholar]
- 33.Bordon A, Bosco N, Du Roure C, Bartholdy B, Kohler H, Matthias G, Rolink AG, Matthias P. Enforced expression of the transcriptional coactivator OBF1 impairs B cell differentiation at the earliest stage of development. PLoS One. 2008;3:e4007. doi: 10.1371/journal.pone.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belyantseva IA, Boger ET, Friedman TB. Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci U S A. 2003;100:13958–13963. doi: 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bok J. Building the mammalian cochlea: an overview. Genes Genomics. 2010;32:1–7. [Google Scholar]
- 36.Sobkowicz HM, Slapnick SM, August BK. The kinocilium of auditory hair cells and evidence for its morphogenetic role during the regeneration of stereocilia and cuticular plates. J Neurocytol. 1995;24:633–653. doi: 10.1007/BF01179815. [DOI] [PubMed] [Google Scholar]
- 37.Shimasaki S, Yamamoto E, Murayama E, Kurio H, Kaneko T, Shibata Y, Inai T, Iida H. Subcellular localization of Tektin2 in rat sperm flagellum. Zoolog Sci. 2010;27:755–761. doi: 10.2108/zsj.27.755. [DOI] [PubMed] [Google Scholar]
- 38.Holzmann D, Ott PM, Felix H. Diagnostic approach to primary ciliary dyskinesia: a review. Eur J Pediatr. 2000;159:95–98. doi: 10.1007/pl00013813. [DOI] [PubMed] [Google Scholar]