Abstract
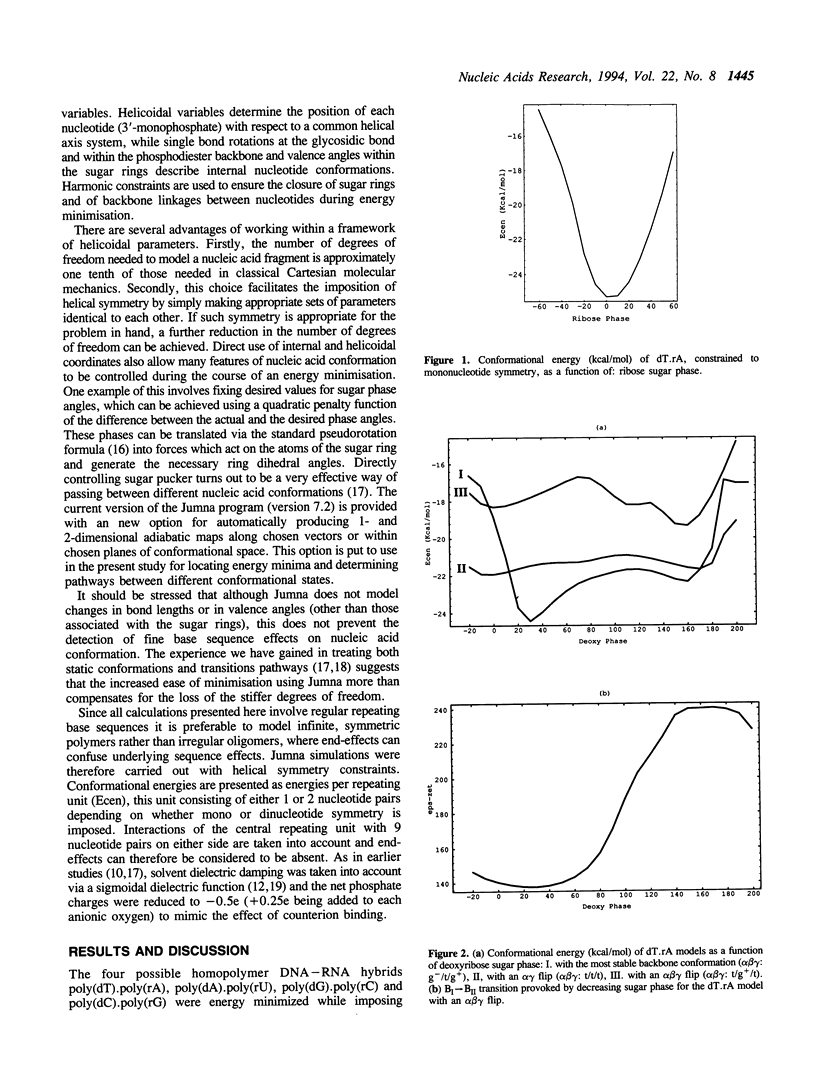
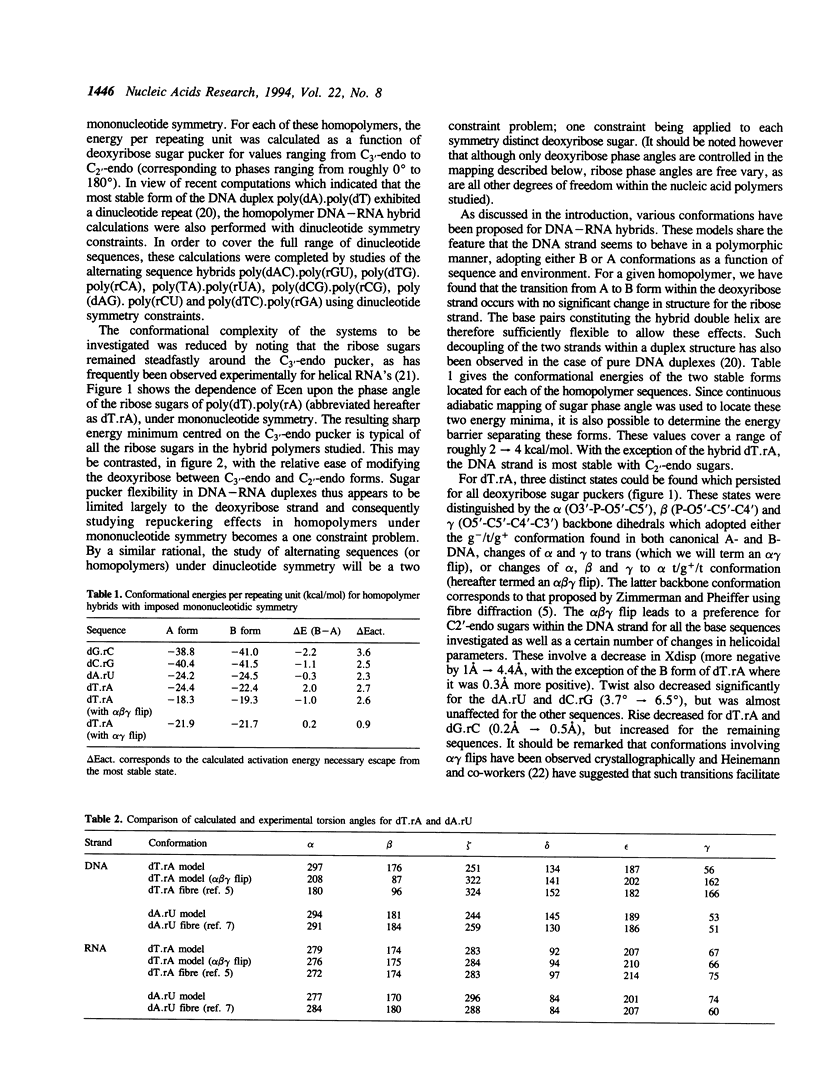
Molecular modelling has been used to probe the conformational preferences of double stranded DNA-RNA hybrids. As might be expected, the sugars of the DNA strand have higher conformational flexibility, but, for the majority of the repetitive sequences studied, these sugars prefer a C2-endo pucker, while ribose sugars uniformly adopt a C3-endo pucker. This gives rise to a strongly heteronomous duplex conformation. One exception to this rule involves the thymidine strand of poly(dT).poly(rA), which marginally prefers a C3-endo pucker. Our study further indicates that the DNA strands of the hybrids favour backbone torsions in the canonical B domain, rather than the modified values proposed on the basis of fibre diffraction studies. Backbone conformational transitions can nevertheless be induced leading to an alpha gamma-flip (alpha:gamma, g-/g(+)-->t/t) or to the alpha beta gamma-flip form proposed from fibre studies (alpha:beta:gamma, g-/t/g(+)-->t/g+/t). The latter transition is also found to be linked to BI-->BII transitions (epsilon:zeta, t/g(-)-->g-/t).
Full text
PDF
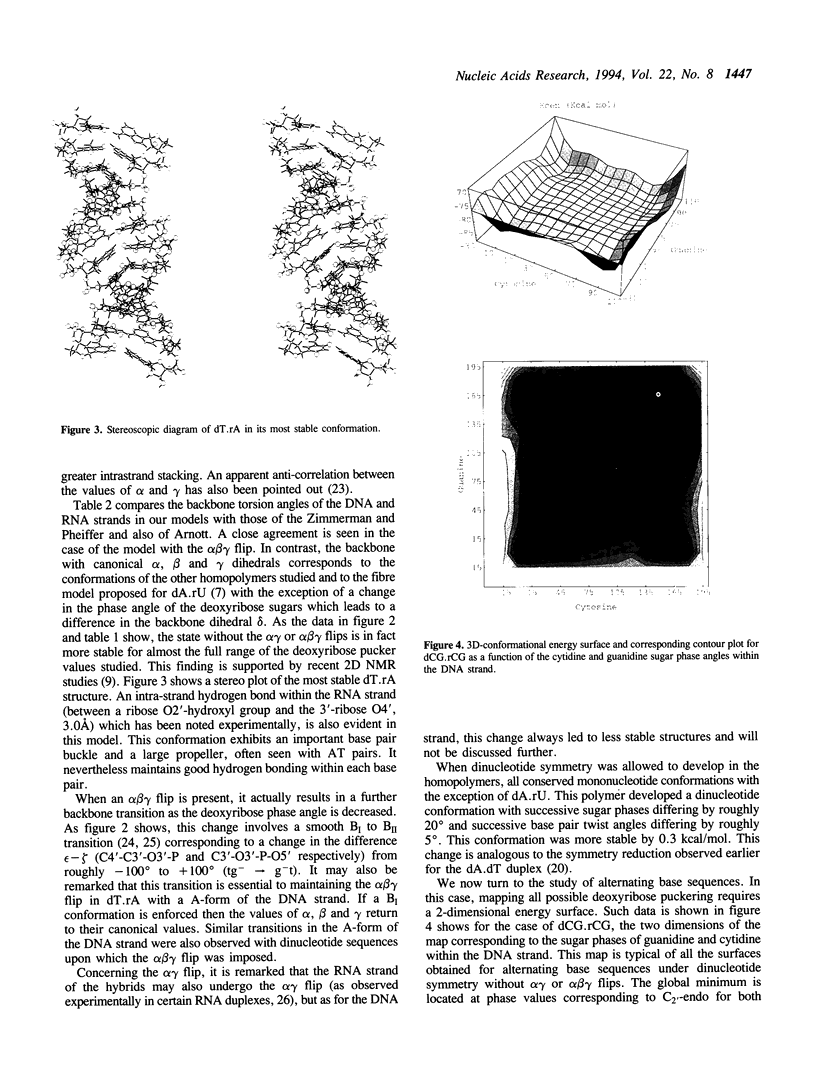
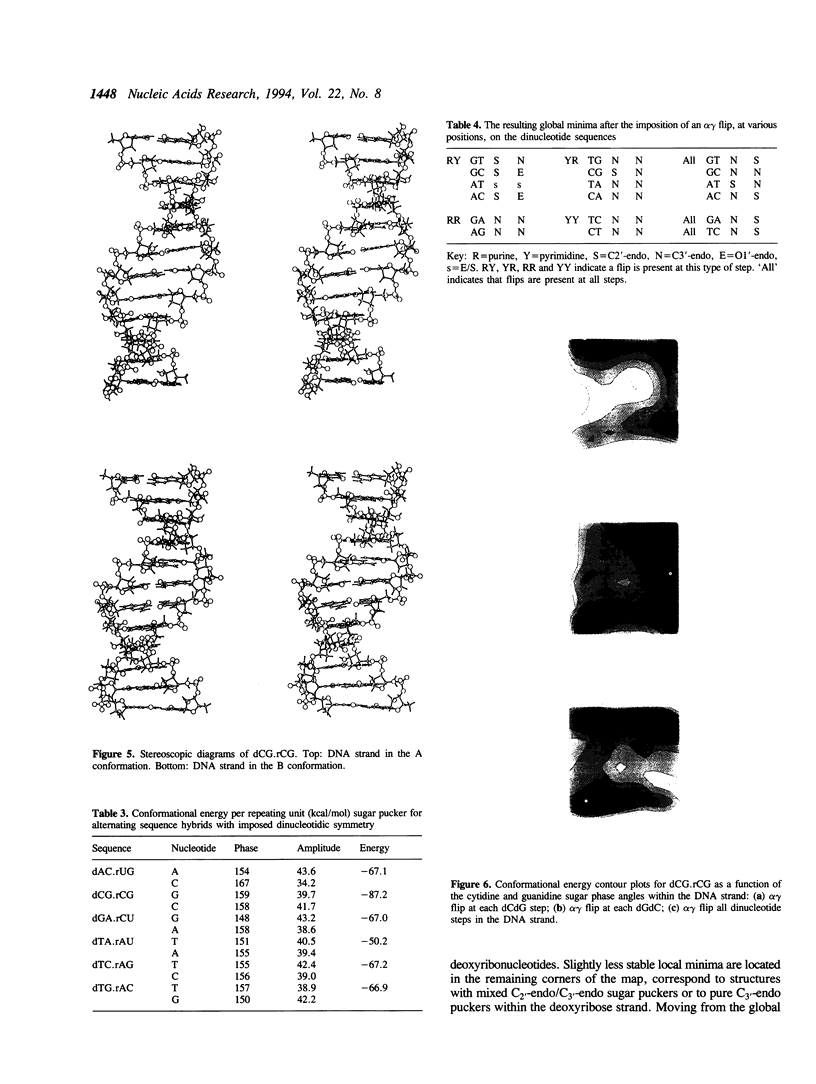

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Millane R. P., Park H. S. DNA-RNA hybrid secondary structures. J Mol Biol. 1986 Apr 20;188(4):631–640. doi: 10.1016/s0022-2836(86)80011-0. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. High-resolution NMR study of a synthetic DNA-RNA hybrid dodecamer containing the consensus pribnow promoter sequence: d(CGTTATAATGCG).r(CGCAUUAUAACG). Biochemistry. 1989 Mar 21;28(6):2435–2443. doi: 10.1021/bi00432a014. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Chevrier B., Podjarny A., Johnson J., de Bear J. S., Gough G. R., Gilham P. T., Moras D. Crystallographic structure of an RNA helix: [U(UA)6A]2. J Mol Biol. 1989 Oct 5;209(3):459–474. doi: 10.1016/0022-2836(89)90010-7. [DOI] [PubMed] [Google Scholar]
- Fedoroff OYu, Salazar M., Reid B. R. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J Mol Biol. 1993 Oct 5;233(3):509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Hartmann B., Piazzola D., Lavery R. BI-BII transitions in B-DNA. Nucleic Acids Res. 1993 Feb 11;21(3):561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Parker I., Kendrick J. A general approach to the optimization of the conformation of ring molecules with an application to valinomycin. J Biomol Struct Dyn. 1986 Dec;4(3):443–462. doi: 10.1080/07391102.1986.10506361. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H., Zakrzewska K., Pullman B. The flexibility of the nucleic acids: (II). The calculation of internal energy and applications to mononucleotide repeat DNA. J Biomol Struct Dyn. 1986 Apr;3(5):989–1014. doi: 10.1080/07391102.1986.10508478. [DOI] [PubMed] [Google Scholar]
- Milman G., Langridge R., Chamberlin M. J. The structure of a DNA-RNA hybrid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1804–1810. doi: 10.1073/pnas.57.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie K. K., Usman N., Nicoghosian K., Cedergren R. J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncin M., Hartmann B., Lavery R. Conformational sub-states in B-DNA. J Mol Biol. 1992 Aug 5;226(3):775–794. doi: 10.1016/0022-2836(92)90632-t. [DOI] [PubMed] [Google Scholar]
- Poncin M., Piazzola D., Lavery R. DNA flexibility as a function of allomorphic conformation and of base sequence. Biopolymers. 1992 Aug;32(8):1077–1103. doi: 10.1002/bip.360320817. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Salazar M., Fedoroff O. Y., Miller J. M., Ribeiro N. S., Reid B. R. The DNA strand in DNA.RNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry. 1993 Apr 27;32(16):4207–4215. doi: 10.1021/bi00067a007. [DOI] [PubMed] [Google Scholar]
- Shindo H., Matsumoto U. Direct evidence for a bimorphic structure of a DNA-RNA hybrid, poly(rA).poly(dT), at high relative humidity. J Biol Chem. 1984 Jul 25;259(14):8682–8684. [PubMed] [Google Scholar]
- Usman N., Cedergren R. Exploiting the chemical synthesis of RNA. Trends Biochem Sci. 1992 Sep;17(9):334–339. doi: 10.1016/0968-0004(92)90306-t. [DOI] [PubMed] [Google Scholar]
- Vorlícková M., Kypr J., Sklenár V. Salt-induced conformational transition of poly[d(A-T)] X poly[d(A-T)]. J Mol Biol. 1983 May 5;166(1):85–92. doi: 10.1016/s0022-2836(83)80052-7. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. A RNA.DNA hybrid that can adopt two conformations: an x-ray diffraction study of poly(rA).poly(dT) in concentrated solution or in fibers. Proc Natl Acad Sci U S A. 1981 Jan;78(1):78–82. doi: 10.1073/pnas.78.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]