Abstract
The conversion of an anonymous DNA sample into numerous oligonucleotides is enzymatically feasible using an unusual restriction endonuclease, CviJI. Depending on reaction conditions, CviJI is capable of digesting DNA at a two or three base recognition sequence. CviJI normally cleaves RGCY sites between the G and C to leave blunt ends. Under 'relaxed' conditions CviJI* cleaves RGCY, and RGCR/YGCY, but not YGCR sites. In theory, CviJI* restriction of pUC19 (2686 bp) should produce 157 fragments, 75% of which are smaller than 20 bp. Instead, 96% of the CviJI* fragments were 18-56 bp long and none of the fragments were smaller than 18 bp. Thermal denaturation of these fragments generates sequence specific oligonucleotides homologous for the cognate template. The enzymatic conversion of anonymous DNA into sequence specific oligomers has implications for several conventional and novel molecular biology procedures.
Full text
PDF

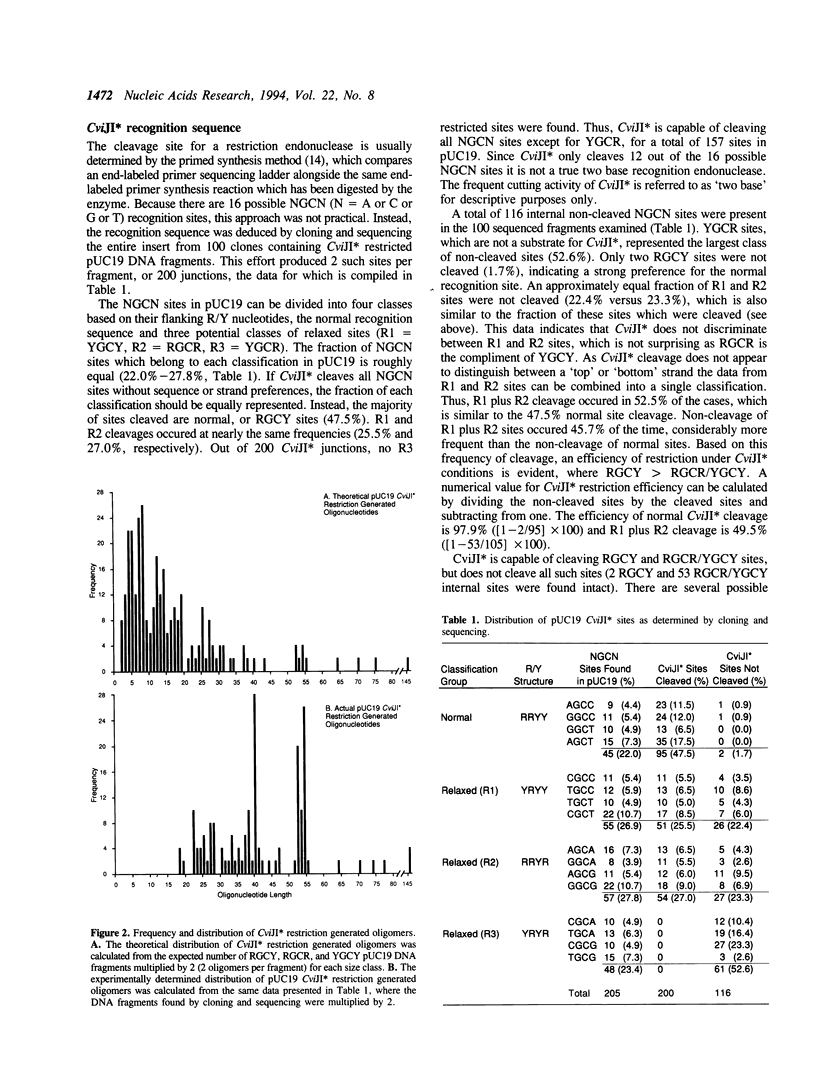



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barany F. The TaqI 'star' reaction: strand preferences reveal hydrogen-bond donor and acceptor sites in canonical sequence recognition. Gene. 1988 May 30;65(2):149–165. doi: 10.1016/0378-1119(88)90452-0. [DOI] [PubMed] [Google Scholar]
- Brooks J. E. Properties and uses of restriction endonucleases. Methods Enzymol. 1987;152:113–129. doi: 10.1016/0076-6879(87)52014-6. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. A general method for defining restriction enzyme cleavage and recognition sites. Methods Enzymol. 1980;65(1):391–404. doi: 10.1016/s0076-6879(80)65050-2. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman D. L., Evans G. A. Restriction endonuclease cleavage at the termini of PCR products. Biotechniques. 1990 Sep;9(3):304–306. [PubMed] [Google Scholar]
- Nelson M., Raschke E., McClelland M. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1993 Jul 1;21(13):3139–3154. doi: 10.1093/nar/21.13.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S. L., Burbank D. E., Grabherr R., van Etten J. L. Cloning and sequencing the cytosine methyltransferase gene M. CviJI from Chlorella virus IL-3A. Virology. 1990 May;176(1):16–24. doi: 10.1016/0042-6822(90)90225-g. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]




