Abstract
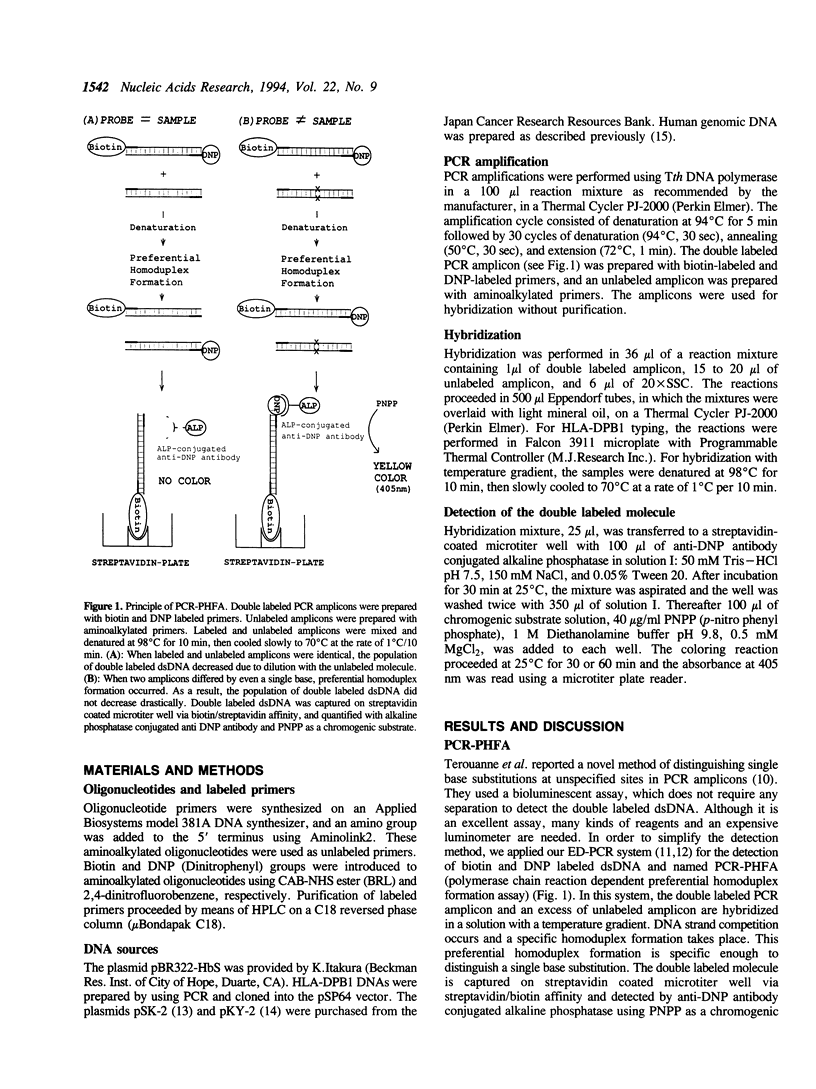
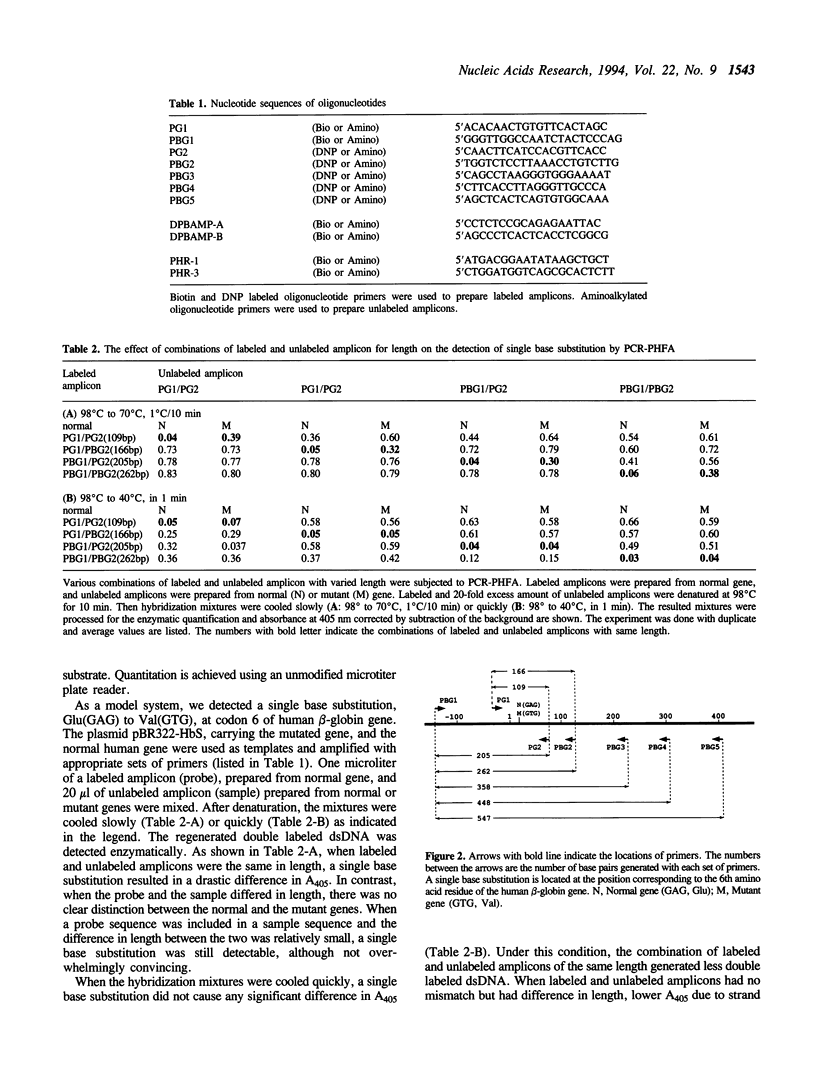
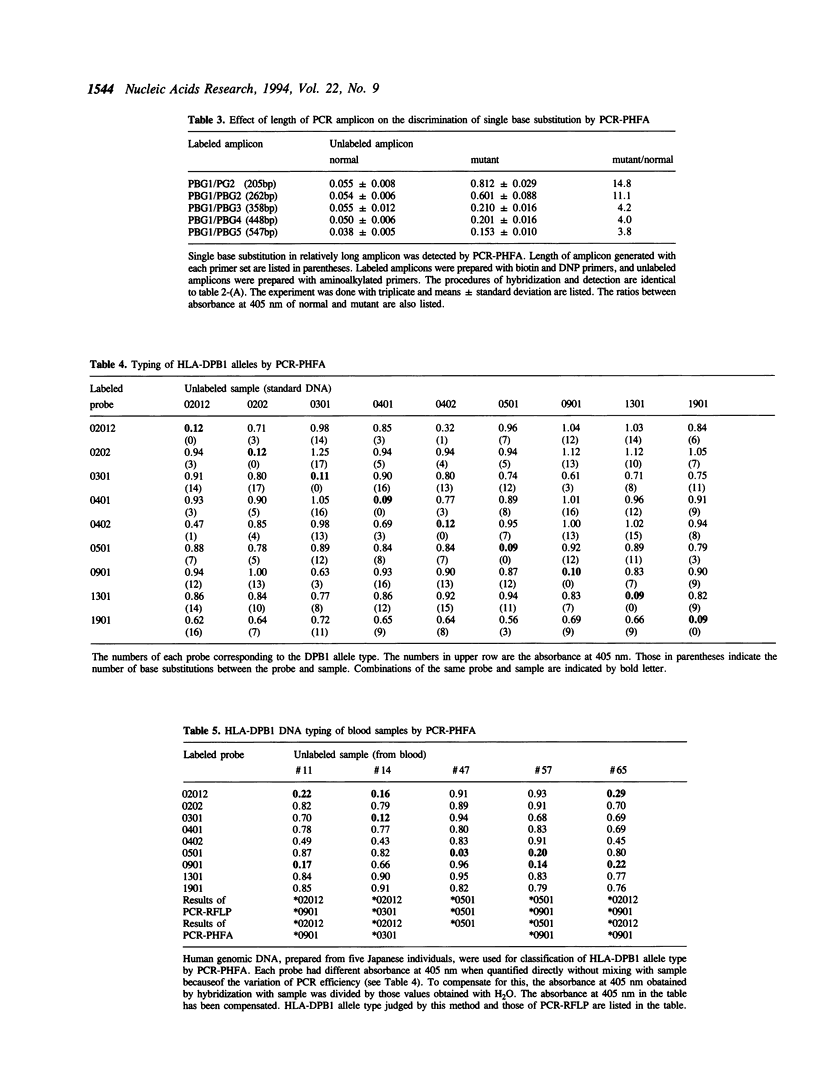
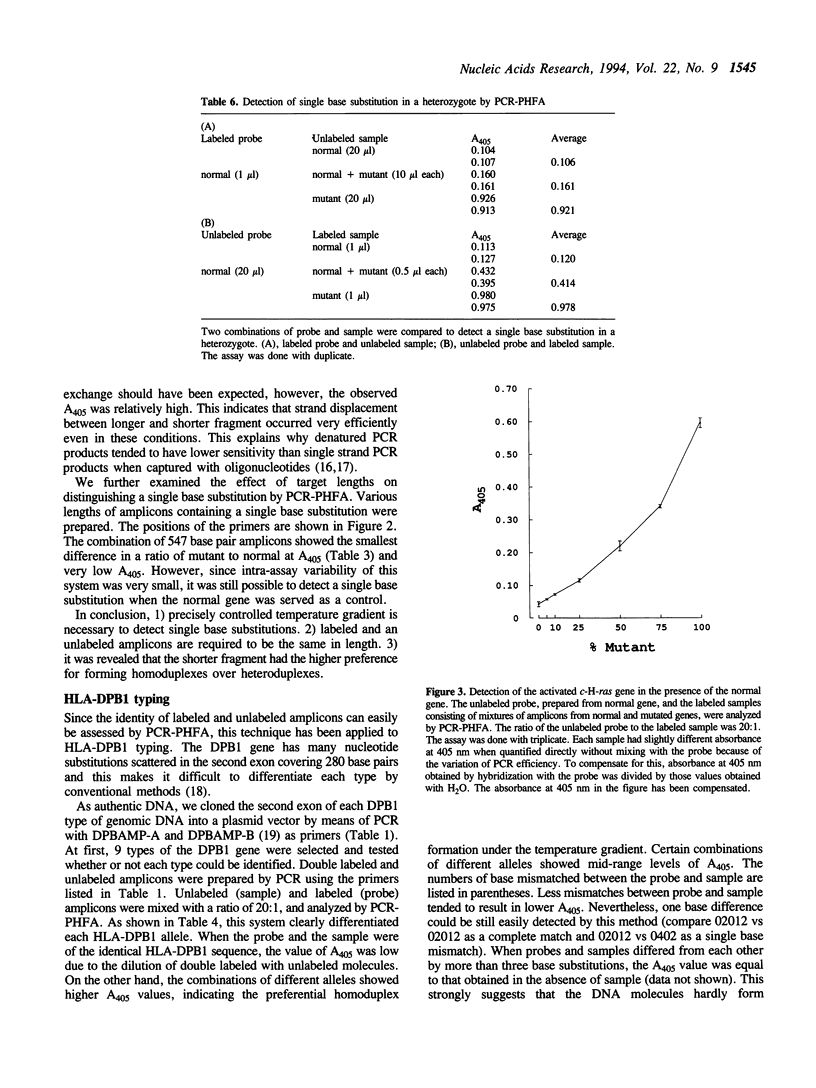
We have developed a simple and reliable method, PCR-PHFA (polymerase chain reaction dependent preferential homoduplex formation assay), for detection of single base substitutions within PCR amplicons. This technique is based upon strand competition during hybridization between a double labeled amplicon, prepared from biotin and DNP labeled primers, and an unlabeled amplicon. Under the precisely controlled temperature gradient, the preferential formation of a homoduplex over a heteroduplex occurs. After annealing, the identical sequence of the double labeled and unlabeled amplicon resulted in a low population of regenerated double labeled dsDNA due to strand exchange between them. Even when the two differed by only a single base substitution, double labeled molecule was regenerated efficiently because of preferential homoduplex formation. The regenerated double labeled molecule was captured onto a streptavidin coated microtiter plate and quantified enzymatically with a chromogenic substrate. The technique has been successfully applied in HLA-DPB1 typing. Furthermore, we detected a mutated gene even in the presence of a large excess of the corresponding normal gene.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bugawan T. L., Begovich A. B., Erlich H. A. Rapid HLA-DPB typing using enzymatically amplified DNA and nonradioactive sequence-specific oligonucleotide probes. Immunogenetics. 1990;32(4):231–241. doi: 10.1007/BF00187094. [DOI] [PubMed] [Google Scholar]
- Conner B. J., Reyes A. A., Morin C., Itakura K., Teplitz R. L., Wallace R. B. Detection of sickle cell beta S-globin allele by hybridization with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1983 Jan;80(1):278–282. doi: 10.1073/pnas.80.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B., Erdman D. D., Durigon E. L., Murtagh J. J., Jr An exonuclease-amplification coupled capture technique improves detection of PCR product. Nucleic Acids Res. 1993 Aug 11;21(16):3905–3906. doi: 10.1093/nar/21.16.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Caskey C. T., Hood L. DNA diagnostics--molecular techniques and automation. Science. 1988 Oct 14;242(4876):229–237. doi: 10.1126/science.3051381. [DOI] [PubMed] [Google Scholar]
- Mano H., Ishikawa F., Hirai H., Takaku F. Mutations of N-ras oncogene in myelodysplastic syndromes and leukemias detected by polymerase chain reaction. Jpn J Cancer Res. 1989 Feb;80(2):102–106. doi: 10.1111/j.1349-7006.1989.tb02274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga S., Kuwata S., Tokunaga K., Uchikawa C., Takahashi K., Akaza T., Mitomi Y., Juji T. Family study on HLA-DPB1 polymorphism: linkage analysis with HLA-DR/DQ and two "new" alleles. Hum Immunol. 1992 Jul;34(3):203–211. doi: 10.1016/0198-8859(92)90113-2. [DOI] [PubMed] [Google Scholar]
- Morrison L. E., Halder T. C., Stols L. M. Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal Biochem. 1989 Dec;183(2):231–244. doi: 10.1016/0003-2697(89)90473-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Maniatis T. Recent advances in the development of methods for detecting single-base substitutions associated with human genetic diseases. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):275–284. doi: 10.1101/sqb.1986.051.01.033. [DOI] [PubMed] [Google Scholar]
- Newton C. R., Graham A., Heptinstall L. E., Powell S. J., Summers C., Kalsheker N., Smith J. C., Markham A. F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989 Apr 11;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J. Detecting single-base mutations. Trends Biotechnol. 1993 Jun;11(6):238–246. doi: 10.1016/0167-7799(93)90135-V. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Bugawan T. L., Horn G. T., Mullis K. B., Erlich H. A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986 Nov 13;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Hirohashi S., Nishimura S., Sugimura T. Transforming activity of human melanoma DNA. Gan. 1983 Dec;74(6):794–797. [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Térouanne B., Balaguer P., Boussioux A. M., Nicolas J. C. Quantitative and qualitative analysis of amplified DNA sequences by a competitive hybridization assay. Anal Biochem. 1992 Sep;205(2):193–199. doi: 10.1016/0003-2697(92)90423-5. [DOI] [PubMed] [Google Scholar]
- Ubukata K., Nakagami S., Nitta A., Yamane A., Kawakami S., Sugiura M., Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992 Jul;30(7):1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa Y., Srivastava S. K., Dunn C. Y., Rhim J. S., Reddy E. P., Aaronson S. A. Acquisition of transforming properties by alternative point mutations within c-bas/has human proto-oncogene. Nature. 1983 Jun 30;303(5920):775–779. doi: 10.1038/303775a0. [DOI] [PubMed] [Google Scholar]


