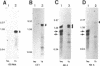Abstract
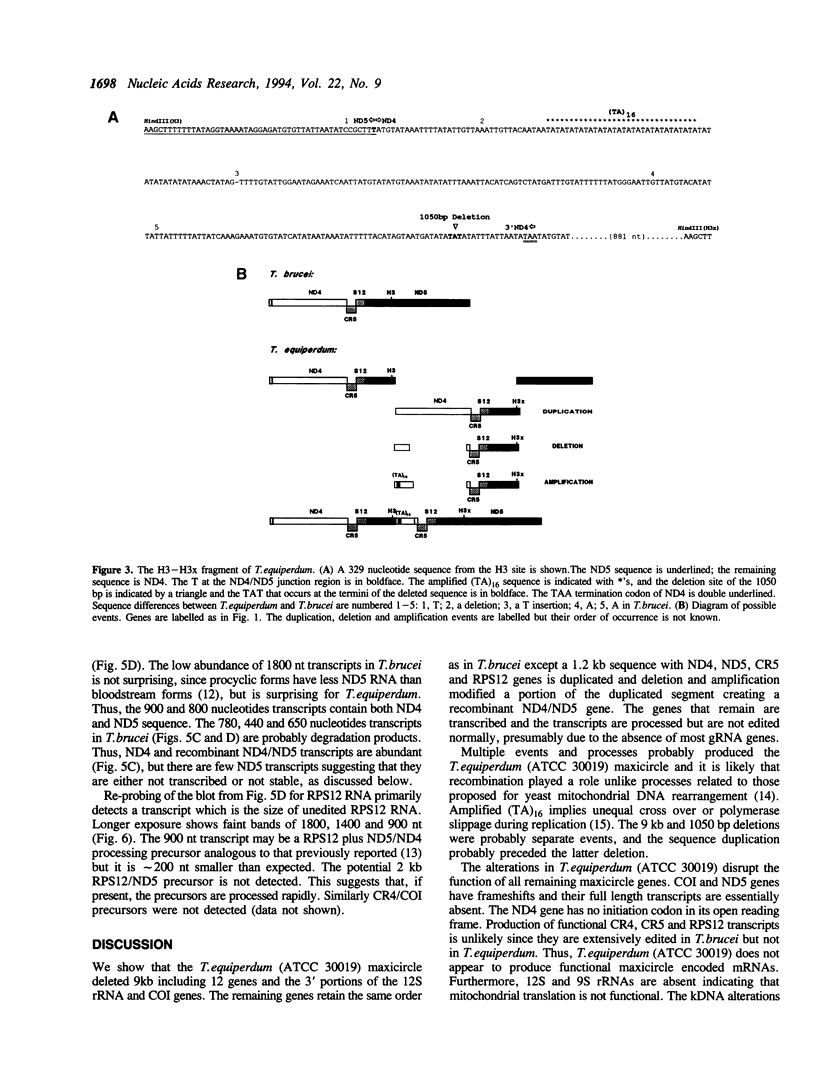
Analyses of the Trypanosoma equiperdum (ATCC 30019) maxicircle reveals deletions, duplications and rearrangement compared to T. brucei. The genes for 9S rRNA and 12 proteins are absent. The 12S rRNA and cytochrome oxidase subunit I (COI) genes lack their 3' ends and are adjacent indicating deletion of intervening genes. The remaining two NADH dehydrogenase subunit genes (ND4 and ND5), the ribosomal protein RPS12 gene and the CR5 gene are duplicated and rearranged. ND4, RPS12 and the CR4 transcripts are abundant in steady state RNA while 12S rRNA and COI transcripts are not detected. Full length ND5 transcripts are rare, if present, but chimeric ND5/ND4 transcripts are abundant. The CR4 and RPS12 transcripts are the size of unedited RNAs suggesting that they are processed. However, they are not edited normally, presumably due to the absence of minicircle gRNA genes.
Full text
PDF



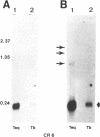
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler B. K., Harris M. E., Bertrand K. I., Hajduk S. L. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3' polyuridine tail formation. Mol Cell Biol. 1991 Dec;11(12):5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrois M., Riou G., Galibert F. Complete nucleotide sequence of minicircle kinetoplast DNA from Trypanosoma equiperdum. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3323–3327. doi: 10.1073/pnas.78.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat G. J., Souza A. E., Feagin J. E., Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992 Jun;52(2):231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- Borst P., Hoeijmakers J. H. Kinetoplast DNA. Plasmid. 1979 Jan;2(1):20–40. doi: 10.1016/0147-619x(79)90003-9. [DOI] [PubMed] [Google Scholar]
- Brun R., Schönenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979 Sep;36(3):289–292. [PubMed] [Google Scholar]
- Clark-Walker G. D. In vivo rearrangement of mitochondrial DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8847–8851. doi: 10.1073/pnas.86.22.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin J. E., Abraham J. M., Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988 May 6;53(3):413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Frasch A. C., Hajduk S. L., Hoeijmakers J. H., Borst P., Brunel E., Davison J. The kinetoplast DNA of Trypanosoma equiperdum. Biochim Biophys Acta. 1980 May 30;607(3):397–410. doi: 10.1016/0005-2787(80)90150-1. [DOI] [PubMed] [Google Scholar]
- Grady R. W., Bienen E. J., Clarkson A. B., Jr p-Alkyloxybenzhydroxamic acids, effective inhibitors of the trypanosome glycerol-3-phosphate oxidase. Mol Biochem Parasitol. 1986 Jun;19(3):231–240. doi: 10.1016/0166-6851(86)90005-8. [DOI] [PubMed] [Google Scholar]
- Jasmer D. P., Feagin J. E., Stuart K. Diverse patterns of expression of the cytochrome c oxidase subunit I gene and unassigned reading frames 4 and 5 during the life cycle of Trypanosoma brucei. Mol Cell Biol. 1985 Nov;5(11):3041–3047. doi: 10.1128/mcb.5.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler P. J., Glick D., Feagin J. E., Morales T. H., Stuart K. D. Structural organization of the maxicircle variable region of Trypanosoma brucei: identification of potential replication origins and topoisomerase II binding sites. Nucleic Acids Res. 1993 Feb 11;21(3):687–694. doi: 10.1093/nar/21.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Hajduk S. L. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol Cell Biol. 1991 Mar;11(3):1668–1675. doi: 10.1128/mcb.11.3.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read L. K., Myler P. J., Stuart K. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J Biol Chem. 1992 Jan 15;267(2):1123–1128. [PubMed] [Google Scholar]
- Riou G. F., Saucier J. M. Characterization of the molecular components in kinetoplast-mitochondrial DNA of Trypanosoma equiperdum. Comparative study of the dyskinetoplastic and wild strains. J Cell Biol. 1979 Jul;82(1):248–263. doi: 10.1083/jcb.82.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou G., Barrois M. Restriction cleavage map of kinetoplast DNA minicircles from Trypanosoma equiperdum. Biochem Biophys Res Commun. 1979 Sep 27;90(2):405–409. doi: 10.1016/0006-291x(79)91249-x. [DOI] [PubMed] [Google Scholar]
- Shu H. H., Wise C. A., Clark-Walker G. D., Martin N. C. A gene required for RNase P activity in Candida (Torulopsis) glabrata mitochondria codes for a 227-nucleotide RNA with homology to bacterial RNase P RNA. Mol Cell Biol. 1991 Mar;11(3):1662–1667. doi: 10.1128/mcb.11.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L. E., Torri A. F., Hajduk S. L. Organized packaging of kinetoplast DNA networks. Cell. 1986 Nov 21;47(4):537–543. doi: 10.1016/0092-8674(86)90618-5. [DOI] [PubMed] [Google Scholar]
- Simpson A. M., Neckelmann N., de la Cruz V. F., Muhich M. L., Simpson L. Mapping and 5' end determination of kinetoplast maxicircle gene transcripts from Leishmania tarentolae. Nucleic Acids Res. 1985 Aug 26;13(16):5977–5993. doi: 10.1093/nar/13.16.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloof P., de Haan A., Eier W., van Iersel M., Boel E., van Steeg H., Benne R. The nucleotide sequence of the variable region in Trypanosoma brucei completes the sequence analysis of the maxicircle component of mitochondrial kinetoplast DNA. Mol Biochem Parasitol. 1992 Dec;56(2):289–299. doi: 10.1016/0166-6851(92)90178-m. [DOI] [PubMed] [Google Scholar]
- Souza A. E., Myler P. J., Stuart K. Maxicircle CR1 transcripts of Trypanosoma brucei are edited and developmentally regulated and encode a putative iron-sulfur protein homologous to an NADH dehydrogenase subunit. Mol Cell Biol. 1992 May;12(5):2100–2107. doi: 10.1128/mcb.12.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. D., Gelvin S. B. Localization of kinetoplast DNA maxicircle transcripts in bloodstream and procyclic form Trypanosoma brucei. Mol Cell Biol. 1982 Jul;2(7):845–852. doi: 10.1128/mcb.2.7.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Feagin J. E. Mitochondrial DNA of kinetoplastids. Int Rev Cytol. 1992;141:65–88. doi: 10.1016/s0074-7696(08)62063-x. [DOI] [PubMed] [Google Scholar]
- Stuart K. Kinetoplast DNA OF Trypanosoma brucei: physical map of the maxicircle. Plasmid. 1979 Oct;2(4):520–528. doi: 10.1016/0147-619x(79)90051-9. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature. 1965 Nov 20;208(5012):762–766. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]